Optimizing Precision Probiotics for Mitigating Graft-Versus-Host Disease
Abstract
1. Introduction
2. Materials and Methods
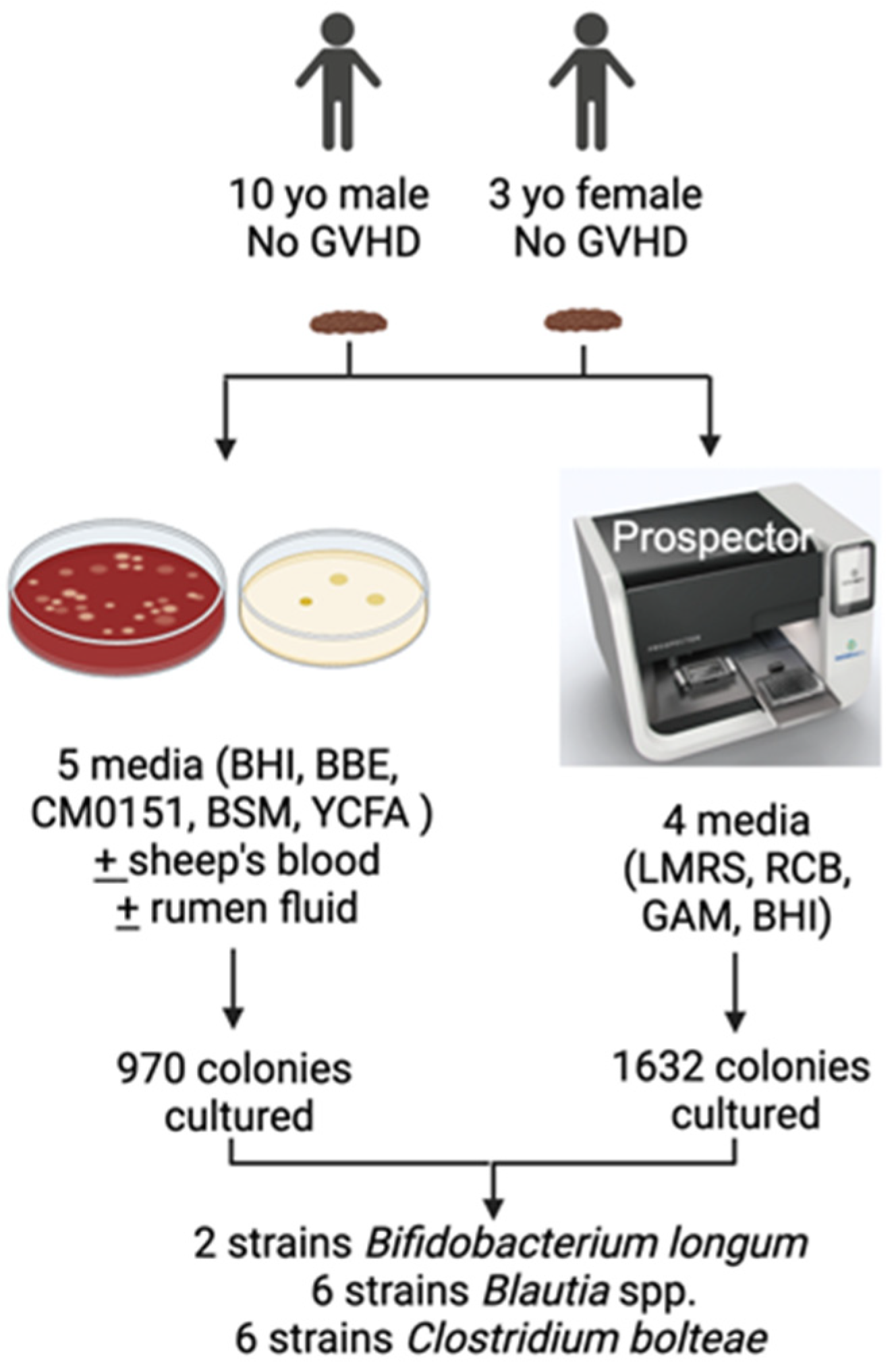
2.1. Isolation of Gut Microbiota
2.2. Gut Microbiota Co-Culture Studies
2.3. SCFA Quantification in Individual and Co-Culture Supernatants
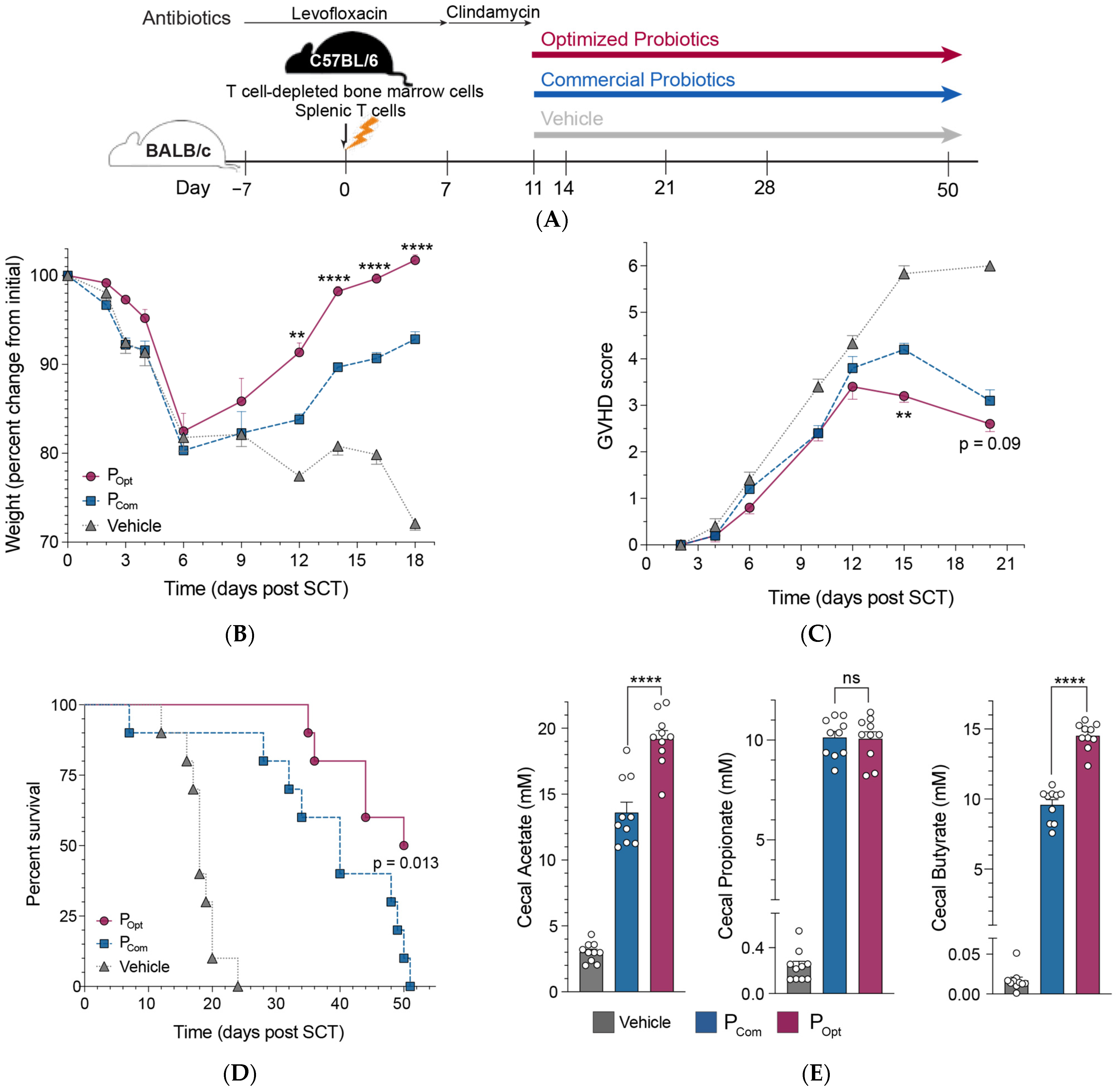
2.4. GVHD Murine Mouse Model
2.5. Quantification of SCFA in Mouse Cecal Contents
2.6. qPCR for Microbiota Analysis
2.7. Statistical Analysis
3. Results
3.1. Isolation of Gut Bacterial Strains
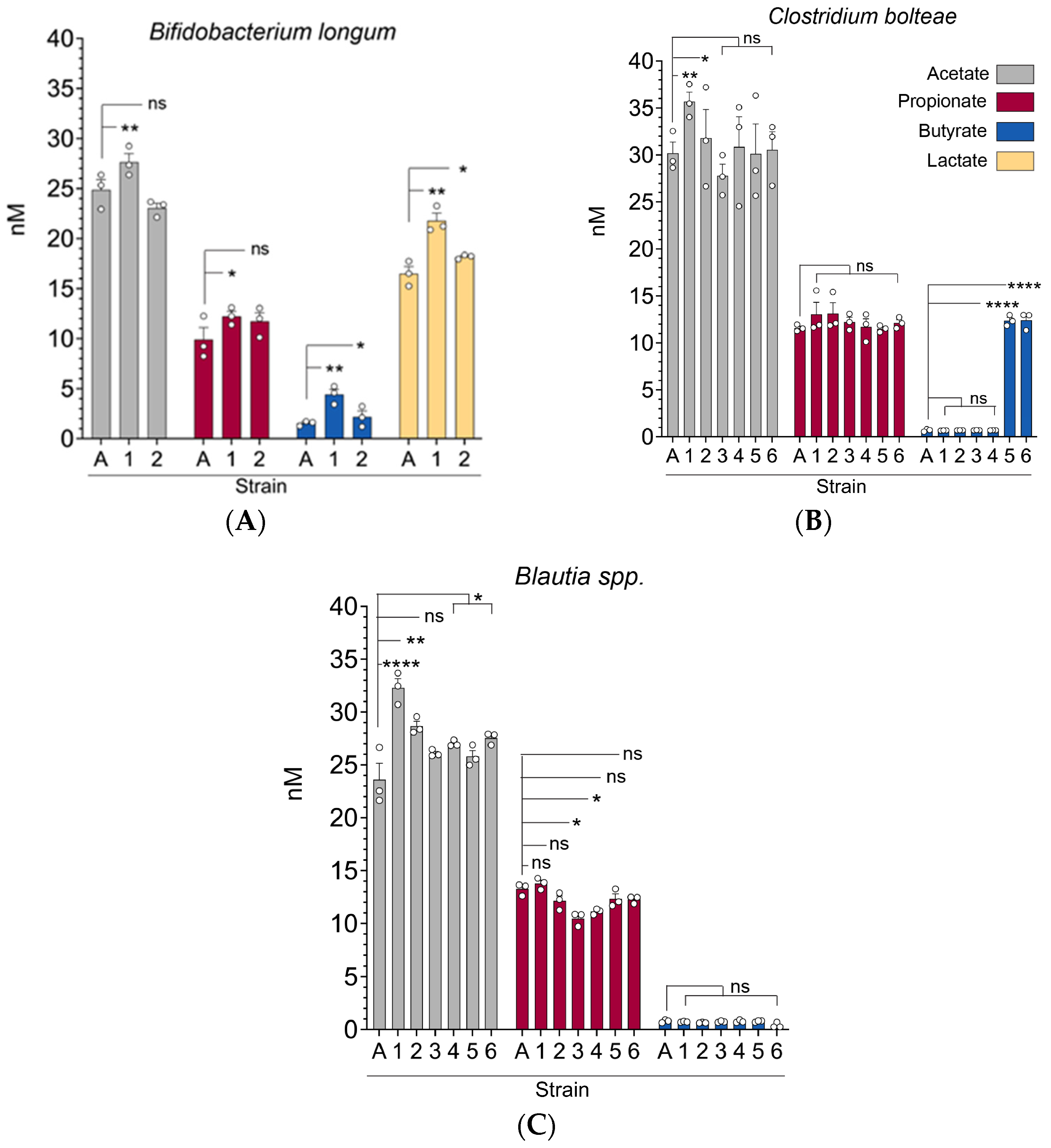
3.2. Gut Microbiota (C. bolteae, Blautia spp., and B. longum) Strains Exhibit Significant Differences in Their Capacity to Produce Short-Chain Fatty Acids In Vitro
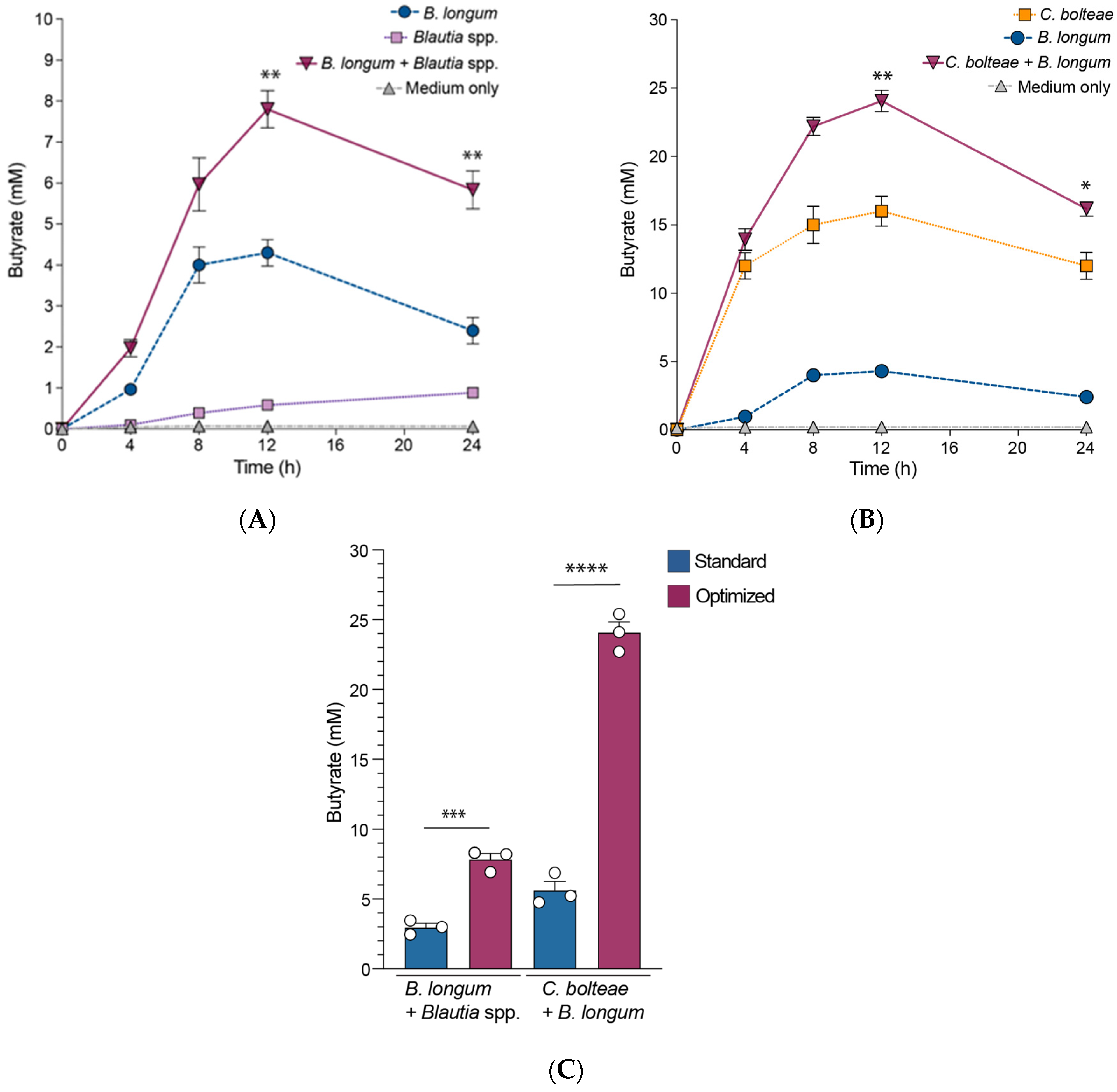
3.3. B. longum Augments Butyrate Production by C. bolteae and Blautia spp. When Co-Cultured In Vitro
3.4. An Optimized Precision Probiotic Therapy Enhances GVHD Mitigation, Prolongs Survival, and Increases SCFA Production in the Murine Gut
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SCT | Stem cell Transplant |
| SCFA | Short-chain fatty acids |
| GVHD | Graft-versus-host disease |
References
- Butto, L.F.; Haller, D. Functional relevance of microbiome signatures: The correlation era requires tools for consolidation. J. Allergy Clin. Immunol. 2017, 139, 1092–1098. [Google Scholar] [CrossRef]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Veiga, P.; Suez, J.; Derrien, M.; Elinav, E. Moving from probiotics to precision probiotics. Nat. Microbiol. 2020, 5, 878–880. [Google Scholar] [CrossRef]
- Simms-Waldrip, T.R.; Sunkersett, G.; Coughlin, L.A.; Savani, M.R.; Arana, C.; Kim, J.; Kim, M.; Zhan, X.; Greenberg, D.E.; Xie, Y.; et al. Antibiotic-Induced Depletion of Anti-inflammatory Clostridia Is Associated with the Development of Graft-versus-Host Disease in Pediatric Stem Cell Transplantation Patients. Biol. Blood Marrow Transplant. 2017, 23, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Gillis, C.C.; Hughes, E.R.; Spiga, L.; Winter, M.G.; Zhu, W.; Furtado de Carvalho, T.; Chanin, R.B.; Behrendt, C.L.; Hooper, L.V.; Santos, R.L.; et al. Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe 2018, 23, 570. [Google Scholar] [CrossRef]
- Fan, D.; Coughlin, L.A.; Neubauer, M.M.; Kim, J.; Kim, M.S.; Zhan, X.; Simms-Waldrip, T.R.; Xie, Y.; Hooper, L.V.; Koh, A.Y. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med. 2015, 21, 808–814. [Google Scholar] [CrossRef]
- Belenguer, A.; Duncan, S.H.; Calder, A.G.; Holtrop, G.; Louis, P.; Lobley, G.E.; Flint, H.J. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 2006, 72, 3593–3599. [Google Scholar] [CrossRef]
- Button, J.E.; Autran, C.A.; Reens, A.L.; Cosetta, C.M.; Smriga, S.; Ericson, M.; Pierce, J.V.; Cook, D.N.; Lee, M.L.; Sun, A.K.; et al. Dosing a synbiotic of human milk oligosaccharides and B. infantis leads to reversible engraftment in healthy adult microbiomes without antibiotics. Cell Host Microbe 2022, 30, 712–725.e717. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Fitzgerald, G.F.; van Sinderen, D. Carbohydrate metabolism in Bifidobacteria. Genes. Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef]
- Chia, L.W.; Mank, M.; Blijenberg, B.; Bongers, R.S.; van Limpt, K.; Wopereis, H.; Tims, S.; Stahl, B.; Belzer, C.; Knol, J. Cross-feeding between Bifidobacterium infantis and Anaerostipes caccae on lactose and human milk oligosaccharides. Benef. Microbes 2021, 12, 69–83. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef]
- Falony, G.; Vlachou, A.; Verbrugghe, K.; De Vuyst, L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 2006, 72, 7835–7841. [Google Scholar] [CrossRef] [PubMed]
- Riviere, A.; Gagnon, M.; Weckx, S.; Roy, D.; De Vuyst, L. Mutual Cross-Feeding Interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 Explain the Bifidogenic and Butyrogenic Effects of Arabinoxylan Oligosaccharides. Appl. Environ. Microbiol. 2015, 81, 7767–7781. [Google Scholar] [CrossRef] [PubMed]
- Shono, Y.; Docampo, M.D.; Peled, J.U.; Perobelli, S.M.; Velardi, E.; Tsai, J.J.; Slingerland, A.E.; Smith, O.M.; Young, L.F.; Gupta, J.; et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci. Transl. Med. 2016, 8, 339ra371. [Google Scholar] [CrossRef] [PubMed]
- Moreira de Gouveia, M.I.; Bernalier-Donadille, A.; Jubelin, G. Enterobacteriaceae in the Human Gut: Dynamics and Ecological Roles in Health and Disease. Biology 2024, 13, 142. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef]
- Shono, Y.; Docampo, M.D.; Peled, J.U.; Perobelli, S.M.; Jenq, R.R. Intestinal microbiota-related effects on graft-versus-host disease. Int. J. Hematol. 2015, 101, 428–437. [Google Scholar] [CrossRef]
- Martin, R.; Chain, F.; Miquel, S.; Motta, J.P.; Vergnolle, N.; Sokol, H.; Langella, P. Using murine colitis models to analyze probiotics-host interactions. FEMS Microbiol. Rev. 2017, 41, S49–S70. [Google Scholar] [CrossRef]
- Biagioli, M.; Carino, A.; Di Giorgio, C.; Marchiano, S.; Bordoni, M.; Roselli, R.; Distrutti, E.; Fiorucci, S. Discovery of a Novel Multi-Strains Probiotic Formulation with Improved Efficacy toward Intestinal Inflammation. Nutrients 2020, 12, 1945. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef]
- Mazzone, L.; Dooling, S.W.; Volpe, E.; Uljarevic, M.; Waters, J.L.; Sabatini, A.; Arturi, L.; Abate, R.; Riccioni, A.; Siracusano, M.; et al. Precision microbial intervention improves social behavior but not autism severity: A pilot double-blind randomized placebo-controlled trial. Cell Host Microbe 2024, 32, 106–116.e106. [Google Scholar] [CrossRef]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Mathewson, N.D.; Jenq, R.; Mathew, A.V.; Koenigsknecht, M.; Hanash, A.; Toubai, T.; Oravecz-Wilson, K.; Wu, S.R.; Sun, Y.; Rossi, C.; et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 2016, 17, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, P.; Araujo, J.R.; Di Santo, J.P. A Cross-Talk Between Microbiota-Derived Short-Chain Fatty Acids and the Host Mucosal Immune System Regulates Intestinal Homeostasis and Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2018, 24, 558–572. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef]
- Sela, D.A.; Mills, D.A. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef]
- Engfer, M.B.; Stahl, B.; Finke, B.; Sawatzki, G.; Daniel, H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 2000, 71, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Martoni, C.J.; Ganopolsky, J.G.; Labbe, A.; Prakash, S. The human microbiome and bile acid metabolism: Dysbiosis, dysmetabolism, disease and intervention. Expert. Opin. Biol. Ther. 2014, 14, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.A.; Ryu, J.H.; Jo, Y.; Hong, C. The role of gut microbiota in T cell immunity and immune mediated disorders. Int. J. Biol. Sci. 2023, 19, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebigbo, N.; Long, A.; Do, P.; Coughlin, L.; Poulides, N.; Jewell, T.; Gan, S.; Zhan, X.; Koh, A.Y. Optimizing Precision Probiotics for Mitigating Graft-Versus-Host Disease. Microorganisms 2025, 13, 706. https://doi.org/10.3390/microorganisms13040706
Ebigbo N, Long A, Do P, Coughlin L, Poulides N, Jewell T, Gan S, Zhan X, Koh AY. Optimizing Precision Probiotics for Mitigating Graft-Versus-Host Disease. Microorganisms. 2025; 13(4):706. https://doi.org/10.3390/microorganisms13040706
Chicago/Turabian StyleEbigbo, Nonyelum, Apple Long, Phinga Do, Laura Coughlin, Nicole Poulides, Talia Jewell, Shuheng Gan, Xiaowei Zhan, and Andrew Y. Koh. 2025. "Optimizing Precision Probiotics for Mitigating Graft-Versus-Host Disease" Microorganisms 13, no. 4: 706. https://doi.org/10.3390/microorganisms13040706
APA StyleEbigbo, N., Long, A., Do, P., Coughlin, L., Poulides, N., Jewell, T., Gan, S., Zhan, X., & Koh, A. Y. (2025). Optimizing Precision Probiotics for Mitigating Graft-Versus-Host Disease. Microorganisms, 13(4), 706. https://doi.org/10.3390/microorganisms13040706





