The Genomic Characteristics of Potential Probiotics: Two Streptococcus salivarius Isolates from a Healthy Individual in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Saliva Sampling
2.2. Bacterial Strains and Culture
2.3. Isolation and Identification of S. salivarius in Saliva Samples
2.3.1. Isolation of S. salivarius
2.3.2. Molecular Biology Identification of S. salivarius
2.4. Plasmid Extraction
2.5. Detection of Salivaricin A2 and Salivaricin B
2.6. Simultaneous Antagonism Assay
2.7. Scanning Electron Microscopy Images
2.8. The Next-Generation Sequencing and Third-Generation Sequencing
2.9. Data Processing and Genome Assembly
2.10. Taxonomic Identification
2.11. Genome Annotation and Functional Prediction
2.12. RNA Extraction
2.13. RNA Sequencing and Transcriptome Analysis
3. Results
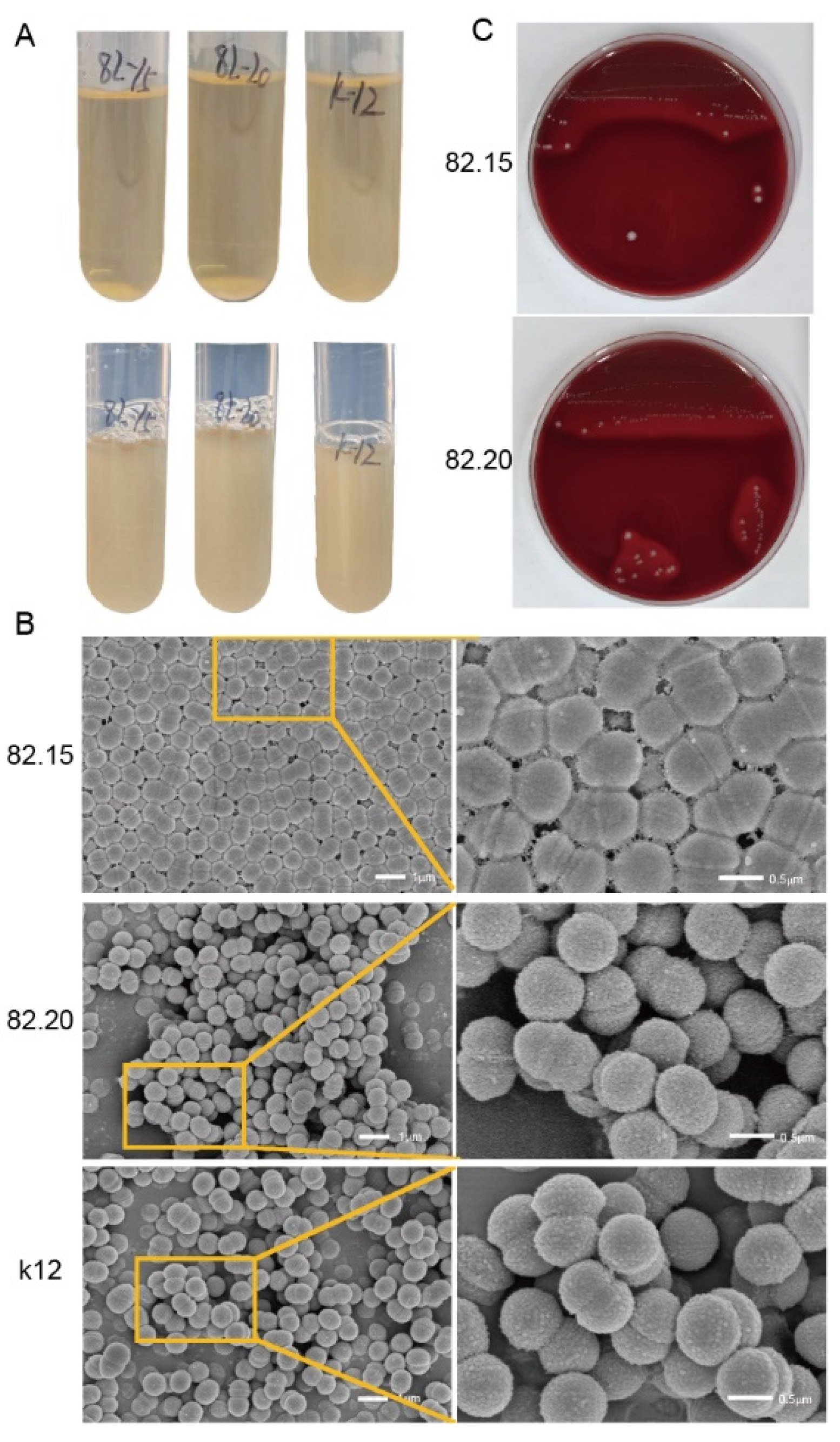
3.1. Morphology of Isolated S. salivarius
3.2. Antibacterial Activity of the Isolated Strains
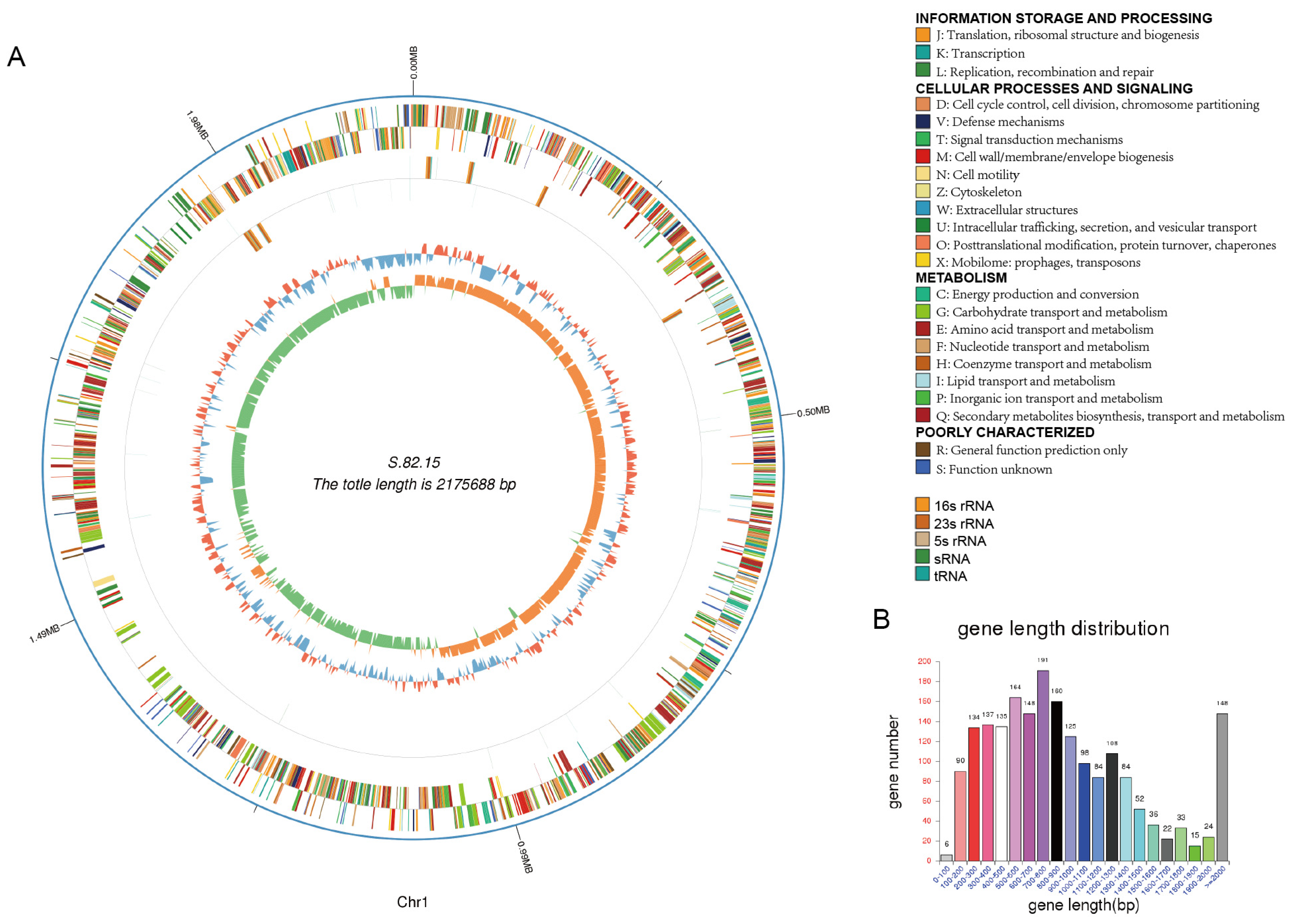
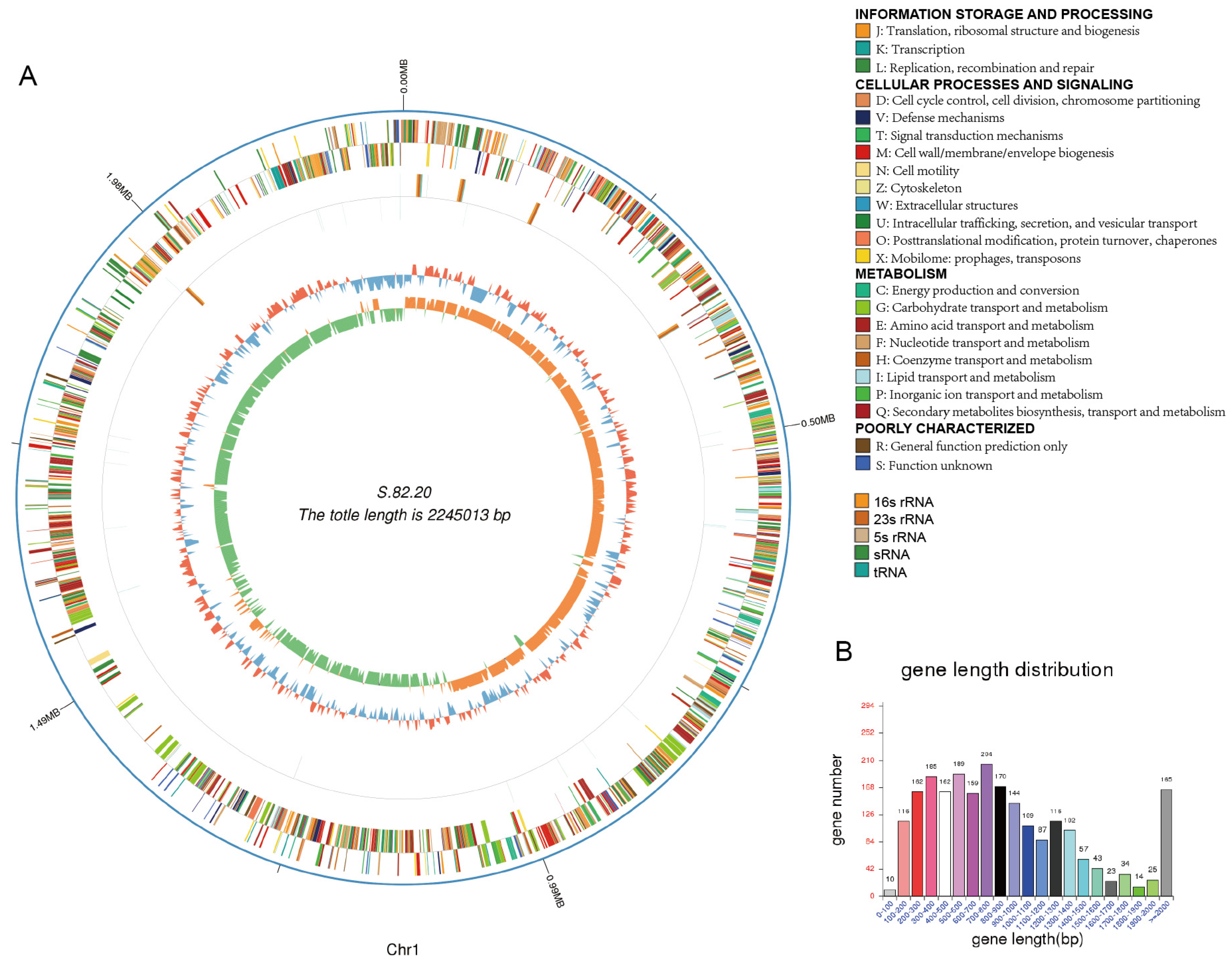
3.3. Genomic Analysis of S.82.15 and S.82.20
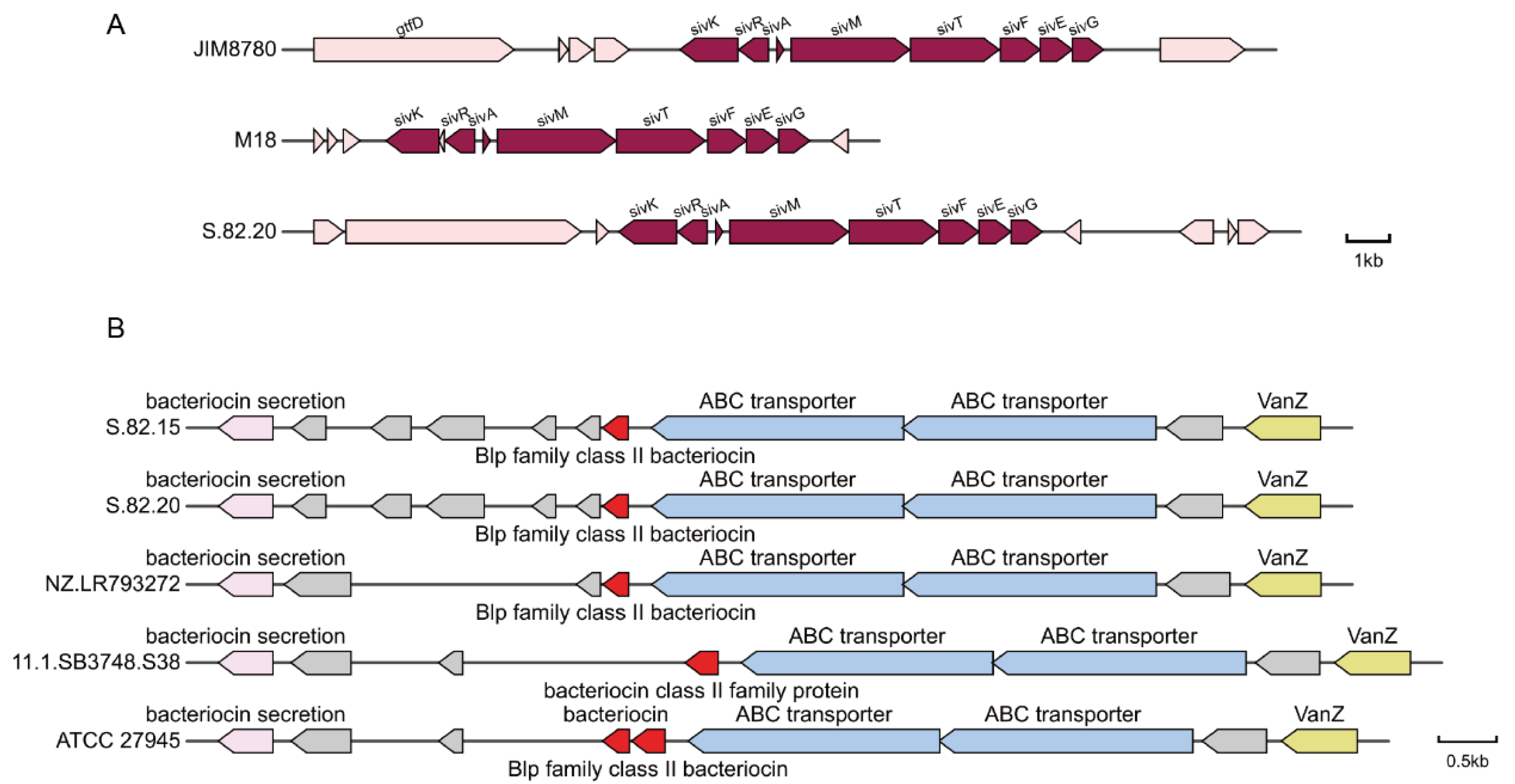
3.4. Detection of Potential Antibacterial Proteins
3.5. Transcriptomic Screen of S.82.20 Against GAS
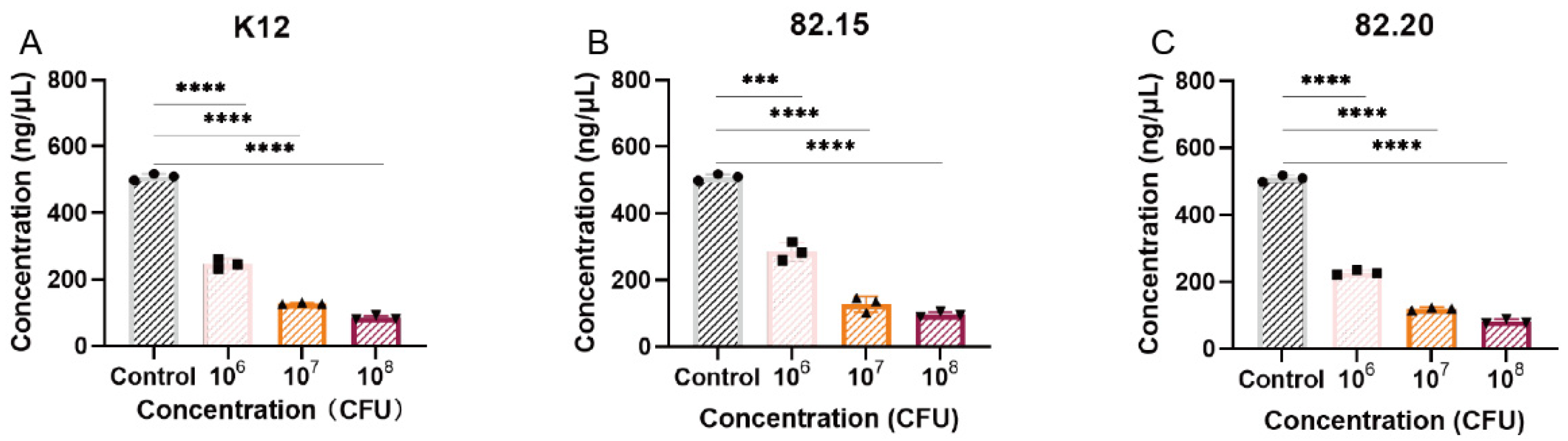
3.6. Toxicity and Biosafety Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaci, G.; Goudercourt, D.; Dennin, V.; Pot, B.; Doré, J.; Ehrlich, S.D.; Renault, P.; Blottière, H.M.; Daniel, C.; Delorme, C. Anti-Inflammatory Properties of Streptococcus salivarius, a Commensal Bacterium of the Oral Cavity and Digestive Tract. Appl. Environ. Microbiol. 2014, 80, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Welch, J.L.M.; Kauffman, K.M.; McLean, J.S.; He, X. The oral microbiome: Diversity, biogeography and human health. Nat. Rev. Microbiol. 2023, 22, 89–104. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of Oral Streptococci. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Laws, G.L.; Hale, J.D.F.; Kemp, R.A. Human Systemic Immune Response to Ingestion of the Oral Probiotic Streptococcus salivarius BLIS K12. Probiotics Antimicrob. Proteins 2021, 13, 1521–1529. [Google Scholar] [CrossRef]
- Hyink, O.; Wescombe, P.A.; Upton, M.; Ragland, N.; Burton, J.P.; Tagg, J.R. Salivaricin A2 and the Novel Lantibiotic Salivaricin B Are Encoded at Adjacent Loci on a 190-Kilobase Transmissible Megaplasmid in the Oral Probiotic Strain Streptococcus salivarius K12. Appl. Environ. Microbiol. 2007, 73, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Yoo, H.-J. Inhibitory effects of Streptococcus salivarius K12 on formation of cariogenic biofilm. J. Dent. Sci. 2023, 18, 65–72. [Google Scholar] [CrossRef]
- Do, H.; Li, Z.-R.; Tripathi, P.K.; Mitra, S.; Guerra, S.; Dash, A.; Weerasekera, D.; Makthal, N.; Shams, S.; Aggarwal, S.; et al. Engineered probiotic overcomes pathogen defences using signal interference and antibiotic production to treat infection in mice. Nat. Microbiol. 2024, 9, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Tunçer, S.; Karaçam, S. Cell-free supernatant of Streptococcus salivarius M18 impairs the pathogenic properties of Pseudomonas aeruginosa and Klebsiella pneumonia. Arch. Microbiol. 2020, 202, 2825–2840. [Google Scholar] [CrossRef]
- Tagg, J.R.; Harold, L.K.; Jain, R.; Hale, J.D.F. Beneficial modulation of human health in the oral cavity and beyond using bacteriocin-like inhibitory substance-producing streptococcal probiotics. Front. Microbiol. 2023, 14, 1161155. [Google Scholar] [CrossRef]
- Reichardt, E.; Shyp, V.; Alig, L.; Verna, C.; Kulik, E.M.; Bornstein, M.M. Antimicrobial effect of probiotic bacteriocins on Streptococcus mutans biofilm in a dynamic oral flow chamber model—An in vitro study. J. Oral Microbiol. 2024, 16, 2304971. [Google Scholar] [CrossRef]
- Nie, Q.; Wan, X.; Tao, H.; Yang, Q.; Zhao, X.; Liu, H.; Hu, J.; Luo, Y.; Shu, T.; Geng, R.; et al. Multi-function screening of probiotics to improve oral health and evaluating their efficacy in a rat periodontitis model. Front. Cell. Infect. Microbiol. 2023, 13, 1261189. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Romero, A.; Zepeda-Hernández, A.; Cárdenas-Rangel, J.; Aguilar-Márquez, R.; Garcia-Amezquita, L.E.; Carrillo-Nieves, D.; García-Cayuela, T. Frozen Fermented Dairy Snacks with Probiotics and Blueberry Bagasse: Stability, Bioactivity, and Digestive Viability. Microorganisms 2025, 13, 86. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Wang, Q.; Jin, Q.; Xu, S.; Tang, L.; Zeng, Z.; Fu, A.; Zhu, J.; Zhang, Q.; et al. Screening of Lactic Acid Bacteria Strains from Chinese Fermented Food (Suanshui) and its Protective Effect on Acute Liver Injury in Mice. Probiotics Antimicrob. Proteins 2024. [Google Scholar] [CrossRef]
- Peng, L.; Yin, Q.; Wang, X.; Zhong, Y.; Wang, Y.; Cai, W.; Zhou, R.; Chen, Y.; Hu, Y.; Cheng, Z.; et al. Pasteurized Akkermansia muciniphila Ameliorates Preeclampsia in Mice by Enhancing Gut Barrier Integrity, Improving Endothelial Function, and Modulating Gut Metabolic Dysregulation. Microorganisms 2024, 12, 2483. [Google Scholar] [CrossRef] [PubMed]
- Torres-Maravilla, E.; Reyes-Pavón, D.; Benítez-Cabello, A.; González-Vázquez, R.; Ramírez-Chamorro, L.M.; Langella, P.; Bermúdez-Humarán, L.G. Strategies for the Identification and Assessment of Bacterial Strains with Specific Probiotic Traits. Microorganisms 2022, 10, 1389. [Google Scholar] [CrossRef]
- Sufaru, I.-G.; Lazar, L.; Sincar, D.-C.; Martu, M.-A.; Pasarin, L.; Luca, E.-O.; Stefanescu, A.; Froicu, E.-M.; Solomon, S.-M. Clinical Effects of Locally Delivered Lactobacillus reuteri as Adjunctive Therapy in Patients with Periodontitis: A Split-Mouth Study. Appl. Sci. 2022, 12, 2470. [Google Scholar] [CrossRef]
- Mendonça, F.H.B.P.; dos Santos, S.S.F.; Faria, I.d.S.d.; e Silva, C.R.G.; Jorge, A.O.C.; Leão, M.V.P. Effects of probiotic bacteria on Candida presence and IgA anti-Candida in the oral cavity of elderly. Braz. Dent. J. 2012, 23, 534–538. [Google Scholar] [CrossRef]
- Peng, X.; Li, Z.; Pei, Y.; Zheng, S.; Liu, J.; Wang, J.; Li, R.; Xu, X. Streptococcus salivarius K12 Alleviates Oral Mucositis in Patients Undergoing Radiotherapy for Malignant Head and Neck Tumors: A Randomized Controlled Trial. J. Clin. Oncol. 2024, 42, 1426–1435. [Google Scholar] [CrossRef]
- Sarlin, S.; Koskela, U.; Honkila, M.; Tähtinen, P.A.; Pokka, T.; Renko, M.; Tapiainen, T. Streptococcus salivarius Probiotics to Prevent Acute Otitis Media in Children. JAMA Netw. Open 2023, 6, e2340608. [Google Scholar] [CrossRef]
- Di Pierro, F.; Colombo, M.; Zanvit, A.; Risso, P.; Rottoli, A.S. Use of Streptococcus salivarius K12 to reduce the incidence of pharyngo-tonsillitis and acute otitis media in children: A retrospective analysis in not-recurrent pediatric subjects. Minerva Pediatr. 2018, 70, 240–245. [Google Scholar] [CrossRef]
- Li, X.; Fields, F.R.; Ho, M.; Marshall-Hudson, A.; Gross, R.; Casser, M.E.; Naito, M. Safety assessment of Streptococcus salivarius DB-B5 as a probiotic candidate for oral health. Food Chem. Toxicol. 2021, 153, 112277. [Google Scholar] [CrossRef]
- Masdea, L.; Kulik, E.; Hauser-Gerspach, I.; Ramseier, A.; Filippi, A.; Waltimo, T. Antimicrobial activity of Streptococcus salivarius K12 on bacteria involved in oral malodour. Arch. Oral Biol. 2012, 57, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Birol, I. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Na, S.-I.; Kim, Y.O.; Yoon, S.-H.; Ha, S.-m.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; E Augustijn, H.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2014, 43, D261–D269. [Google Scholar] [CrossRef]
- Urban, M.; Pant, R.; Raghunath, A.; Irvine, A.G.; Pedro, H.; Hammond-Kosack, K.E. The Pathogen-Host Interactions database (PHI-base): Additions and future developments. Nucleic Acids Res. 2014, 43, D645–D655. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiong, Z.; Sun, L.; Yang, J.; Jin, Q. VFDB 2012 update: Toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2011, 40, D641–D645. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2016, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- McClure, R.; Balasubramanian, D.; Sun, Y.; Bobrovskyy, M.; Sumby, P.; Genco, C.A.; Vanderpool, C.K.; Tjaden, B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013, 41, e140. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Burton, J.P.; Chilcott, C.N.; Wescombe, P.A.; Tagg, J.R. Extended Safety Data for the Oral Cavity Probiotic Streptococcus salivarius K12. Probiotics Antimicrob. Proteins 2010, 2, 135–144. [Google Scholar] [CrossRef]
- Budala, D.G.; Martu, M.-A.; Maftei, G.-A.; Diaconu-Popa, D.A.; Danila, V.; Luchian, I. The Role of Natural Compounds in Optimizing Contemporary Dental Treatment—Current Status and Future Trends. J. Funct. Biomater. 2023, 14, 273. [Google Scholar] [CrossRef]
- González-Vázquez, R.; Zúñiga-León, E.; Torres-Maravilla, E.; Leyte-Lugo, M.; Mendoza-Pérez, F.; Hernández-Delgado, N.C.; Pérez-Pastén-Borja, R.; Azaola-Espinosa, A.; Mayorga-Reyes, L. Genomic and Biochemical Characterization of Bifidobacterium pseudocatenulatum JCLA3 Isolated from Human Intestine. Microorganisms 2022, 10, 2100. [Google Scholar] [CrossRef]
- Reyes-Castillo, P.A.; González-Vázquez, R.; Torres-Maravilla, E.; Bautista-Hernández, J.I.; Zúñiga-León, E.; Leyte-Lugo, M.; Mateos-Sánchez, L.; Mendoza-Pérez, F.; Gutiérrez-Nava, M.A.; Reyes-Pavón, D.; et al. Bifidobacterium longum LBUX23 Isolated from Feces of a Newborn; Potential Probiotic Properties and Genomic Characterization. Microorganisms 2023, 11, 1648. [Google Scholar] [CrossRef] [PubMed]
- Begić, G.; Badovinac, I.J.; Karleuša, L.; Kralik, K.; Peloza, O.C.; Kuiš, D.; Gobin, I. Streptococcus salivarius as an Important Factor in Dental Biofilm Homeostasis: Influence on Streptococcus mutans and Aggregatibacter actinomycetemcomitans in Mixed Biofilm. Int. J. Mol. Sci. 2023, 24, 7249. [Google Scholar] [CrossRef] [PubMed]
- Couvigny, B.; Kulakauskas, S.; Pons, N.; Quinquis, B.; Abraham, A.-L.; Meylheuc, T.; Delorme, C.; Renault, P.; Briandet, R.; Lapaque, N.; et al. Identification of New Factors Modulating Adhesion Abilities of the Pioneer Commensal Bacterium Streptococcus salivarius. Front. Microbiol. 2018, 9, 273. [Google Scholar] [CrossRef]
- Couvigny, B.; Lapaque, N.; Rigottier-Gois, L.; Guillot, A.; Chat, S.; Meylheuc, T.; Kulakauskas, S.; Rohde, M.; Mistou, M.-Y.; Renault, P.; et al. Three glycosylated serine-rich repeat proteins play a pivotal role in adhesion and colonization of the pioneer commensal bacterium, Streptococcus salivarius. Environ. Microbiol. 2017, 19, 3579–3594. [Google Scholar] [CrossRef]
- Spacova, I.; O’neill, C.; Lebeer, S. Lacticaseibacillus rhamnosus GG inhibits infection of human keratinocytes by Staphylococcus aureus through mechanisms involving cell surface molecules and pH reduction. Benef. Microbes 2020, 11, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Wescombe, P.A.; Burton, J.P.; Cadieux, P.A.; Klesse, N.A.; Hyink, O.; Heng, N.C.K.; Chilcott, C.N.; Reid, G.; Tagg, J.R. Megaplasmids encode differing combinations of lantibiotics in Streptococcus salivarius. Antonie Van Leeuwenhoek 2006, 90, 269–280. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Upton, M.; Renault, P.; Wirawan, R.E.; Power, D.; Burton, J.P.; Chilcott, C.N.; Tagg, J.R. Salivaricin 9, a new lantibiotic produced by Streptococcus salivarius. Microbiology 2011, 157, 1290–1299. [Google Scholar] [CrossRef]
- Barbour, A.; Philip, K. Enhanced Production, Purification, Characterization and Mechanism of Action of Salivaricin 9 Lantibiotic Produced by Streptococcus salivarius NU10. PLoS ONE 2013, 8, e77751. [Google Scholar] [CrossRef]
- Barbour, A.; Wescombe, P.; Smith, L. Evolution of Lantibiotic Salivaricins: New Weapons to Fight Infectious Diseases. Trends Microbiol. 2020, 28, 578–593. [Google Scholar] [CrossRef]
- Dawid, S.; Roche, A.M.; Weiser, J.N. The blp Bacteriocins of Streptococcus pneumoniae Mediate Intraspecies Competition both In Vitro and In Vivo. Infect. Immun. 2007, 75, 443–451. [Google Scholar] [CrossRef]
- Valente, C.; Dawid, S.; Pinto, F.R.; Hinds, J.; Simões, A.S.; Gould, K.A.; Mendes, L.A.; de Lencastre, H.; Sá-Leão, R. The blp Locus of Streptococcus pneumoniae Plays a Limited Role in the Selection of Strains That Can Cocolonize the Human Nasopharynx. Appl. Environ. Microbiol. 2016, 82, 5206–5215. [Google Scholar] [CrossRef] [PubMed]
- Wescombe, P.A.; Dyet, K.H.; Dierksen, K.P.; Power, D.A.; Jack, R.W.; Burton, J.P.; Inglis, M.A.; Wescombe, A.L.; Tagg, J.R. Salivaricin G32, a Homolog of the PrototypeStreptococcus pyogenesNisin-Like Lantibiotic SA-FF22, Produced by the Commensal SpeciesStreptococcus salivarius. Int. J. Microbiol. 2012, 2012, 738503. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.V.; Heng, N.C.K.; Carne, A.; Tagg, J.R.; Wescombe, P.A. Salivaricin E and abundant dextranase activity may contribute to the anti-cariogenic potential of the probiotic candidate Streptococcus salivarius JH. Microbiology 2016, 162, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Birri, D.J.; Brede, D.A.; Nes, I.F. Salivaricin D, a Novel Intrinsically Trypsin-Resistant Lantibiotic from Streptococcus salivarius 5M6c Isolated from a Healthy Infant. Appl. Environ. Microbiol. 2012, 78, 402–410. [Google Scholar] [CrossRef]
- Tompkins, G.R.; Tagg, J.R. Bacteriocin-like Inhibitory Activity Associated with Beta-hemolytic Strains of Streptococcus salivarius. J. Dent. Res. 1987, 66, 5. [Google Scholar] [CrossRef]



| GAS a | M. luteusa | |||
|---|---|---|---|---|
| Aerobic | Anaerobic | Aerobic | Anaerobic | |
| 82.15 | +++ | ++ | − | ++ |
| 82.20 | ++ | ++ | ++ | ++ |
| K12 | + | ++ | + | ++ |
| S.82.15 | S.82.20 | |
|---|---|---|
| Genome Size | 2,175,688 | 2,414,610 |
| Gene Number | 1994 | 2276 |
| Gene Length | 1,900,044 | 2,119,581 |
| GC Content | 40.93 | 40.62 |
| % of Genome (Genes) | 87.33 | 87.78 |
| Gene Average Length | 953 | 931 |
| Gene Internal Length | 275,644 | 295,029 |
| Gene Internal GC Content | 34.93 | 33.99 |
| % of Genome (Internal) | 12.67 | 12.22 |
| Functional Class | Class Description | S.82.15 | S.82.20 |
|---|---|---|---|
| C | Energy production and conversion | 46 | 47 |
| D | Cell cycle control, cell division, and chromosome partitioning | 30 | 32 |
| E | Amino acid transport and metabolism | 175 | 177 |
| F | Nucleotide transport and metabolism | 87 | 87 |
| G | Carbohydrate transport and metabolism | 91 | 93 |
| H | Coenzyme transport and metabolism | 92 | 92 |
| I | Lipid transport and metabolism | 54 | 54 |
| J | Translation, ribosomal structure, and biogenesis | 201 | 203 |
| K | Transcription | 109 | 115 |
| L | Replication, recombination, and repair | 90 | 105 |
| M | Cell wall/membrane/envelope biogenesis | 113 | 117 |
| N | Cell motility | 14 | 16 |
| O | Post-translational modification, protein turnover, and chaperones | 58 | 59 |
| P | Inorganic ion transport and metabolism | 72 | 73 |
| Q | Secondary metabolites biosynthesis, transport, and catabolism | 12 | 12 |
| R | General function prediction only | 110 | 111 |
| S | Function unknown | 76 | 78 |
| T | Signal transduction mechanisms | 82 | 86 |
| U | Intracellular trafficking, secretion, and vesicular transport | 25 | 25 |
| V | Defense mechanisms | 56 | 65 |
| W | Extracellular structures | 6 | 6 |
| X | Mobilome: prophages and transposons | 29 | 42 |
| Z | Cytoskeleton | 1 | 1 |
| Sample ID | Type | Number | Average Length (bps) | Total Length (bps) |
|---|---|---|---|---|
| S.82.15 | tRNA | 78 | 75 | 5891 |
| S.82.15 | 5s | 7 | 136 | 949 |
| S.82.15 | 16s | 7 | 1536 | 10,753 |
| S.82.15 | 23s | 7 | 2899 | 20,293 |
| S.82.15 | sRNA | 10 | 108 | 1087 |
| S.82.20 | tRNA | 58 | 75 | 4379 |
| S.82.20 | 5s | 5 | 132 | 660 |
| S.82.20 | 16s | 5 | 1537 | 7684 |
| S.82.20 | 23s | 5 | 2901 | 14,504 |
| S.82.20 | sRNA | 12 | 121 | 1457 |
| Strain | Assembly Accession | Genome Size (bps) | ANI of S.82.15 Chromosome (%) | ANI of S.82.20 Chromosome (%) | Plasmid Size (bps) | ANI of S.82.20 Plasmid (%) |
|---|---|---|---|---|---|---|
| M18 | GCA_000225385.2 | 2,142,944 | 95.64 | 95.45 | 183,037 | 95.98 |
| K12 | GCA_000286295.1 | 2,241,314 | 95.02 | 94.99 | 185,045 | 92.47 |
| SALI-10 | GCA_022936265.1 | 2,096,969 | 96.55 | 96.57 | 164,439 | 93.2 |
| ATCC 25975 | GCA_002094975.1 | 2,199,793 | 95.90 | 95.79 | 126,555 | 93.24 |
| LAB813 | GCA_008305695.1 | 2,242,557 | 95.87 | 95.77 | 183,700 | 92.86 |
| Query ID | Subject ID | Query Cover (%) | Identity (%) | Description |
|---|---|---|---|---|
| S.82.20.Plas | NZ_MN480762.1 | 62.23 | 93.06 | NU10 plasmid pSsal-NU10 |
| S.82.20.Plas | NZ_CP018188.1 | 63.08 | 95.33 | ICDC2 plasmid |
| S.82.20.Plas | NZ_CP040803.1 | 50.4 | 94.93 | LAB813 plasmid pSAL813 |
| S.82.20.Plas | NZ_CP090008.1 | 50.4 | 94.92 | SALI-10 plasmid pSaLI10 |
| S.82.20.Plas | NZ_CP015284.1 | 46.84 | 94.29 | ATCC 25975 plasmid unnamed |
| Gene ID | Gene Name | Accession Number | Identity | E Value | Description |
|---|---|---|---|---|---|
| S.82.20_GM000144 | SivK | ACX68641.1 | 99.341 | 0 | SivK [Streptococcus salivarius] |
| S.82.20_GM000145 | SivR | ACX68642.1 | 99.569 | 2.74 × 10−169 | SivR [Streptococcus salivarius] |
| S.82.20_GM000146 | SivA | ABI54434.1 | 100 | 1.51 × 10−36 | SivA (plasmid) [Streptococcus salivarius] |
| S.82.20_GM000147 | SivM | WP_013990309.1 | 99.569 | 0 | Salivaricin 9 biosynthesis lanthionine synthetase SivM [Streptococcus salivarius] |
| S.82.20_GM000148 | SivT | ACX68645.1 | 99.422 | 0 | Putative ABC transporter [Streptococcus salivarius] |
| S.82.20_GM000149 | SivF | EGX29340.1 | 100 | 0 | Transport ATP-binding protein (plasmid) [Streptococcus salivarius] |
| S.82.20_GM000150 | SivE | ACX68647.1 | 99.602 | 2.72 × 10−179 | Putative transporter [Streptococcus salivarius] |
| S.82.20_GM000151 | SivG | ACX68648.1 | 100 | 3.86 × 10−178 | Putative transporter [Streptococcus salivarius] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, M.; Li, Q.; Zhang, F.; Yao, D.; Huang, W.; Lv, Q.; Jiang, H.; Kong, D.; Ren, Y.; Chen, S.; et al. The Genomic Characteristics of Potential Probiotics: Two Streptococcus salivarius Isolates from a Healthy Individual in China. Microorganisms 2025, 13, 694. https://doi.org/10.3390/microorganisms13030694
Sun M, Li Q, Zhang F, Yao D, Huang W, Lv Q, Jiang H, Kong D, Ren Y, Chen S, et al. The Genomic Characteristics of Potential Probiotics: Two Streptococcus salivarius Isolates from a Healthy Individual in China. Microorganisms. 2025; 13(3):694. https://doi.org/10.3390/microorganisms13030694
Chicago/Turabian StyleSun, Mingyue, Qian Li, Feiran Zhang, Ding Yao, Wenhua Huang, Qingyu Lv, Hua Jiang, Decong Kong, Yuhao Ren, Shaolong Chen, and et al. 2025. "The Genomic Characteristics of Potential Probiotics: Two Streptococcus salivarius Isolates from a Healthy Individual in China" Microorganisms 13, no. 3: 694. https://doi.org/10.3390/microorganisms13030694
APA StyleSun, M., Li, Q., Zhang, F., Yao, D., Huang, W., Lv, Q., Jiang, H., Kong, D., Ren, Y., Chen, S., Jiang, Y., & Liu, P. (2025). The Genomic Characteristics of Potential Probiotics: Two Streptococcus salivarius Isolates from a Healthy Individual in China. Microorganisms, 13(3), 694. https://doi.org/10.3390/microorganisms13030694






