Genomic Insights into Selenate Reduction by Anaerobacillus Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Inoculum and Preparation of Enrichment Cultures
2.2. Isolation of Se-Reducing Microorganisms
2.3. DNA Extraction and Genome Sequencing
2.4. Genome Assembly, Annotation, and Analysis
3. Results and Discussion
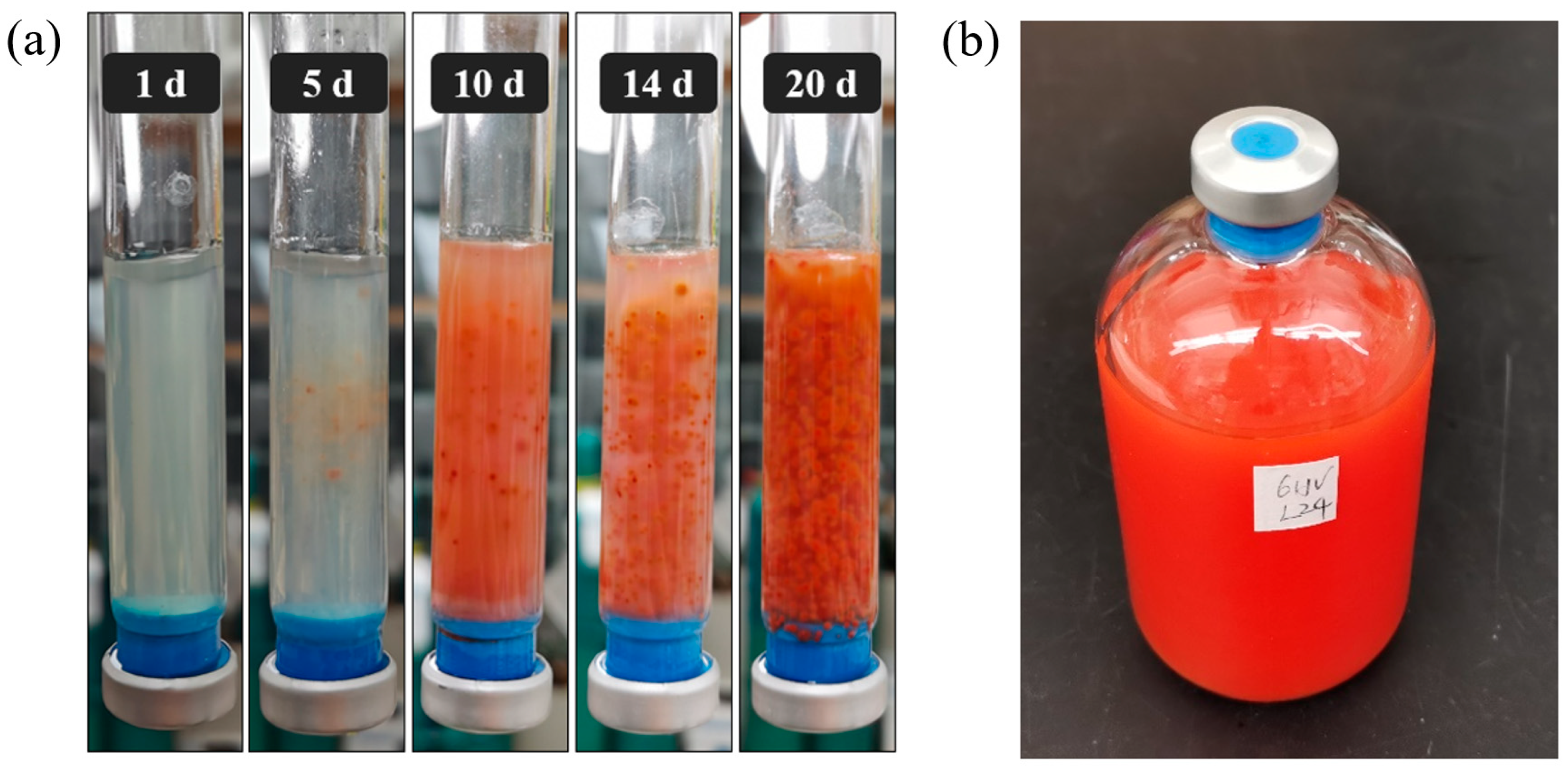
3.1. Growth Characteristic and Metabolic Diversity
3.2. Comprehensive Genome Profiling of Anaerobacillus Species
3.2.1. Overall Genome Characteristic
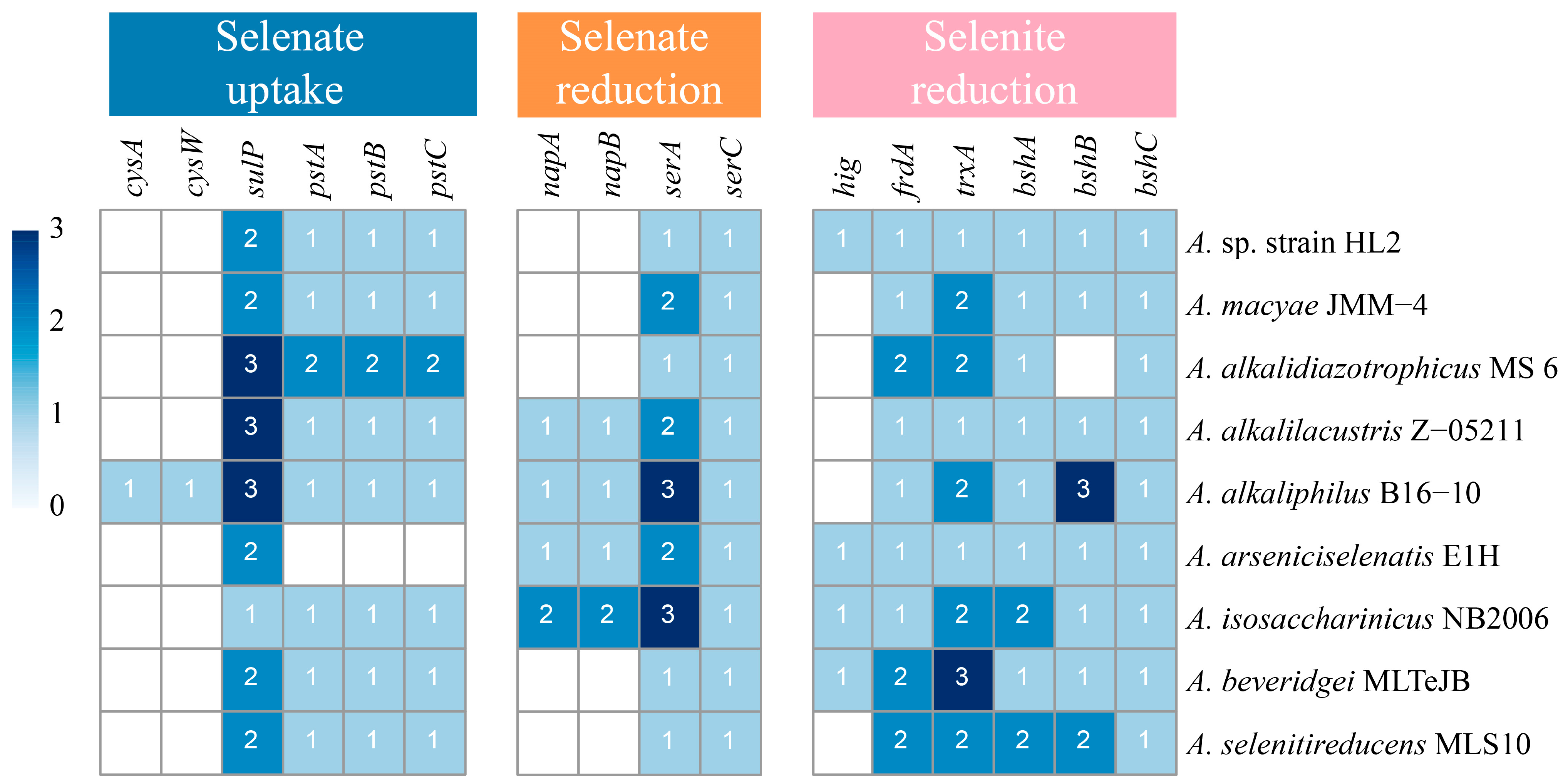
3.2.2. Identification of Genes Related to Se Transport and Transformation
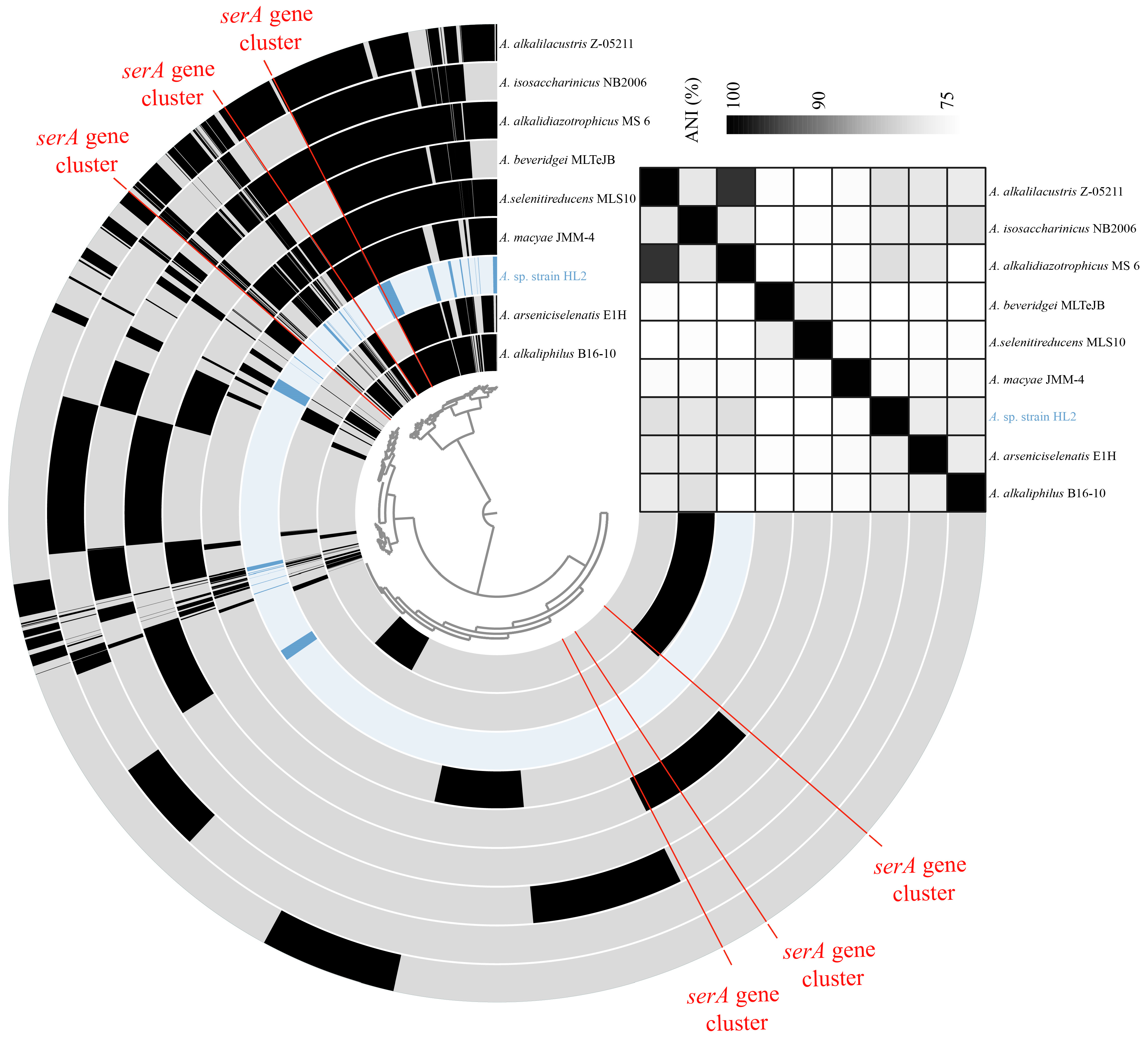
3.2.3. Comparative Analysis of serA Gene Cluster in Anaerobacillus Species
3.2.4. Metabolic Pathway Linking Se Reduction in Anaerobacillus sp. Strain HL2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santesmasses, D.; Mariotti, M.; Gladyshev, V.N. Bioinformatics of Selenoproteins. Antioxid. Redox Signal. 2020, 33, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Reich, H.J.; Hondal, R.J. Why Nature Chose Selenium. ACS Chem. Biol. 2016, 11, 821–841. [Google Scholar] [CrossRef] [PubMed]
- Ekumah, J.-N.; Ma, Y.; Akpabli-Tsigbe, N.D.K.; Kwaw, E.; Ma, S.M.; Hu, J. Global soil distribution, dietary access routes, bioconversion mechanisms and the human health significance of selenium: A review. Food Biosci. 2021, 41, 100960. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.; Zeng, W.; Liu, Y.; Zhao, D.; Zhang, Y.; Jia, X. Screening and Identification of Soil Selenium-Enriched Strains and Application in Auricularia auricula. Microorganisms 2024, 12, 1136. [Google Scholar] [CrossRef]
- Wang, L.; Ju, J.; Xie, H.; Qiao, F.; Luo, Q.; Zhou, L. Comparative Study on Growth and Metabolomic Profiles of Six Lactobacilli Strains by Sodium Selenite. Microorganisms 2024, 12, 1937. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.M.; Ghazy, M.; El-Sayed, H.; El-Hameed, R.M.; Khalil, R.G.; Korany, S.M.; Aloufi, A.S.; Hammam, O.A.; Morad, M.Y. Histopathological, Immunohistochemical, Biochemical, and In Silico Molecular Docking Study of Fungal-Mediated Selenium Oxide Nanoparticles on Biomphalaria alexandrina (Ehrenberg, 1831) Snails. Microorganisms 2023, 11, 811. [Google Scholar] [CrossRef] [PubMed]
- Rauschenbach, I.; Narasingarao, P.; Häggblom, M.M. Desulfurispirillum indicum sp. nov., a selenate- and selenite-respiring bacterium isolated from an estuarine canal. Int. J. Syst. Evol. Microbiol. 2011, 61, 654–658. [Google Scholar] [CrossRef]
- Wadgaonkar, S.L.; Nancharaiah, Y.V.; Jacob, C.; Esposito, G.; Lens, P.N.L. Microbial transformation of Se oxyanions in cultures of Delftia lacustris grown under aerobic conditions. J. Microbiol. 2019, 57, 362–371. [Google Scholar] [CrossRef]
- Staicu, L.C.; Barton, L.L. Selenium respiration in anaerobic bacteria: Does energy generation pay off? J. Inorg. Biochem. 2021, 222, 111509. [Google Scholar] [CrossRef]
- Hageman, S.P.W.; van der Weijden, R.D.; Weijma, J.; Buisman, C.J.N. Microbiological selenate to selenite conversion for selenium removal. Water Res. 2013, 47, 2118–2128. [Google Scholar] [CrossRef]
- von Wintzingerode, F.; Gobel, U.B.; Siddiqui, R.A.; Rösick, U.; Schumann, P.; Frühling, A.; Rohde, M.; Pukall, R.; Stackebrandt, E. Salana multivorans gen. nov., sp. nov., a novel actinobacterium isolated from an anaerobic bioreactor and capable of selenate reduction. Int. J. Syst. Evol. Microbiol. 2001, 51, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Rauschenbach, I.; Posternak, V.; Cantarella, P.; McConnell, J.; Starovoytov, V.; Häggblom, M.M. Seleniivibrio woodruffii gen. nov., sp. nov., a selenate- and arsenate-respiring bacterium in the Deferribacteraceae. Int. J. Syst. Evol. Microbiol. 2013, 63, 3659–3665. [Google Scholar] [CrossRef]
- Narasingarao, P.; Häggblom, M.M. Pelobacter seleniigenes sp. nov., a selenate-respiring bacterium. Int. J. Syst. Evol. Microbiol. 2007, 57, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Iino, T.; Suzuki, K.-I.; Harayama, S. Ferrimonas futtsuensis sp. nov. and Ferrimonas kyonanensis sp. nov., selenate-reducing bacteria belonging to the Gammaproteobacteria isolated from Tokyo Bay. Int. J. Syst. Evol. Microbiol. 2006, 56, 2639–2645. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Yamashita, M.; Fujimoto, N.; Kuroda, M.; Kashiwa, M.; Sei, K.; Fujita, M.; Ike, M. Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int. J. Syst. Evol. Microbiol. 2007, 57, 1060–1064. [Google Scholar] [CrossRef]
- Ike, M.; Kazuaki, T.M.; Tomoko, F.M.; Kashiwa, M.; Masanori, F.M. Selenate reduction by bacteria isolated from aquatic environment free from selenium contamination. Water Res. 2000, 34, 3019–3025. [Google Scholar] [CrossRef]
- Hunter, W.J.; Manter, D.K. Reduction of Selenite to Elemental Red Selenium by Rhizobium sp. Strain B1. Curr. Microbiol. 2007, 55, 344–349. [Google Scholar] [CrossRef]
- Kagami, T.; Narita, T.; Kuroda, M.; Notaguchi, E.; Yamashita, M.; Sei, K.; Soda, S.; Ike, M. Effective selenium volatilization under aerobic conditions and recovery from the aqueous phase by NT-I. Water Res. 2013, 47, 1361–1368. [Google Scholar] [CrossRef]
- Shi, L.-D.; Lv, P.-L.; McIlroy, S.J.; Wang, Z.; Dong, X.-L.; Kouris, A.; Lai, C.-Y.; Tyson, G.W.; Strous, M.; Zhao, H.-P. Methane-dependent selenate reduction by a bacterial consortium. ISME J. 2021, 15, 3683–3692. [Google Scholar] [CrossRef]
- Wen, L.-L.; Lai, C.-Y.; Yang, Q.; Chen, J.-X.; Zhang, Y.; Ontiveros-Valencia, A.; Zhao, H.-P. Quantitative detection of selenate-reducing bacteria by real-time PCR targeting the selenate reductase gene. Enzym. Microb. Technol. 2016, 85, 19–24. [Google Scholar] [CrossRef]
- Kuroda, M.; Yamashita, M.; Miwa, E.; Imao, K.; Fujimoto, N.; Ono, H.; Nagano, K.; Sei, K.; Ike, M. Molecular Cloning and Characterization of the srdBCA Operon, Encoding the Respiratory Selenate Reductase Complex, from the Selenate-Reducing Bacterium Bacillus selenatarsenatis SF-1. J. Bacteriol. 2011, 193, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Sabaty, M.; Avazeri, C.; Pignol, D.; Vermeglio, A. Characterization of the Reduction of Selenate and Tellurite by Nitrate Reductases. Appl. Environ. Microbiol. 2001, 67, 5122–5126. [Google Scholar] [CrossRef] [PubMed]
- Theisen, J.; Yee, N. The Molecular Basis for Selenate Reduction in Citrobacter freundii. Geomicrobiol. J. 2014, 31, 875–883. [Google Scholar] [CrossRef]
- Ridley, H.; Watts, C.A.; Richardson, D.J.; Butler, C.S. Resolution of Distinct Membrane-Bound Enzymes from Enterobacter cloacae SLD1a-1 That Are Responsible for Selective Reduction of Nitrate and Selenate Oxyanions. Appl. Environ. Microbiol. 2006, 72, 5173–5180. [Google Scholar] [CrossRef]
- Wells, M.; McGarry, J.; Gaye Maissa, M.; Basu, P.; Oremland Ronald, S.; Stolz, J.F. Respiratory Selenite Reductase from Bacillus selenitireducens Strain MLS10. J. Bacteriol. 2019, 201, e00614-18. [Google Scholar] [CrossRef]
- Li, D.-B.; Cheng, Y.-Y.; Wu, C.; Li, W.-W.; Li, N.; Yang, Z.-C.; Tong, Z.-H.; Yu, H.-Q. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci. Rep. 2014, 4, 3735. [Google Scholar] [CrossRef]
- Basaglia, M.; Toffanin, A.; Baldan, E.; Bottegal, M.; Shapleigh, J.P.; Casella, S. Selenite-reducing capacity of the copper-containing nitrite reductase of Rhizobium sullae. FEMS Microbiol. Lett. 2010, 269, 124–130. [Google Scholar] [CrossRef]
- Wang, D.; Xia, X.; Wu, S.; Zheng, S.; Wang, G. The essentialness of glutathione reductase GorA for biosynthesis of Se(0)-nanoparticles and GSH for CdSe quantum dot formation in Pseudomonas stutzeri TS44. J. Hazard. Mater. 2019, 366, 301–310. [Google Scholar] [CrossRef]
- Yasir, M.; Zhang, Y.; Xu, Z.; Luo, M.; Wang, G. NAD(P)H-dependent thioredoxin-disulfide reductase TrxR is essential for tellurite and selenite reduction and resistance in Bacillus sp. Y3. FEMS Microbiol. Ecol. 2020, 96, fiaa126. [Google Scholar] [CrossRef]
- Xia, X.; Wu, S.J.; Li, N.H.; Wang, D.; Zheng, S.X.; Wang, G. Novel bacterial selenite reductase CsrF responsible for Se(IV) and Cr(VI) reduction that produces nanoparticles in Alishewanella sp. WH16-1. J. Hazard. Mater. 2018, 342, 499–509. [Google Scholar] [CrossRef]
- Yanke, L.J.; Bryant, R.D.; Laishley, E.J. Hydrogenase I of Clostridium pasteurianum functions as a novel selenite reductase. Anaerobe 1995, 1, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, Y.; Tang, C.; Jia, H.; Wu, L. Speeding up selenite bioremediation using the highly selenite-tolerant strain Providencia rettgeri HF16-A novel mechanism of selenite reduction based on proteomic analysis. J. Hazard. Mater. 2021, 406, 124690. [Google Scholar] [CrossRef]
- Tan, Y.; Wang, Y.; Wang, Y.; Xu, D.; Huang, Y.; Wang, D.; Wang, G.; Rensing, C.; Zheng, S. Novel mechanisms of selenate and selenite reduction in the obligate aerobic bacterium Comamonas testosteroni S44. J. Hazard. Mater. 2018, 359, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Song, D.G.; Li, X.X.; Cheng, Y.Z.; Xiao, X.; Lu, Z.Q.; Wang, Y.; Wang, F. Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci. Rep. 2017, 7, 3239. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.; Zhou, S.F.; Li, D.B.; Song, D.; Yang, S.; Xu, M. Reduction of selenite to selenium nanoparticles by highly selenite-tolerant bacteria isolated from seleniferous soil. J. Hazard. Mater. 2024, 472, 134491. [Google Scholar] [CrossRef]
- Zhu, T.-T.; Tian, L.-J.; Yu, H.-Q. Phosphate-Suppressed Selenite Biotransformation by Escherichia coli. Environ. Sci. Technol. 2020, 54, 10703–10711. [Google Scholar] [CrossRef]
- Mataigne, V.; Vannier, N.; Vandenkoornhuyse, P.; Hacquard, S. Multi-genome metabolic modeling predicts functional inter-dependencies in the Arabidopsis root microbiome. Microbiome 2022, 10, 217. [Google Scholar] [CrossRef]
- Joshi, A.; Thite, S.; Karodi, P.; Joseph, N.; Lodha, T. Alkalihalobacterium elongatum gen. nov. sp. nov.: An Antibiotic-Producing Bacterium Isolated From Lonar Lake and Reclassification of the Genus Alkalihalobacillus Into Seven Novel Genera. Front. Microbiol. 2022, 13, 722369, Erratum in Front. Microbiol. 2022, 13, 871596. [Google Scholar] [CrossRef]
- Fisher, J.C.; Hollibaugh, J.T. Selenate-Dependent Anaerobic Arsenite Oxidation by a Bacterium from Mono Lake, California. Appl. Environ. Microbiol. 2008, 74, 2588–2594. [Google Scholar] [CrossRef]
- Baesman, S.M.; Stolz, J.F.; Kulp, T.R.; Oremland, R.S. Enrichment and isolation of Bacillus beveridgei sp. nov., a facultative anaerobic haloalkaliphile from Mono Lake, California, that respires oxyanions of tellurium, selenium, and arsenic. Extremophiles 2009, 13, 695–705. [Google Scholar] [CrossRef]
- Blum, J.S.; Bindi, A.B.; Buzzelli, J.; Stolz, J.F.; Oremland, R.S. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: Two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 1998, 171, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Santini, J.M. Bacillus macyae sp. nov., an arsenate-respiring bacterium isolated from an Australian gold mine. Int. J. Syst. Evol. Microbiol. 2004, 54, 2241–2244. [Google Scholar] [CrossRef] [PubMed]
- Knight, V.K.; Nijenhuis, I.; Kerkhof, L.J.; Häggblom, M.M. Degradation of aromatic compounds coupled to selenate reduction. Geomicrobiol. J. 2002, 19, 77–86. [Google Scholar] [CrossRef]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2020, 19, 225–240. [Google Scholar] [CrossRef]
- Janabi, A.H.D.; Kerkhof, L.J.; McGuinness, L.R.; Biddle, A.S.; McKeever, K.H. Comparison of a modified phenol/chloroform and commercial-kit methods for extracting DNA from horse fecal material. J. Microbiol. Methods 2016, 129, 14–19. [Google Scholar] [CrossRef]
- Chen, W.; Karton, A.; Hussian, T.; Javaid, S.; Wang, F.; Pang, Y.; Jia, G. Spontaneous shape and phase control of colloidal ZnSe nanocrystals by tailoring Se precursor reactivity. CrystEngComm 2019, 21, 2955–2961. [Google Scholar] [CrossRef]
- Nie, X.L.; Yang, X.R.; He, J.Y.; Liu, P.; Shi, H.; Wang, T.; Zhang, D. Bioconversion of inorganic selenium to less toxic selenium forms by microbes: A review. Front. Bioeng. Biotechnol. 2023, 11, 1167123. [Google Scholar] [CrossRef]
- Fesharaki, P.J.; Nazari, P.; Shakibaie, M.; Rezaie, S.; Banoee, M.; Abdollahi, M.; Shahverdi, A.R. Biosynthesis of selenium nanoparticles using Klebsiella pneumoniae and their recovery by a simple sterilization process. Braz. J. Microbiol. 2010, 41, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Nayak, V.; Singh, K.R.; Singh, A.K.; Singh, R.P. Potentialities of selenium nanoparticles in biomedical science. New J. Chem. 2021, 45, 2849–2878. [Google Scholar] [CrossRef]
- Sorokin, I.D.; Kravchenko, I.K.; Tourova, T.P.; Kolganova, T.V.; Boulygina, E.S.; Sorokin, D.Y. Bacillus alkalidiazotrophicus sp. nov., a diazotrophic, low salt-tolerant alkaliphile isolated from Mongolian soda soil. Int. J. Syst. Evol. Microbiol. 2008, 58, 2459–2464. [Google Scholar] [CrossRef]
- Zavarzina, D.G.; Tourova, T.P.; Kolganova, T.V.; Boulygina, E.S.; Zhilina, T.N. Description of Anaerobacillus alkalilacustre gen. nov., sp. nov.—Strictly anaerobic diazotrophic bacillus isolated from soda lake and transfer of Bacillus arseniciselenatis, Bacillus macyae, and Bacillus alkalidiazotrophicus to Anaerobacillus as the new combinations A. arseniciselenatis comb. nov., A. macyae comb. nov., and A. alkalidiazotrophicus comb. nov. Microbiology 2009, 78, 723–731. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Sukhacheva, M.V.; Muyzer, G. Desulfuribacillus alkaliarsenatis gen. nov. sp. nov., a deep-lineage, obligately anaerobic, dissimilatory sulfur and arsenate-reducing, haloalkaliphilic representative of the order Bacillales from soda lakes. Extremophiles 2012, 16, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Borsodi, A.K.; Aszalós, J.M.; Bihari, P.; Nagy, I.; Schumann, P.; Spröer, C.; Kovács, A.L.; Bóka, K.; Dobosy, P.; Óvári, M.; et al. Anaerobacillus alkaliphilus sp. nov., a novel alkaliphilic and moderately halophilic bacterium. Int. J. Syst. Evol. Microbiol. 2019, 69, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Bassil, N.M.; Lloyd, J.R. Anaerobacillus isosaccharinicus sp. nov., an alkaliphilic bacterium which degrades isosaccharinic acid. Int. J. Syst. Evol. Microbiol. 2018, 69, 631–637. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Gunturu, S.; Harvey, W.T.; Rosselló-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Barajas, E.; Díaz-Pérez, C.; Ramírez-Díaz, M.I.; Riveros-Rosas, H.; Cervantes, C. Bacterial transport of sulfate, molybdate, and related oxyanions. BioMetals 2011, 24, 687–707. [Google Scholar] [CrossRef]
- Stoeva, M.K.; Coates, J.D. Specific inhibitors of respiratory sulfate reduction: Towards a mechanistic understanding. Microbiology 2018, 165, 254–269. [Google Scholar] [CrossRef]
- Cypionka, H. Characterization of sulfate transport in Desulfovibrio desulfuricans. Arch. Microbiol. 1989, 152, 237–243. [Google Scholar] [CrossRef]
- Shibagaki, N.; Rose, A.; McDermott, J.; Fujiwara, T.; Hayashi, H.; Yoneyama, T.; Davies, J.P. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. Cell Mol. Biol. 2002, 29, 475–486. [Google Scholar] [CrossRef]
- Guymer, D.; Maillard, J.; Sargent, F. A genetic analysis of in vivo selenate reduction by Salmonella enterica serovar Typhimurium LT2 and Escherichia coli K12. Arch. Microbiol. 2009, 191, 519–528. [Google Scholar] [CrossRef]
- Xu, R.; Kolton, M.; Tao, W.; Sun, X.X.; Su, P.; Huang, D.; Zhang, M.; Yang, Z.; Guo, Z.; Gao, H.; et al. Anaerobic selenite-reducing bacteria and their metabolic potentials in Se-rich sediment revealed by the combination of DNA-stable isotope probing, metagenomic binning, and metatranscriptomics. J. Hazard. Mater. 2023, 457, 131834. [Google Scholar] [CrossRef] [PubMed]
- McEwan, A.G.; Ridge, J.P.; McDevitt, C.A.; Hugenholtz, P. The DMSO Reductase Family of Microbial Molybdenum Enzymes; Molecular Properties and Role in the Dissimilatory Reduction of Toxic Elements. Geomicrobiol. J. 2002, 19, 3–21. [Google Scholar] [CrossRef]
- Krafft, T.; Bowen, A.; Theis, F.; Macy, J.M. Cloning and Sequencing of the Genes Encoding the Periplasmic-Cytochrome B-Containing Selenate Reductase of Thauera selenatis. DNA Seq. 2000, 10, 365–377. [Google Scholar] [CrossRef]
- Septer, A.N.; Bose, J.L.; Dunn, A.K.; Stabb, E.V. FNR-mediated regulation of bioluminescence and anaerobic respiration in the light-organ symbiont Vibrio fischeri. FEMS Microbiol. Lett. 2010, 306, 72–81. [Google Scholar] [CrossRef]
- Salzberg, L.I.; Luo, Y.; Hachmann, A.-B.; Mascher, T.; Helmann, J.D. The Bacillus subtilis GntR Family Repressor YtrA Responds to Cell Wall Antibiotics. J. Bacteriol. 2011, 193, 5793–5801. [Google Scholar] [CrossRef]
- Jha, B.; Kumar, D.; Sharma, A.; Dwivedy, A.; Singh, R.; Biswal, B.K. Identification and structural characterization of a histidinol phosphate phosphatase from Mycobacterium tuberculosis. J. Biol. Chem. 2018, 293, 10102–10118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dobson, C.M.; Wu, X.; Lerner-Ellis, J.; Rosenblatt, D.S.; Gravel, R.A. Impact of cblB mutations on the function of ATP:cob(I)alamin adenosyltransferase in disorders of vitamin B12 metabolism. Mol. Genet. Metab. 2006, 87, 315–322. [Google Scholar] [CrossRef]
- Llamas, A.; Mendel, R.R.; Schwarz, G. Synthesis of adenylated molybdopterin: An essential step for molybdenum insertion. J. Biol. Chem. 2004, 279, 55241–55246. [Google Scholar] [CrossRef]
- Wolfgang, B.; Thauer, R.K. Flavin-Based Electron Bifurcation, Ferredoxin, Flavodoxin, and Anaerobic Respiration With Protons (Ech) or NAD+ (Rnf) as Electron Acceptors: A Historical Review. Front. Microbiol. 2018, 9, 401. [Google Scholar] [CrossRef]


| Metabolism | 1 | 2 a | 3 b | 4 c | 5 d | 6 e,f | 7 a | 8 g | 9 h |
|---|---|---|---|---|---|---|---|---|---|
| Fermentation | − | − | + | + | + | + | + | − | + |
| Electron Donors | |||||||||
| Acetate | + | + | + | − | + | + | − | − | − |
| Starch | + | − | + | + | + | + | + | − | + |
| Pyruvate | − | + | + | + | n.a. | + | − | − | + |
| Electron Acceptors | |||||||||
| Oxygen | − | − | − | + | − | − | − | + | − |
| Nitrate | − | − | + | + | − | − | + | + | + |
| Selenate | + | − | + | + | − | + | + | n.a. | n.a. |
| Selenite | + | − | + | + | − | + | + | n.a. | n.a. |
| Arsenate | + | + | + | + | − | + | + | − | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhang, J.; Liang, J.; Wang, Y.; Ren, C.; Chen, X.; Cheng, D.; Zhang, H.; Liu, H. Genomic Insights into Selenate Reduction by Anaerobacillus Species. Microorganisms 2025, 13, 659. https://doi.org/10.3390/microorganisms13030659
Wang Q, Zhang J, Liang J, Wang Y, Ren C, Chen X, Cheng D, Zhang H, Liu H. Genomic Insights into Selenate Reduction by Anaerobacillus Species. Microorganisms. 2025; 13(3):659. https://doi.org/10.3390/microorganisms13030659
Chicago/Turabian StyleWang, Qidong, Jian Zhang, Jinhui Liang, Yanlong Wang, Chongyang Ren, Xinhan Chen, Dongle Cheng, Huanxin Zhang, and Huaqing Liu. 2025. "Genomic Insights into Selenate Reduction by Anaerobacillus Species" Microorganisms 13, no. 3: 659. https://doi.org/10.3390/microorganisms13030659
APA StyleWang, Q., Zhang, J., Liang, J., Wang, Y., Ren, C., Chen, X., Cheng, D., Zhang, H., & Liu, H. (2025). Genomic Insights into Selenate Reduction by Anaerobacillus Species. Microorganisms, 13(3), 659. https://doi.org/10.3390/microorganisms13030659






