The Response Microbial of the Cucumber Rhizosphere Network Keystone Taxa of the Cucumber Rhizosphere to Continuous Fertilization
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sample Collection
2.3. Evaluation of Soil Physicochemical Properties and Plant Health
2.4. Rhizosphere Soil DNA Extraction and NovaSeq Sequencing
2.5. NovaSeq Sequencing Data Processing
2.6. Network Analysis
2.7. Statistical Analysis
3. Results
3.1. Soil Properties and Cucumber Growth
3.2. Changes in Rhizosphere Microbial Network Structure Caused by Continuous Fertilization
3.3. Predicted Keystone Taxa Related to Fertilizer Treatments
3.4. Structural Equation Model Construction
4. Discussion
4.1. Intensive Fertilization Degraded the Rhizosphere
4.2. Rhizosphere Microbial Networks Were Reshaped by Continuous Fertilization
4.3. Response of Rhizosphere Microbial Keystone Taxa to Fertilizers
4.4. Rhizosphere Microbial Network Variation Unveiled by Structural Equation Models
4.5. Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Li, T.; Li, Y.; Björn, L.O.; Rosendahl, S. Long-term fertilization alters the complexity and stability of soil microbial networks in a wheat-maize cropping system. Soil Biol. Biochem. 2021, 163, 108452. [Google Scholar]
- Domeignoz-Horta, L.A.; Cappelli, S.L.; Shrestha, R.; Gerin, S.; Lohila, A.K.; Heinonsalo, J.; Nelson, D.B.; Kahmen, A.; Duan, P.; Sebag, D.; et al. Plant diversity drives positive microbial associations in the rhizosphere enhancing carbon use efficiency in agricultural soils. Nat. Commun. 2024, 15, 8065. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.; Liu, Y.; Wang, Z.; Zhang, Y.; Chen, S. Rhizosphere microbiomes as a secondary genome: A novel perspective on plant growth and health. Nat. Microbiol. 2023, 8, 2156–2172. [Google Scholar]
- Wang, L.; Zhang, L.; Yang, Q.; Liu, H.; Chen, L.; Zhang, M.; Guo, J. Soil microbiota diversity and the functional roles of key taxa in soil health and plant growth. Environ. Microbiol. 2024, 26, 1903–1921. [Google Scholar]
- Kumar, A.; Singh, R.; Yadav, A.; Yadav, P. Cucumber (Cucumis sativus L.) Phytochemicals and Their Health Promoting Benefits: A Comprehensive Review. J. Food Sci. Technol. 2024, 61, 2279–2292. [Google Scholar]
- Zhang, L.; Yuan, L.; Ai, C.; Zhang, H.; Zhou, W. Maize functional requirements drive the selection of rhizobacteria under long-term fertilization practices. New Phytol. 2024, 243, 854–866. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Zhang, Z.; Zhao, Y.; Chen, L.; Yang, J.; Xu, Y.; He, Y. Effects of fertilization on the rhizosphere microbial community and plant growth in a cucumber cultivation system. Appl. Soil Ecol. 2019, 140, 58–66. [Google Scholar]
- Zhang, Y.; Zhang, X.; Wei, Z.; Yang, H.; Shen, Q.; Zhang, R. Rhizosphere microorganisms as key drivers of nutrient cycling and disease suppression in agroecosystems. Environ. Microbiol. Rep. 2021, 13, 249–257. [Google Scholar]
- Mouquet, N.; Dossantos, M.; Thébault, E.; Guillemette, T.; Loreau, M. Ecological network analysis reveals the impact of biodiversity loss on ecosystem functioning. Nat. Commun. 2019, 10, 1185. [Google Scholar]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Gao, L.; Yang, L.; Xu, J.; Li, X.; Wang, S.; Zhang, R. Microbial network analysis reveals the negative effect of agricultural practices on fungal community structure and connectivity in soil. Appl. Soil Ecol. 2021, 163, 103909. [Google Scholar]
- Yuan, H.; Ge, T.; Zhou, P.; Liu, S.; Roberts, P.; Zhu, H.; Zou, Z.; Tong, C.; Wu, J. Soil microbial biomass and bacterial and fungal community structures responses to long-term fertilization in paddy soils. J. Soils Sediments 2013, 13, 877–886. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Zhang, W.; Li, L.; Xu, S.; Liu, Z.; Guo, S. Spatiotemporal variation in the abundance and distribution of keystone taxa in soil: Implications for soil health and ecosystem services. Soil Biol. Biochem. 2020, 142, 107706. [Google Scholar]
- Herren, C.M.; McMahon, K.D. Keystone taxa predict compositional change in microbial communities. Environ. Microbiol. 2018, 20, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tang, J.; Li, C.; Zhang, H.; Yuan, S. Reducing potential of chemical fertilizers and scientific fertilization countermeasure in vegetable production in China. J. Plant Nutr. Fertil. 2017, 23, 1480–1493. [Google Scholar]
- Cai, F.; Pang, G.; Li, R.-X.; Li, R.; Gu, X.-L.; Shen, Q.-R.; Chen, W. Bioorganic fertilizer maintains a more stable soil microbiome than chemical fertilizer for monocropping. Biol. Fertil. Soils 2017, 53, 861–872. [Google Scholar] [CrossRef]
- Wang, G.; Xu, Y.; Jin, J.; Liu, J.; Zhang, Q.; Liu, X. Effect of soil type and soybean genotype on fungal community in soybean rhizosphere during reproductive growth stages. Plant Soil 2009, 317, 135. [Google Scholar] [CrossRef]
- Caires, E.F.; Filho, R.Z.; Barth, G.; Joris, H.A. Optimizing nitrogen use efficiency for no-till corn production by improving root growth and capturing NO3-N in subsoil. Pedosphere 2016, 26, 474–485. [Google Scholar] [CrossRef]
- Chen, L.; Yang, X.; Raza, W.; Li, J.; Liu, Y.; Qiu, M.; Zhang, F.; Shen, Q. Trichoderma harzianum SQR-T037 rapidly degrades allelochemicals in rhizospheres of continuously cropped cucumbers. Appl. Microbiol. Biotechnol. 2011, 89, 1653–1663. [Google Scholar] [CrossRef]
- Zhang, F.; Zhu, Z.; Yang, X.; Ran, W.; Shen, Q. Trichoderma harzianum T-E5 significantly affects cucumber root exudates and fungal community in the cucumber rhizosphere. Appl. Soil Ecol. 2013, 72, 41–48. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Tscharntke, T.; Clough, Y.; Wanger, T.C.; Jackson, L.; Motzke, I.; Perfecto, I.; Vandermeer, J.; Whitbread, A. Global food security, biodiversity conservation and the future of agricultural intensification. Biol. Conserv. 2012, 151, 53–59. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, H.; Guo, X.; Yang, W.; Liu, J.; Zhang, T.; Huang, X. Soil degradation and its impact on soil microbial communities: A review of recent research. Geoderma 2021, 386, 114854. [Google Scholar]
- Beiying, Z.; Tianlin, C.; Bing, W. Effects of longterm uses of chemical fertilizers on soil quality. Chin. Agric. Sci. Bull. 2010, 26, 182–187. [Google Scholar]
- Zhao, X.; Zhang, W.; Liu, Y.; Zhang, J.; Chen, L. Effects of long-term fertilization on soil acidification and salinization in agroecosystems: A review. Sci. Total Environ. 2019, 668, 254–267. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Z.; Wu, Y.; Sun, H.; Zhang, Y.; Zhou, D. Organic and compound fertilizers improve soil properties and mitigate secondary salinization in arid regions. Agric. Ecosyst. Environ. 2021, 307, 107250. [Google Scholar] [CrossRef]
- Qiu, M.; Zhang, R.; Xue, C.; Zhang, S.; Li, S.; Zhang, N.; Shen, Q. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fertil. Soils 2012, 48, 807–816. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, F.; He, Y.; Liu, S.; Chen, Z.; Xu, Q. Continuous application of bioorganic fertilizer alters microbial community structure and decreases microbial abundance in cucumber rhizosphere soil. J. Soils Sediments 2021, 21, 2794–2805. [Google Scholar]
- Liu, L.; Chen, X.; Zhang, Q.; Wang, C.; Yang, J.; Liu, S. Shifts in the rhizosphere microbiome of tomato in response to soil types and cultivation practices. Microorganisms 2021, 9, 1221. [Google Scholar]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- de Menezes, A.B.; Müller, C.; Clipson, N.; Doyle, E. The soil microbiome at the Gi-FACE experiment responds to a moisture gradient but not to CO2 enrichment. Microbiology 2016, 162, 1572–1582. [Google Scholar] [CrossRef]
- Li, P.; Wang, W.; Li, J.; Zhang, H.; Zhao, Q. Bacteroidetes as a sensitive bioindicator of agricultural soil quality: The influence of long-term fertilization on soil microbiome structure. Soil Biol. Biochem. 2020, 146, 107817. [Google Scholar]
- Bouskill, N.J.; Barker-Finkel, J.; Galloway, T.S.; Handy, R.D.; Ford, T.E. Temporal bacterial diversity associated with metal-contaminated river sediments. Ecotoxicology 2010, 19, 317–328. [Google Scholar] [CrossRef]
- LeBlanc, N.; Crouch, J.A. Prokaryotic taxa play keystone roles in the soil microbiome associated with woody perennial plants in the genus Buxus. Ecol. Evol. 2019, 9, 11102–11111. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Y.; Ma, X.; Li, X.; Wang, Y. Impact of long-term NPK fertilization on fungal communities in agricultural soils: Focusing on the exclusion of Basidiomycota. Soil Biol. Biochem. 2020, 148, 107874. [Google Scholar]
- Oksanen, J. Multivariate analysis of ecological communities in R: Vegan tutorial. R Package Version 2011, 1, 1–43. [Google Scholar]
- Kent, M. Numerical classification and ordination methods in biogeography. Prog. Phys. Geogr. 2006, 30, 399–408. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Bowker, M.A.; Grace, J.B.; Powell, J.R. From patterns to causal understanding: Structural equation modeling (SEM) in soil ecology. Pedobiologia 2015, 58, 65–72. [Google Scholar] [CrossRef]
- Zhu, S.; Vivanco, J.M.; Manter, D.K. Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl. Soil Ecol. 2016, 107, 324–333. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.; Li, X.; Gao, L.; Zhang, Y. Effects of continuous fertilization on soil salinity and microbial community in a rice-wheat rotation system. Environ. Sci. Pollut. Res. 2018, 25, 10056–10065. [Google Scholar]
- Hamlin, R.L.; Barker, A.V. Influence of ammonium and nitrate nutrition on plant growth and zinc accumulation by Indian mustard. J. Plant Nutr. 2006, 29, 1523–1541. [Google Scholar] [CrossRef]
- Mosier, A.; Doran, J.; Freney, J. Managing soil denitrification. J. Soil Water Conserv. 2002, 57, 505–512. [Google Scholar]
- Liu, C.; Yang, X.; Liu, Y.; Zhang, X.; Li, L. Root exudates regulate the rhizosphere microbiome in a maize–wheat intercropping system. ISME J. 2019, 6, 1529–1541. [Google Scholar]
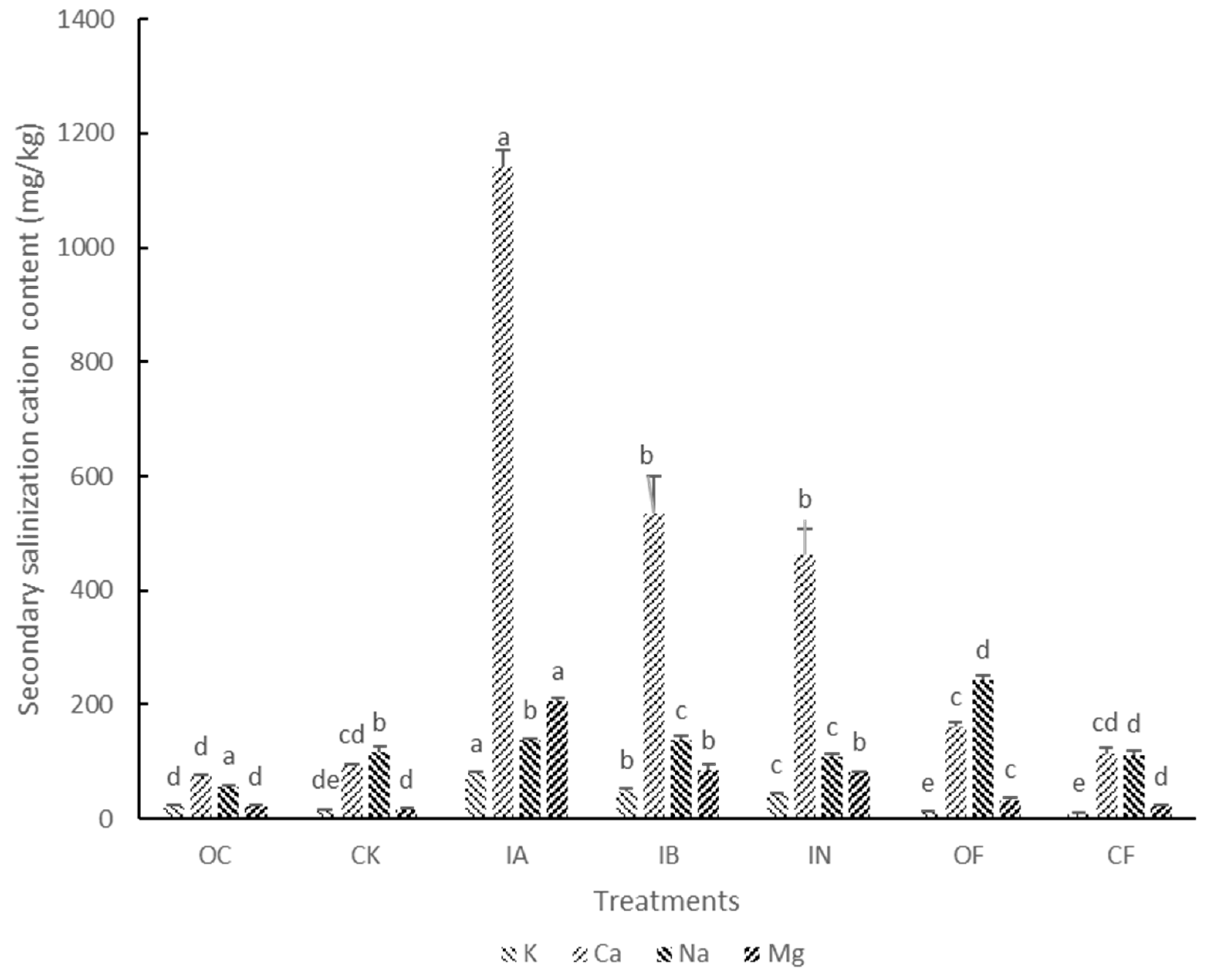
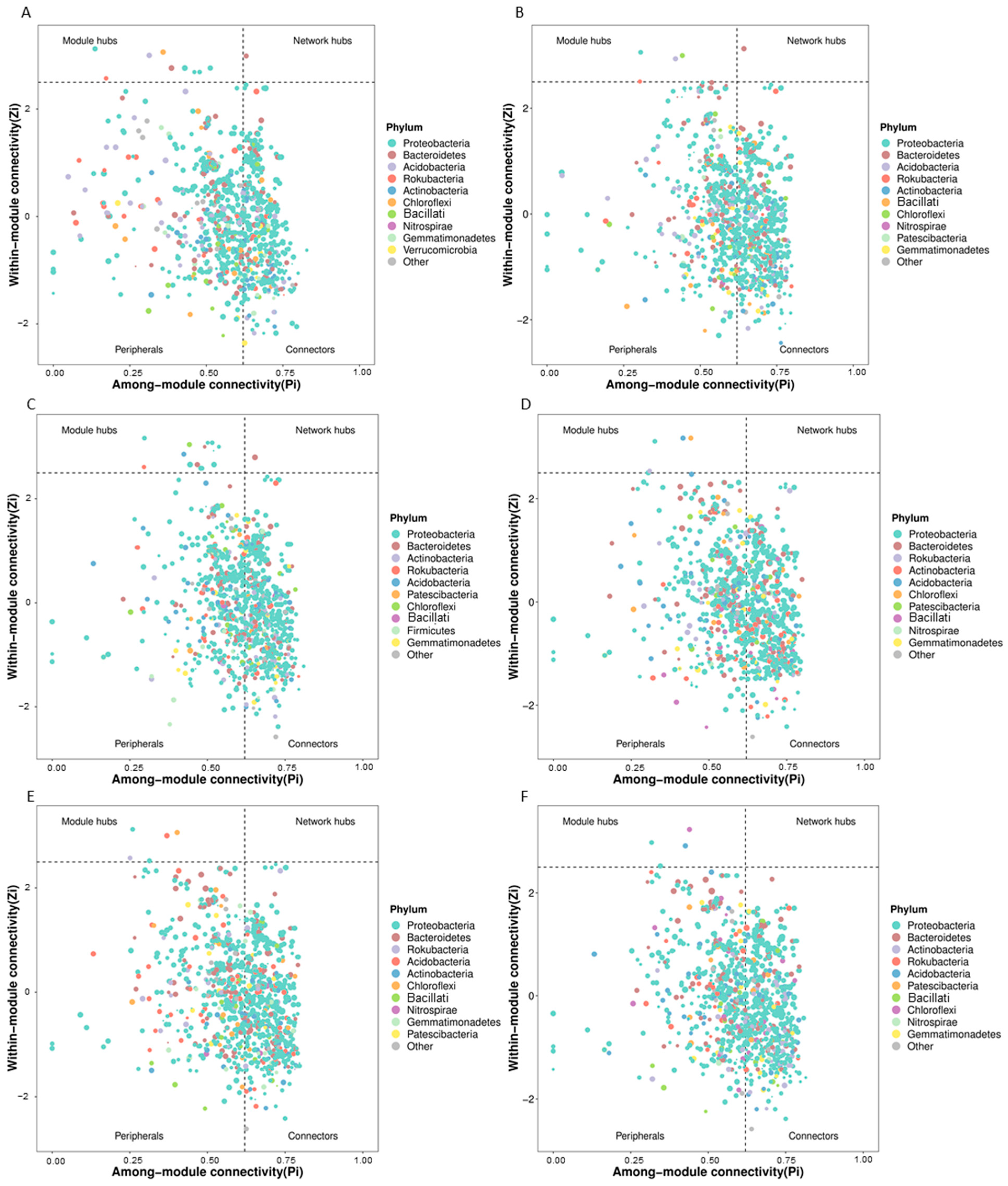
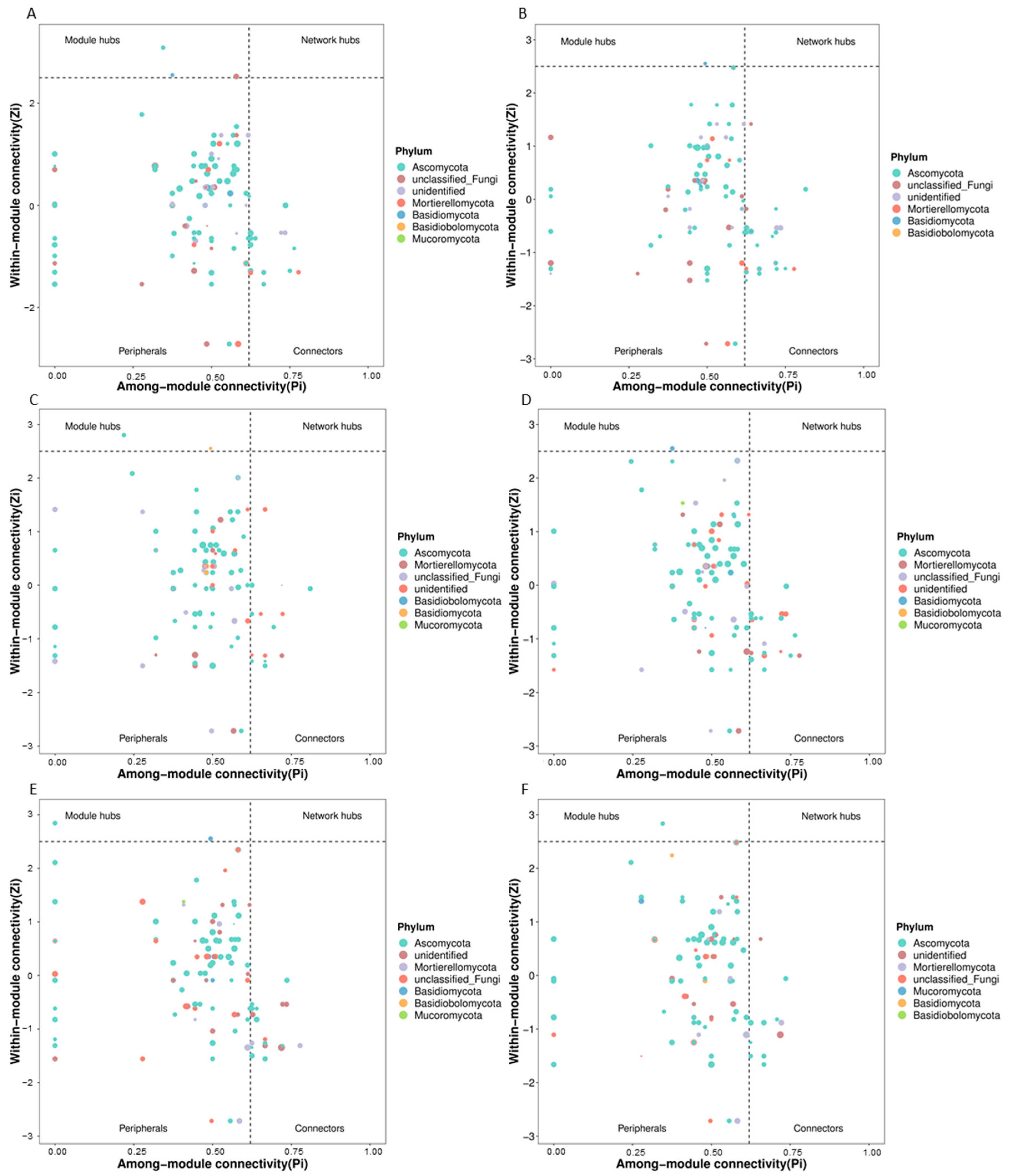

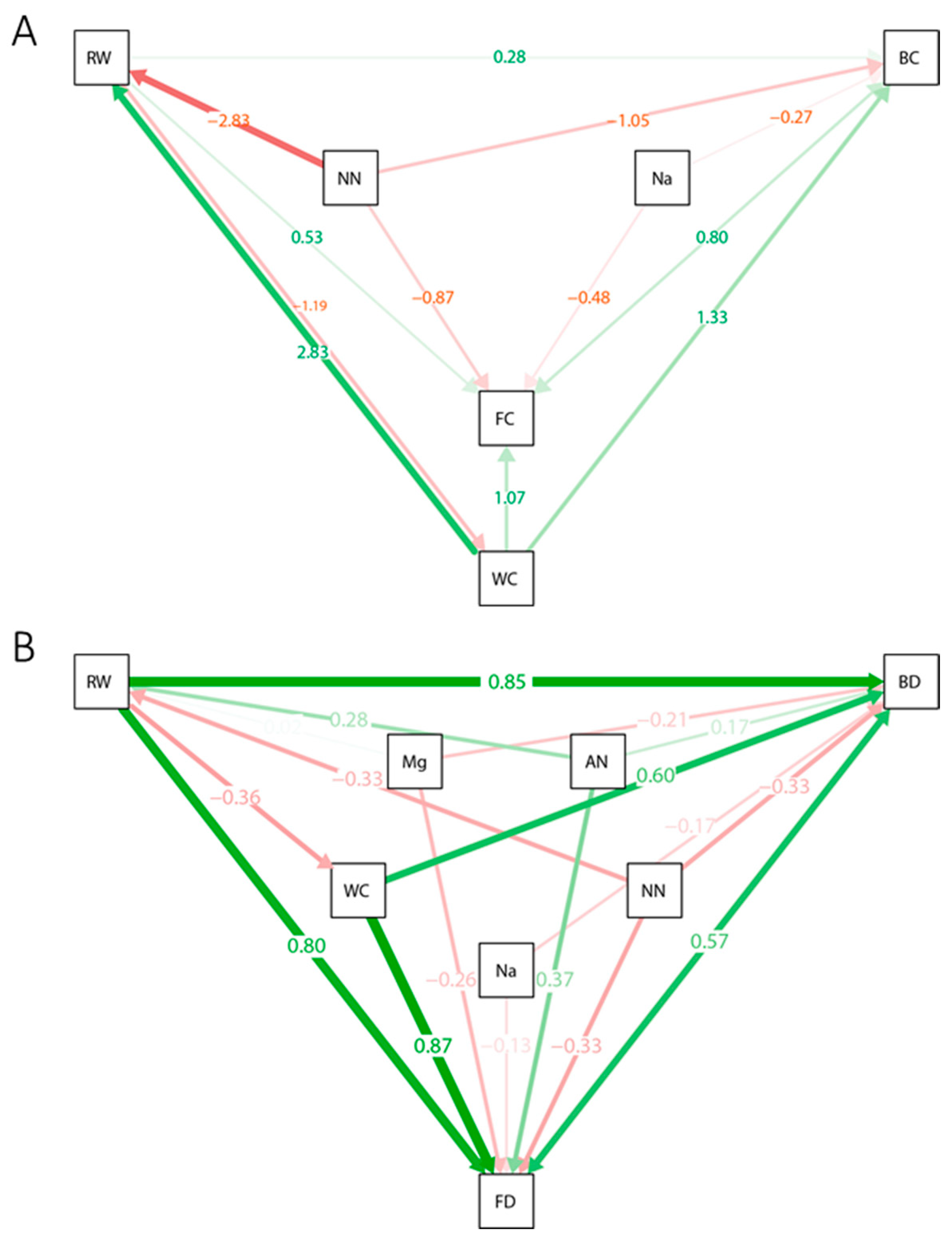
| Sample | Water Content (%) | pH | EC (μS cm−1) | NH4+-N (mg kg−1) | NO3−-N (mg kg−1) | Root Weight (g Plant−1) | Fresh Weight (g Plant−1) | Disease Index |
|---|---|---|---|---|---|---|---|---|
| OC | 30.86 ± 2.72 c | 7.22 ± 0.02 d | 111.33 ± 0.77 f | 6.72 ± 0.79 ab | 89.81 ± 4.31 c | NA | NA | NA |
| CK | 23.8 ± 1.46 d | 7.81 ± 0.05 a | 186.33 ± 14.83 e | 5.61 ± 0.18 cd | 37.58 ± 6.21 c | 0.52 ± 0.07 ab | 6.04 ± 0.43 e | 1.83 ± 0.35 e |
| IA | 38.51 ± 6.94 a | 6.82 ± 0.03 f | 1149.67 ± 15.63 a | 7.07 ± 1.08 ab | 571.82 ± 77.46 b | 0.33 ± 0.08 d | 12.96 ± 2.54 c | 2.67 ± 0.44 b |
| IB | 40.88 ± 3.43 a | 7.03 ± 0.01 e | 689.33 ± 69.81 c | 6.47 ± 0.46 bc | 563.74 ± 92.89 b | 0.22 ± 0.06 e | 7.42 ± 1.25 de | 3.83 ± 0.35 a |
| IN | 35.63 ± 2.07 b | 6.99 ± 0.03 e | 759 ± 10 b | 7.21 ± 0.57 ab | 879.07 ± 59.13 a | 0.27 ± 0.05 e | 12.55 ± 0.98 c | 2.67 ± 0.69 b |
| OF | 18.8 ± 1.1 e | 7.44 ± 0.01 c | 377.67 ± 14.73 d | 5.55 ± 0.71 d | 40.4 ± 2.52 c | 0.61 ± 0.17 a | 18.68 ± 2.96 a | 1.5 ± 0.46 f |
| CF | 14.6 ± 0.48 f | 7.54 ± 0.03 b | 182.87 ± 12.4 e | 7.27 ± 1.49 a | 59.73 ± 8.51 c | 0.4 ± 0.06 c | 14.93 ± 2.58 b | 2.5 ± 0.46 c |
| Sample | BC | BP | Shannon–Wiener (H’) | FC | FP | Shannon–Wiener (H’) |
|---|---|---|---|---|---|---|
| OC | 7519.33 a | 0.887762 a | 6.45 a | 545.747 a | 0.696502 a | 4.12 a |
| CK | 5114.23 c | 0.769883 b | 5.89 b | 508.907 b | 0.601616 b | 3.78 b |
| IA | 4304.61 d | 0.726326 c | 5.45 c | 292.24 d | 0.579451 c | 3.45 c |
| IB | 5271.89 b | 0.700113 d | 5.67 d | 380.1 c | 0.488957 e | 3.12 d |
| IN | 3699.93 e | 0.655445 e | 5.23 e | 259.066 e | 0.496514 d | 2.98 e |
| OF | 2979.23 f | 0.615297 f | 4.89 f | 123.904 g | 0.16346 g | 2.45 f |
| CF | 2215.64 g | 0.598955 g | 4.56 g | 134.658 f | 0.21207 f | 2.23 g |
| Sample | Bacterial | Fungal | Shannon–Wiener (H’) | ||
|---|---|---|---|---|---|
| Vertex Number | Degree Centralization | Vertex Number | Degree Centralization | ||
| OC | 352 d | 15,784 e | 81 c | 804 de | 3.45 b |
| CK | 726 a | 116,520 a | 80 c | 1626 ab | 4.12 a |
| OF | 612 b | 56,234 bc | 52 d | 896 d | 3.78 c |
| CF | 501 c | 39,354 d | 50 d | 742 e | 3.56 d |
| IA | 682 a | 57,138 bc | 83 c | 1531 bc | 4.05 a |
| IB | 711 a | 63,670 b | 80 c | 1682 a | 4.10 a |
| IN | 612 b | 49,478 c | 77 c | 1494 c | 3.98 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Wu, H.; Li, Y.; Chen, J. The Response Microbial of the Cucumber Rhizosphere Network Keystone Taxa of the Cucumber Rhizosphere to Continuous Fertilization. Microorganisms 2025, 13, 451. https://doi.org/10.3390/microorganisms13020451
Tang J, Wu H, Li Y, Chen J. The Response Microbial of the Cucumber Rhizosphere Network Keystone Taxa of the Cucumber Rhizosphere to Continuous Fertilization. Microorganisms. 2025; 13(2):451. https://doi.org/10.3390/microorganisms13020451
Chicago/Turabian StyleTang, Jiaquan, Haoyang Wu, Yaqian Li, and Jie Chen. 2025. "The Response Microbial of the Cucumber Rhizosphere Network Keystone Taxa of the Cucumber Rhizosphere to Continuous Fertilization" Microorganisms 13, no. 2: 451. https://doi.org/10.3390/microorganisms13020451
APA StyleTang, J., Wu, H., Li, Y., & Chen, J. (2025). The Response Microbial of the Cucumber Rhizosphere Network Keystone Taxa of the Cucumber Rhizosphere to Continuous Fertilization. Microorganisms, 13(2), 451. https://doi.org/10.3390/microorganisms13020451








