1. Introduction
The term ‘endophyte’ has been defined as simply referring to microorganisms that inhabit the interior environment of plants during all or part of their life cycle, irrespective of their function [
1]. As natural components of plant microecosystems, endophytes benefit or harm host plants in various ways [
2], and some endophytes that provide beneficial functions for their hosts have been developed into bio-regents and applied in sustainable agricultural practices [
3].
Endophytes are categorized as culturable endophytes (CE) or cultivation-recalcitrant endophytes (CREs) based on their culturability. Early studies primarily focused on the study and application of CEs isolated from surface-sterilized plant tissues. However, advances in culture-independent techniques, such as fatty acid profiling [
4] and DNA sequencing-based metagenomic analysis, have revealed that the majority of plant endophytes are CREs [
5]. Endophytes can also be classified according to their relationship with the host plant as facultative (those requiring host symbiosis only during specific life stages) or obligate endophytes (those entirely dependent on the host throughout their life cycle) [
6]. Obligate endophytes with CRE characteristics exhibit high host eco-niche specificity [
7] and are referred to here as host-dependent endophytes (HDEs). It is evident that HDEs occupy the most intimate microbial environment of plant cells. Due to their dependency on and specificity to their hosts, some HDEs can be vertically transmitted across host generations, potentially serving as a type of heritable plant trait [
8,
9], making them particularly interesting. However, due to methodological limitations, the taxonomic diversity and function of HDEs in plants remain poorly characterized.
Callus, an unorganized cell mass formed through dedifferentiation of explants in in vitro cultures, retains the totipotency to regenerate into a whole plant [
10]. Studies have revealed that in vitro-cultured plant calli are colonized by diverse endophytes [
5,
11,
12]. These endophytes were inherited from the maternal plant, and the majority of the callus-hosted-endophytes appear to be HDEs, making callus an idea material for studying plant HDEs. In this study, we established an approach to study the possible eco-physiological functions of plant HDEs using grapevine (
Vitis vinifera L. ×
V. labrusca L.) callus cultured over a long period as a host material.
4. Discussion
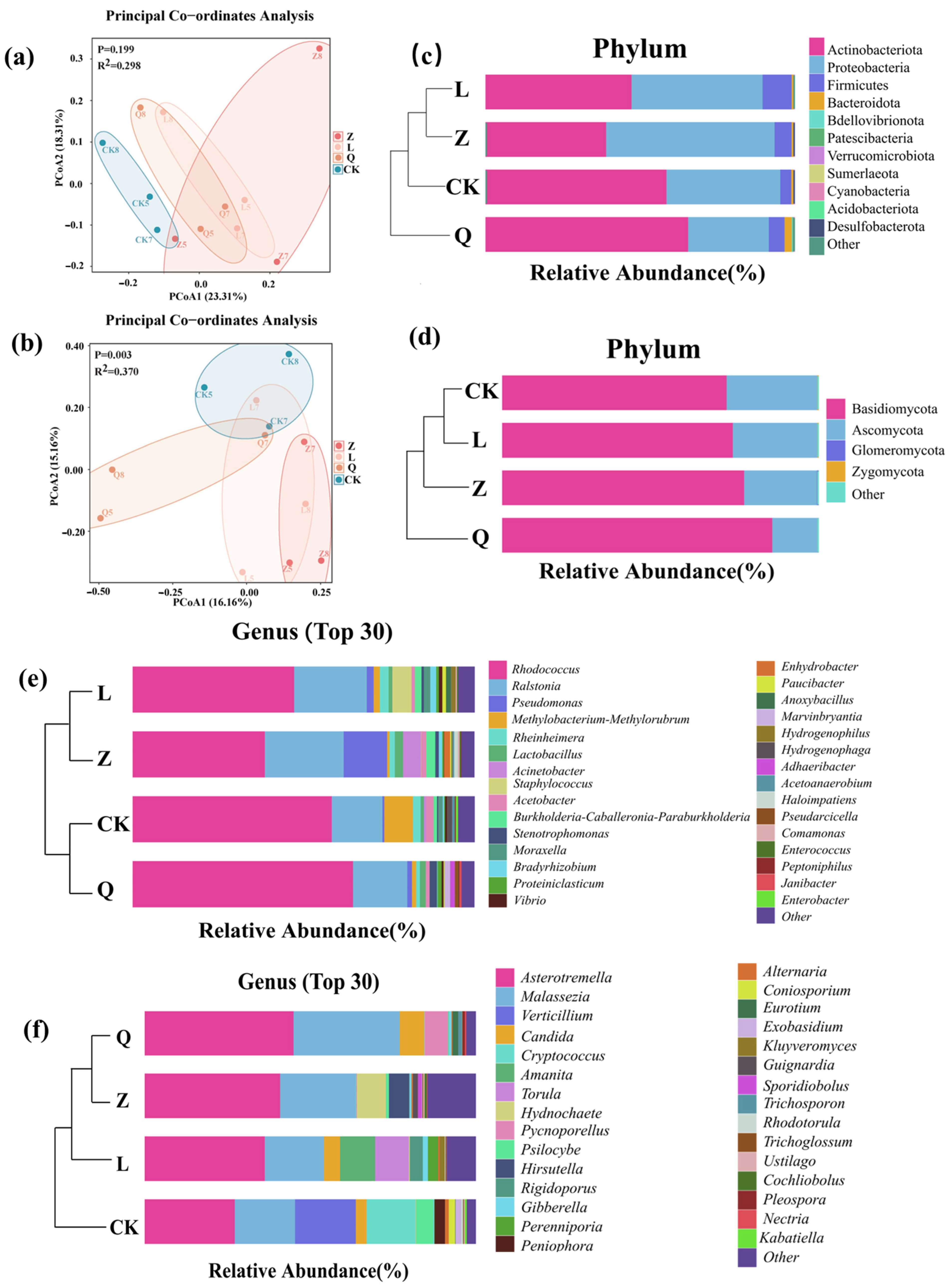
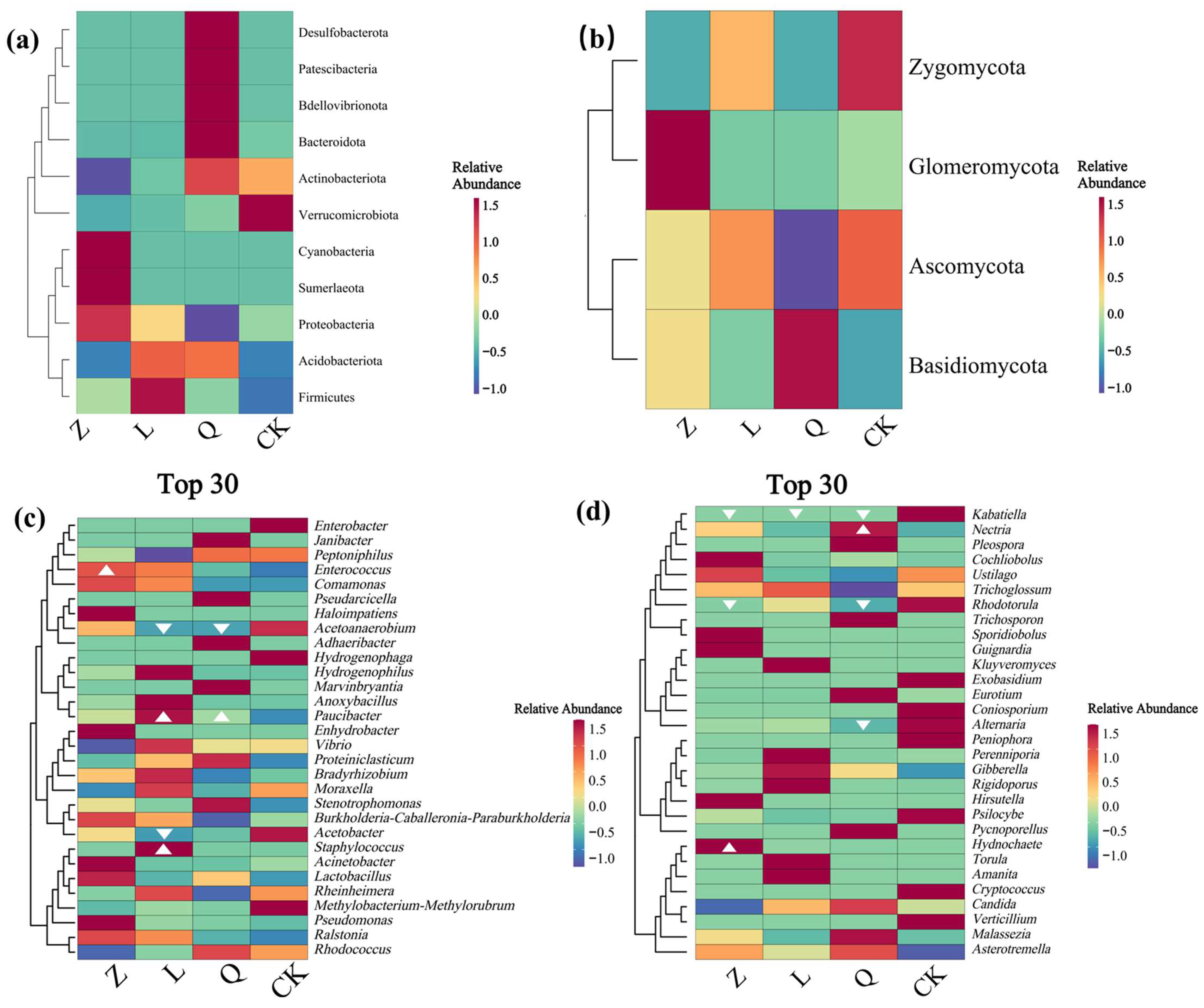
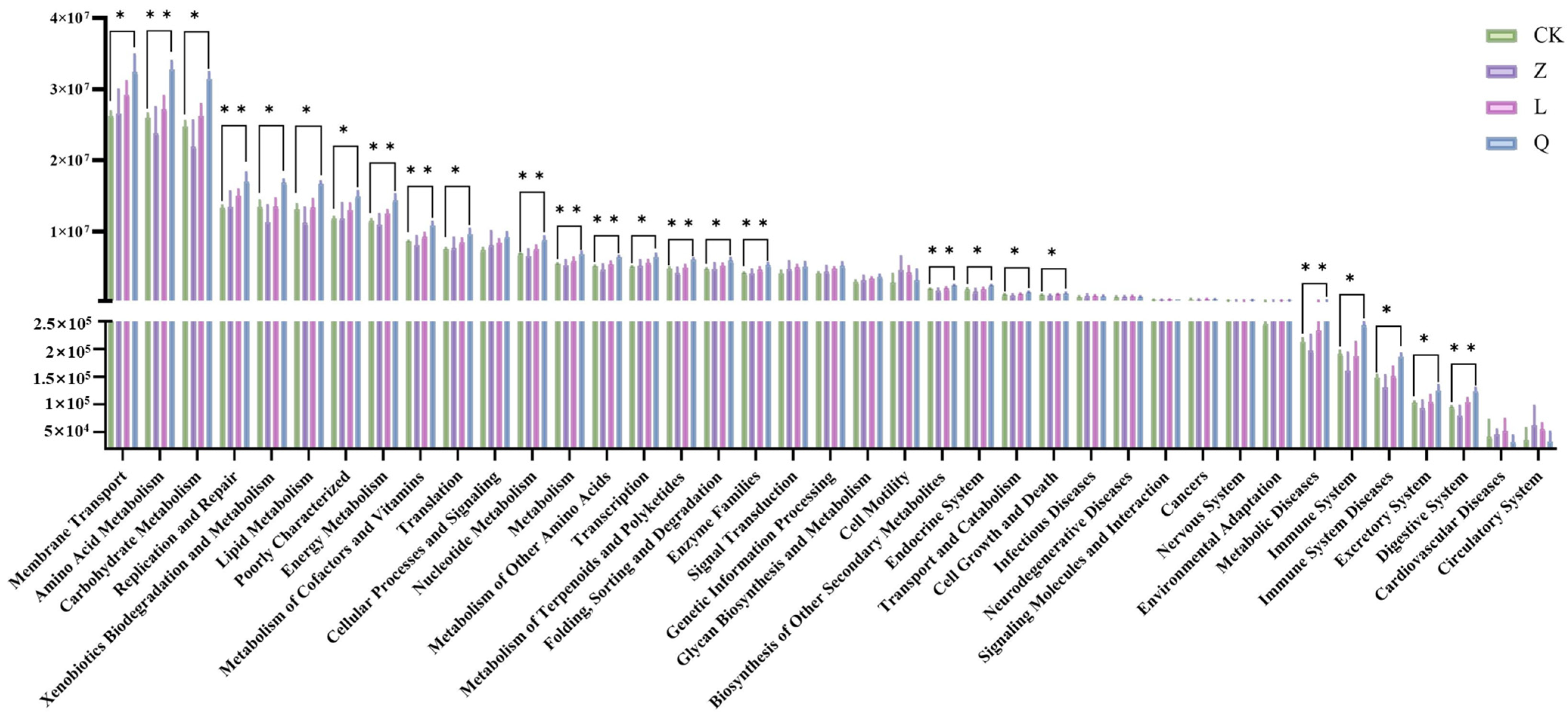
HDEs identified in long-term subcultured grape callus exhibited substantial structural divergence from the endophytic microbiota of field-grown grape plants. Natural grapevine microbiota are typically dominated by
Proteobacteria and
Firmicutes, enriched with functional taxa associated with plant disease resistance and nutrient assimilation, such as
Burkholderia and
Bacillus [
1]. In contrast, the callus-associated communities were primarily composed of Actinobacteriota (52.80%) and Basidiomycota (82.59%), with
Rhodococcus (52.21%) and
Asterotremella (37.39%) emerging as the dominant genera. This disparity likely arises from the unique selective pressures of in vitro culture conditions, where stable carbon sources (e.g., sucrose) and phytohormones (e.g., NAA, KT) may enrich microbiota adapted to high-osmolarity environments and biosynthetic demands [
5]. Additionally, the absence of vascular systems and environmental stress signaling in callus tissues may favor the stable colonization of obligate symbionts or metabolically dependent endophytes. Notably, the abundance of
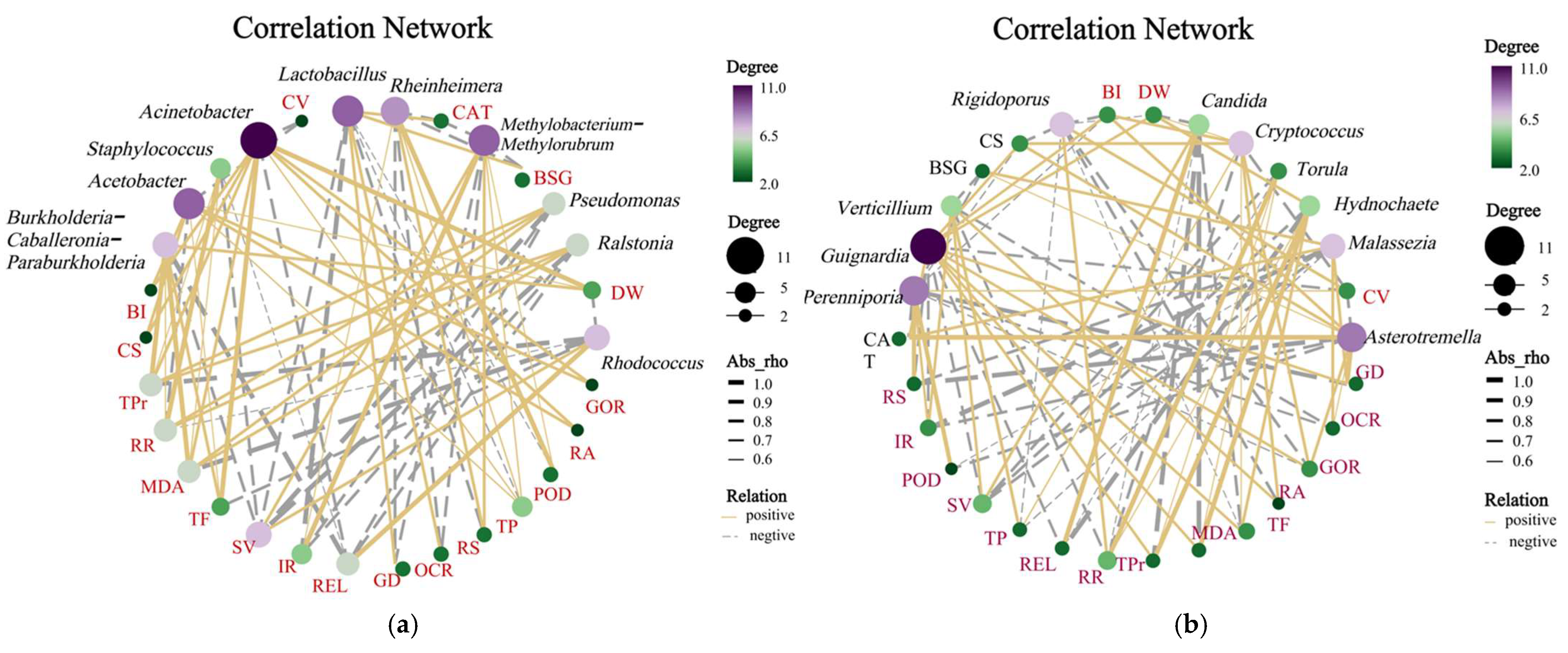
Rhodococcus showed a significant positive correlation with callus dry weight ratio (
Figure 7), suggesting its potential role in modulating host carbon metabolism to maintain cellular homeostasis. These findings underscore how in vitro culture conditions reshape endophytic assemblies, favoring taxa with specialized metabolic adaptations over those optimized for ecological interactions in natural plant systems.
Compared to previous studies on endophytic microbiota in
Vitis vinifera L. ×
V. labrusca L. callus [
12], the bacterial genera
Ralstonia and
Pseudomonas remained dominant in this study, consistent with their roles in nutrient acquisition and stress tolerance. Notably,
Methylobacterium-Methylorubrum and
Rhodococcus emerged as unique keystone taxa in the current system, potentially linked to methanol metabolism and lipid degradation, respectively, suggesting adaptive responses to carbon source shifts or host-derived metabolites during long-term subculture. In contrast,
Acinetobacter and
Halomonas, previously reported as dominant taxa [
12], were undetected here, possibly due to differences in subculture frequency or medium salinity. Xiang et al. identified
Erysiphe and
Sarocladium as predominant fungal genera in Rose Honey tissue-cultured seedlings, alongside bacterial enrichment of
Cupriavidus and
Cellvibrio [
13], which may be associated with phototrophic metabolism, cell wall remodeling, and nutrient cycling. By comparison, the current callus system was dominated by
Rhodococcus and
Methylobacterium in bacteria and
Astrotremella and
Verticillium in fungi, implying adaptations to phenolic degradation, carbon utilization, and oxidative stress. The common presence of
Ralstonia across studies likely reflects its broad adaptability to plant endophytic niches. These disparities underscore the integrated effects of tissue type (callus versus seedlings), physiological state, and subculture-induced selection pressures on endophytic community assembly.
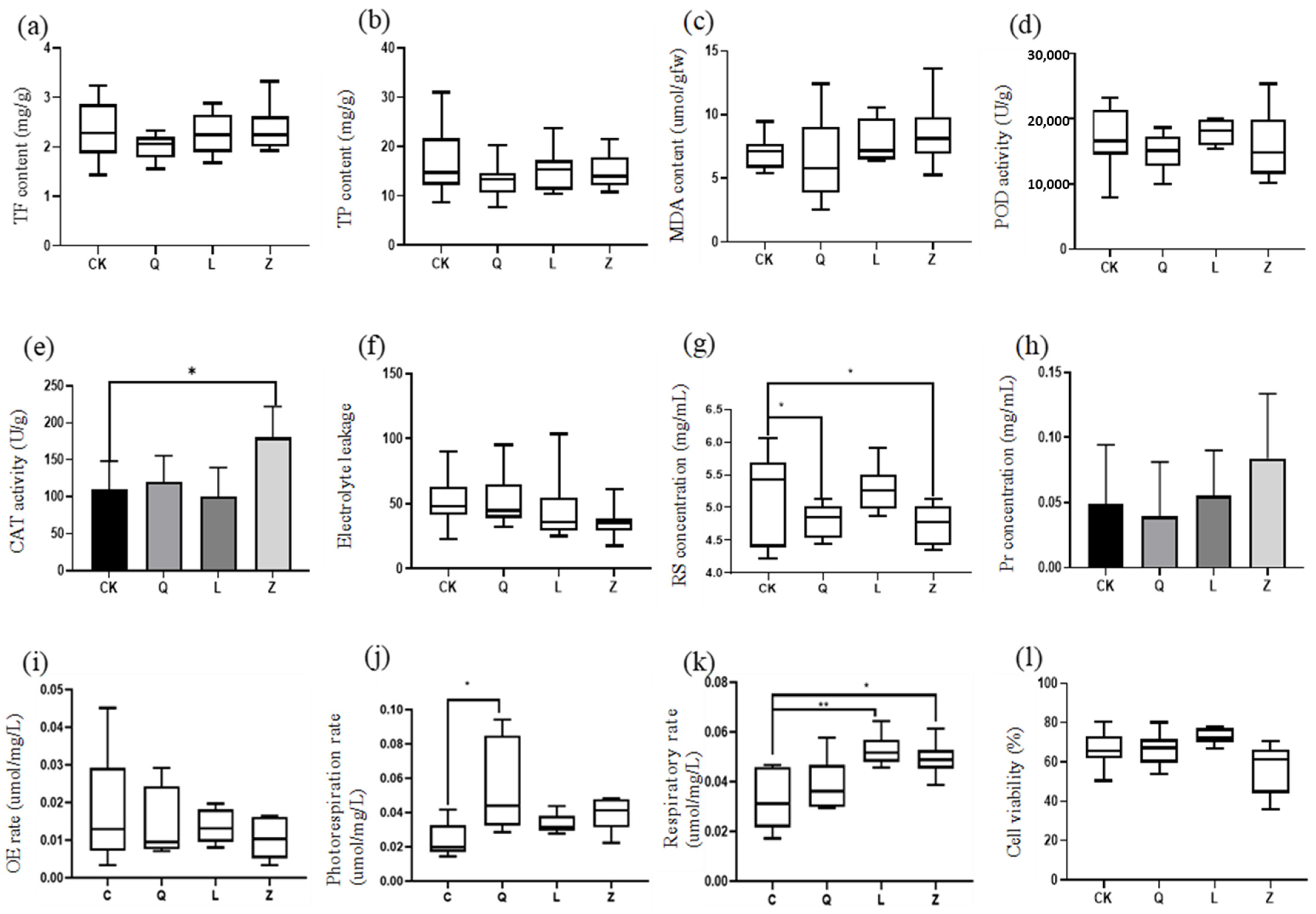
Among the dominant genera identified,
Pseudomonas and
Methylobacterium exhibited functionally distinct roles. The abundance of
Pseudomonas showed significant positive correlations with host peroxidase (POD, r = 0.68) and catalase (CAT, r = 0.72) activities (
p < 0.05), suggesting its potential role in alleviating oxidative stress by enhancing antioxidant defenses, thereby stabilizing callus cell viability [
25].
Methylobacterium, previously implicated in plant nitrogen metabolism via bacterial urease activity [
26,
27,
28]), displayed a significant negative correlation with photosynthetic and respiratory rates in this study (r = −0.61,
p < 0.05), implying its involvement in modulating carbon metabolism to influence host energy allocation. Notably, while
Rhodococcus,
Methylobacterium-Methylorubrum,
Rheinheimera, and
Staphylococcus reduced callus browning indices, these effects did not reach statistical significance, contrasting with prior findings where
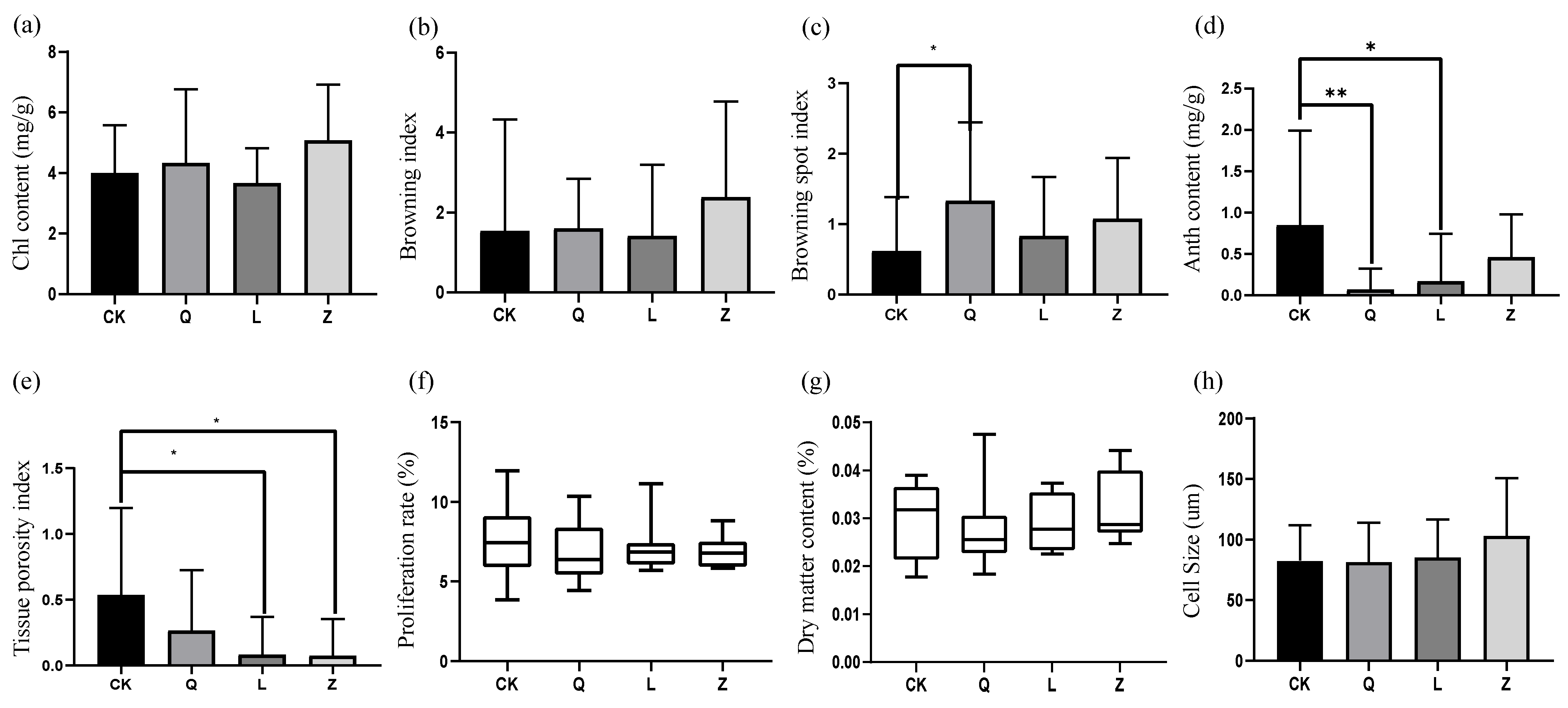
Bacillus sp. strain ST-B1. A, isolated by Wang et al., exacerbated browning in grape callus. Similarly, M. Pirttilä et al. reported delayed tissue browning in
Pinus sylvestris callus inoculated with
M. extorquens,
P. synxantha, and
R. minuta [
29]. Fungal HDEs, particularly
Asterotremella, demonstrated significant positive correlations with total protein content (r = 0.54) and reducing sugar concentrations (r = 0.49,
p < 0.05), potentially mediated by extracellular enzyme secretion to enhance host carbon utilization from the medium. However, the molecular mechanisms underlying these taxa–host interactions, especially direct metabolic crosstalk, require further validation.
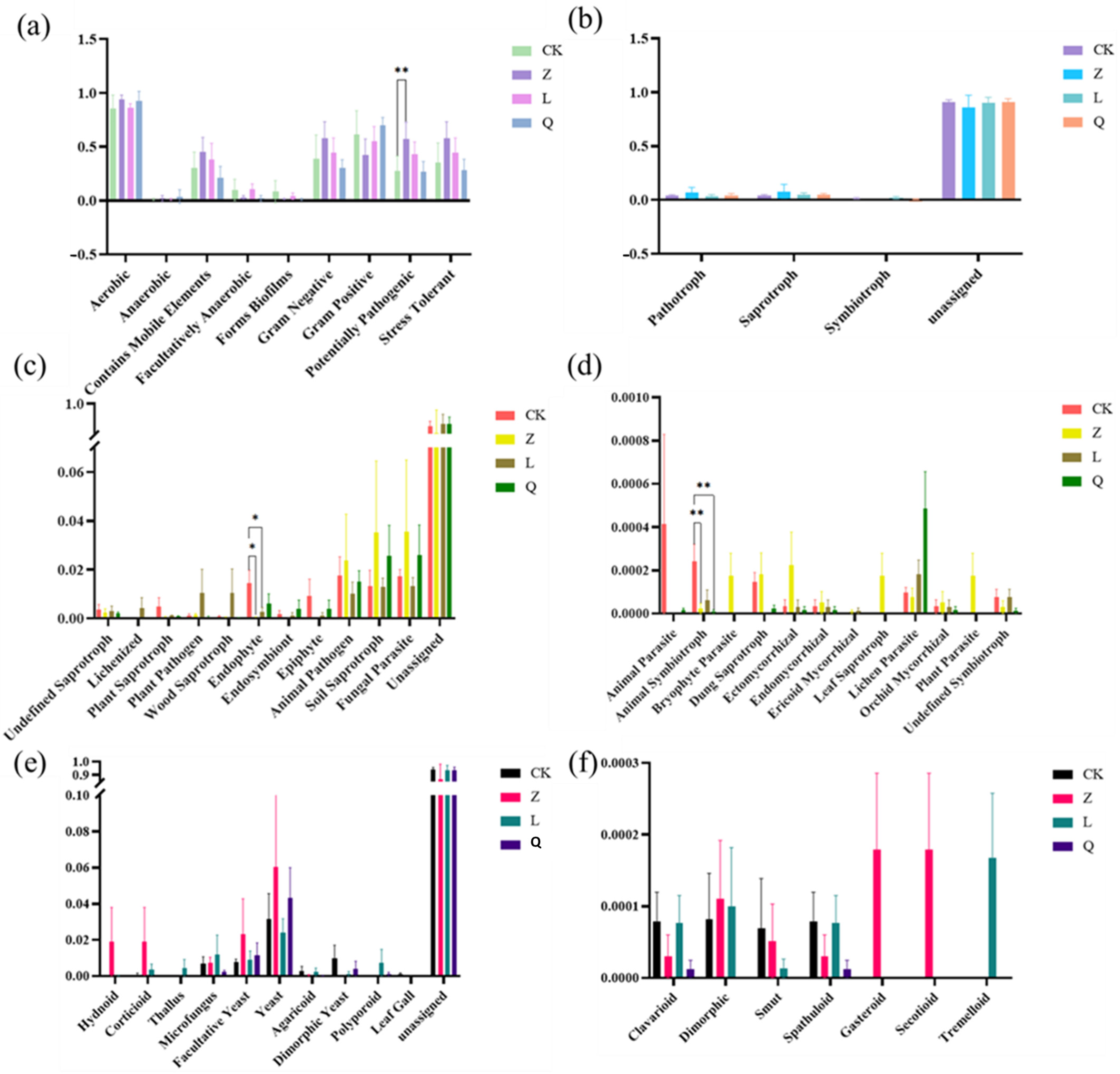
This study establishes grapevine callus as a robust model system for investigating functional relationships between HDEs and plant physiology through antibiotic-mediated community modulation. Antibiotic-specific effects were observed in HDE assembly, with penicillin selectively enriching Gram-positive taxa (e.g., Paucibacter) while nystatin promoted Gram-negative bacteria and suppressed Colidextribacter, demonstrating precision microbial manipulation. Taxon–host phenotype correlations revealed associations of Rheinheimer with phenolic compounds and cellular viability, Pseudomonas with elevated antioxidant enzymes (POD, CAT), and Rhodococcus with biomass accumulation. The fungal endophytes Astrotremella, Malassezia, and Hydnochaete showed positive correlations with total protein content. The persistence of core taxa (Rhodococcus, Astrotremella) validated the callus system’s reliability for studying obligate endophytes. By developing an “antibiotic remodeling–multiomics profiling–functional validation” framework, this research pioneers a new approach to investigating HDEs, enabling future studies on synthetic community reconstruction and molecular interaction mechanisms for crop stress resilience and propagation optimization. These findings position HDEs as dynamic regulators of host physiology through community composition, providing methodological foundations for agricultural microbiome innovation.
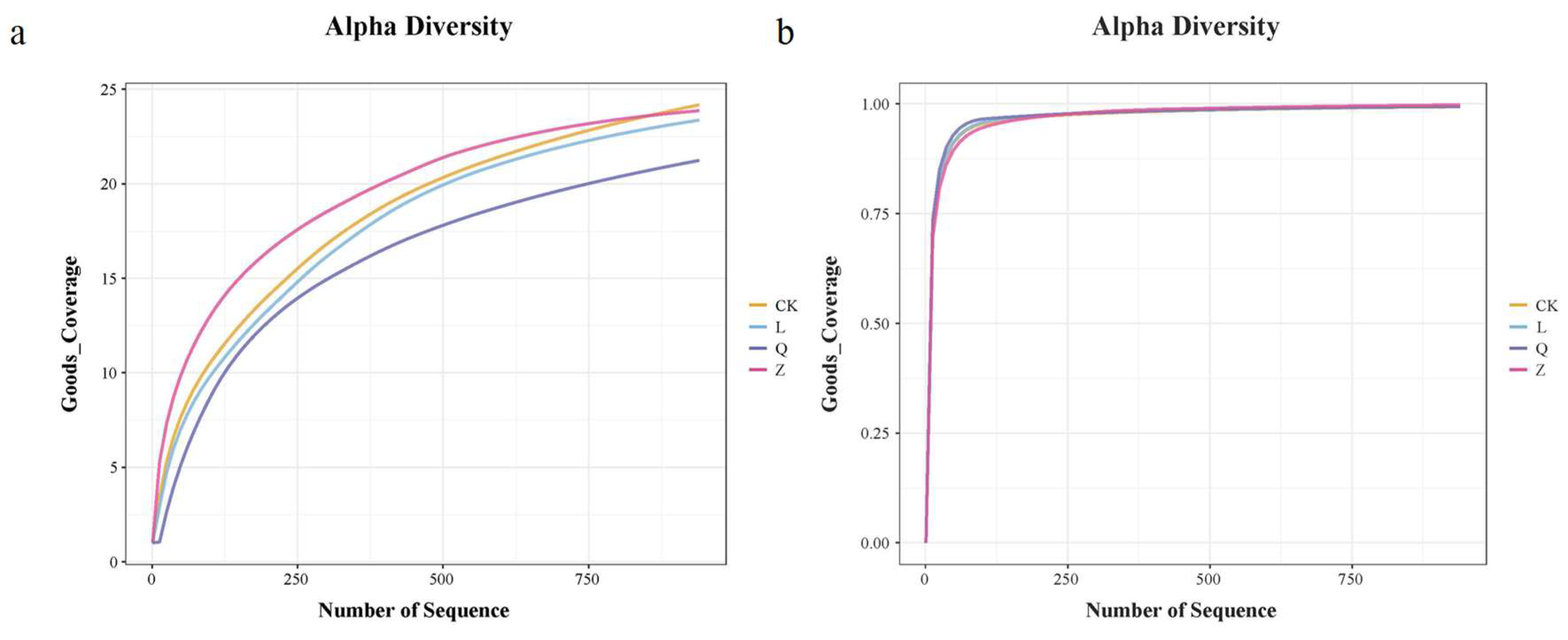
However, functional identification of HDEs using callus tissue present significant challenges when investigating non-core HDE taxa. The spatial heterogeneity of these HDEs within callus tissue leads to random distribution across subcultured samples. This may also account for the substantial variation in alpha diversity observed within treatments.