Microbial Community Analysis and Environmental Association in Cave 6 of the Yungang Grottoes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site Description and Sample Collection
2.2. DNA Extraction and High-Throughput Sequencing
2.3. Bioinformatics and Statistical Analysis
3. Results
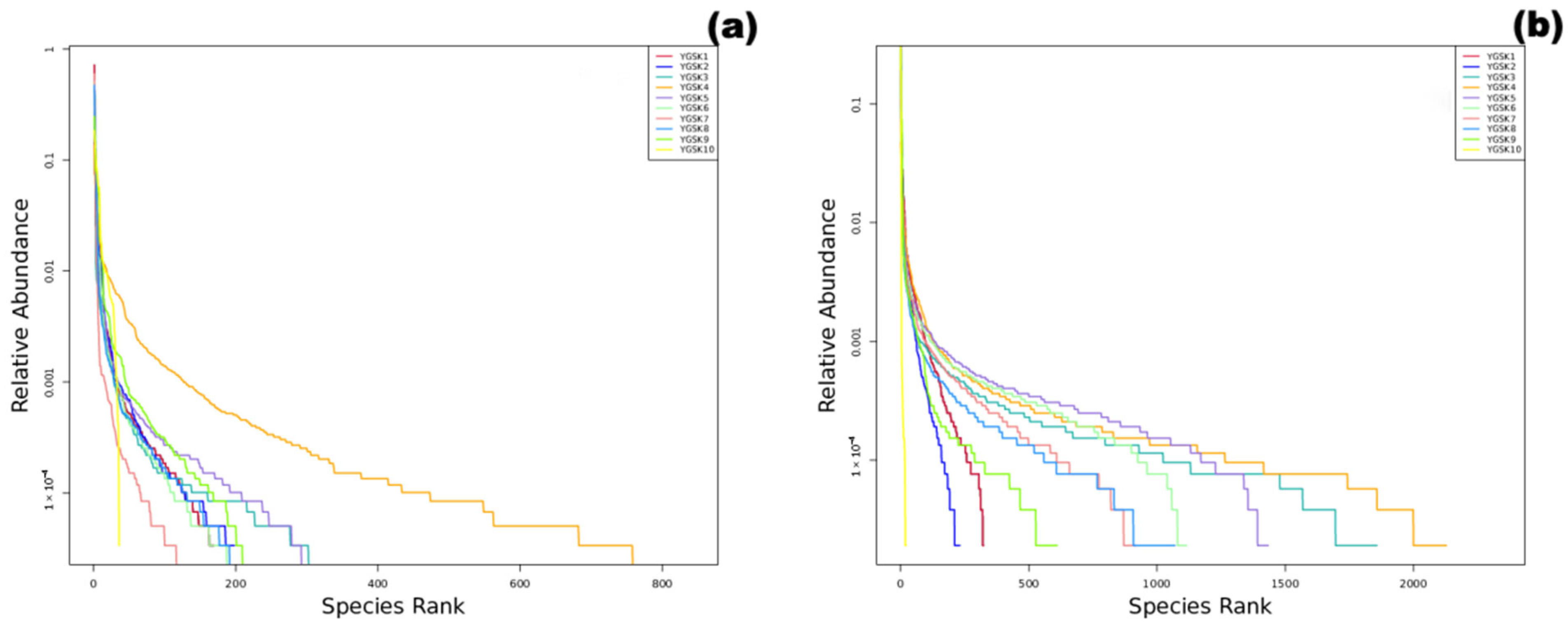
3.1. Microbial Community Composition Structure
3.1.1. Fungal Community
3.1.2. Bacterial Community
3.2. Alpha Diversity
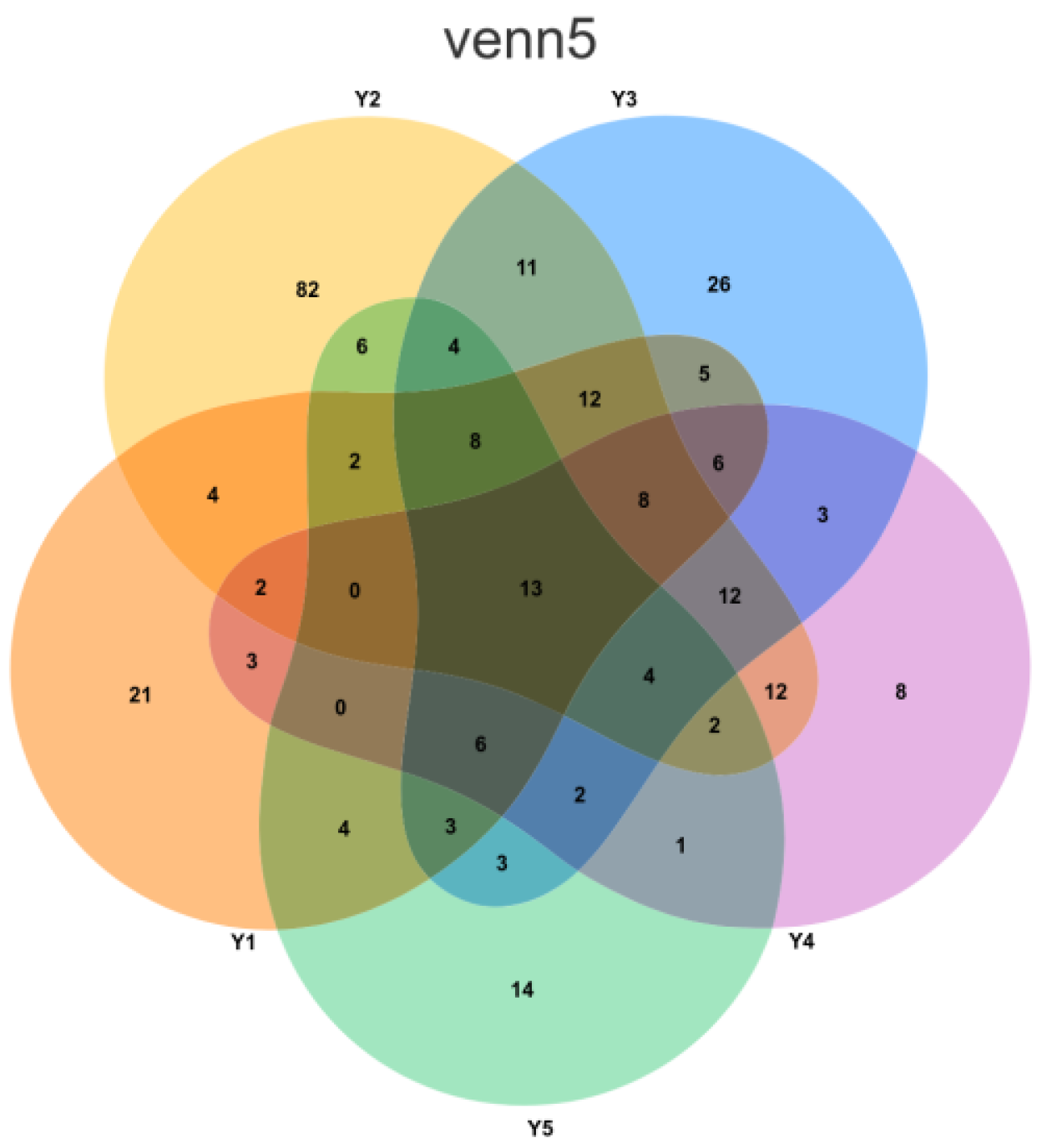
3.3. Core Microbiome Analysis
3.4. Unique Species Analysis by Microenvironment
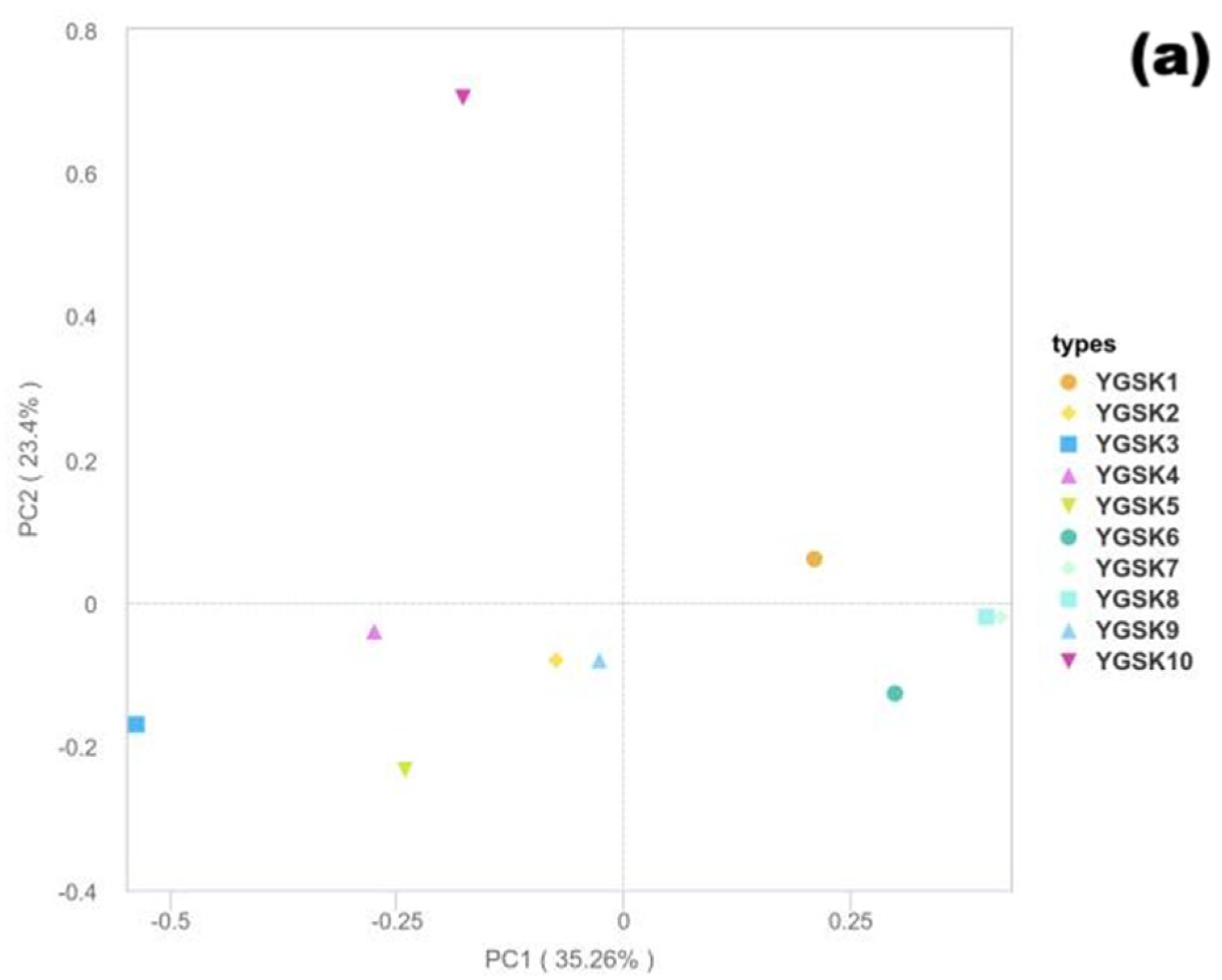
3.5. Beta Diversity Analysis
- (1)
- Fungal Community Structure Characteristics
- (2)
- Bacterial Community Structure Characteristics
- (3)
- Differences in Community Differentiation Mechanisms
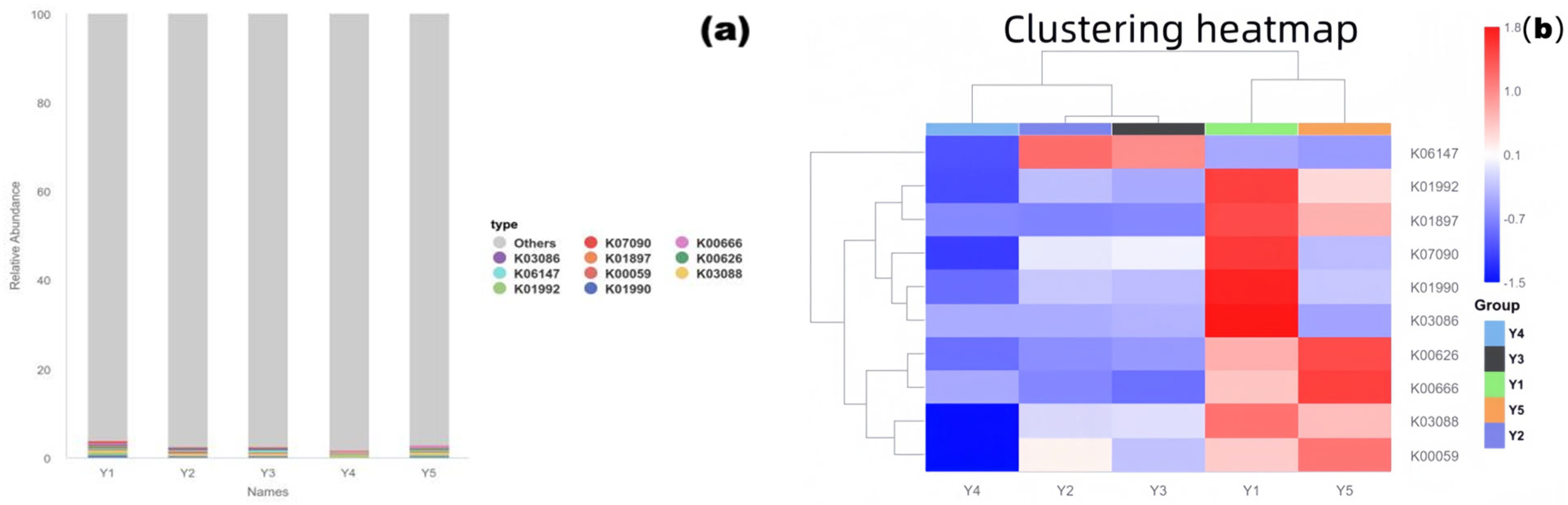
3.6. PICRUSt2 Functional Prediction (KEGG Pathways Focused on Deterioration-Related Functions
- (1)
- Functional Composition Structure
- (2)
- Functional Clustering and Enrichment of Deterioration-Related Pathways
- (3)
- Implications for Heritage Conservation
4. Discussion
4.1. Association Between Microbial Community Structure and Environmental Factors
4.2. Potential Causes of Alpha Diversity Differences
4.3. Biodeterioration Risk Assessment and Conservation Implications
4.4. Beta Diversity Analysis
4.5. Functional Prediction
5. Conclusions
- (1)
- The microbial community structure within the cave exhibited significant microenvironment heterogeneity. Ascomycota and Proteobacteria were the overall dominant phyla for fungi and bacteria, respectively, but genus-level composition varied significantly across different microenvironments, reflecting strong filtering effects of niche conditions on microbial distribution.
- (2)
- Microbial diversity was jointly influenced by environmental openness and external stress factors. Areas with frequent external interaction (e.g., around the lighting window) had the highest species diversity, while the red pigment area, inhibited by heavy metals (e.g., mercury in cinnabar), showed significantly reduced diversity.
- (3)
- Significant signals of human-derived microorganisms, such as Malassezia and Escherichia-Shigella, were detected on Buddha surface communities, clearly indicating biological pollution introduced by visitor contact. Their potential public health risks and biochemical erosion of relic surfaces require high attention.
- (4)
- Multiple microbial taxa with known degradation functions (e.g., Cladosporium, Rubrobacter, and Streptomyces) were significantly enriched in dust accumulation areas (wall junctions/cavities) and open areas (around the lighting window), identifying these as high-risk zones for biodeterioration that should be key targets for future monitoring and protection.
- (5)
- Bacteria and fungi exhibited distinctly different response strategies to environmental factors: bacterial community structure was more susceptible to microenvironment resource heterogeneity and human disturbance intensity, while fungal communities showed stronger substrate-specific adaptation to organic substrate types (e.g., pigment binders, human secretions).
- (6)
- Functional prediction analysis further indicated that microbial communities in dust accumulation areas and areas between Buddha statues were enriched with genes related to organic matter degradation and acid production metabolism, predicting a high potential risk of biodeterioration in these areas. The interventions include removal of dust deposits, installation of breathable air filters or fine mesh screens on windows, and placement of glass barriers or railings for accessible Buddha statues.
- (7)
- This study, from the perspectives of microbial community structure, ecological function, and potential risk, clarified the driving mechanisms of microenvironmental factors on the assembly and functional selection of grotto microbial communities. The research results can provide theoretical basis and practical reference for the scientific conservation and precise prevention and control of the Yungang Grottoes and other similar cultural heritage sites.
6. Research Limitations and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, H.; Bai, B. Cultural inheritance and application of Yungang Grottoes statues in art education. Arts Educ. 2024, 38, 332–343. [Google Scholar]
- Ozawa, M. A Study on the Creation of the Standing Tathagata Buddha in the Upper Niches of Cave 6 at Yungang Grottoes. In Proceedings of the 2005 Yungang International Academic Symposium, Datong, China, 26–30 July 2005; pp. 273–285. [Google Scholar]
- Wei, Z.; Ma, M.Y. Early Grottoes in Hexi from the Perspective of Northern China—Part 2 of the Study on Early Hexi Grottoes. Dunhuang Res. 2022, 5, 97–110. [Google Scholar] [CrossRef]
- Huang, P. An Archaeological Study of Northern Wei Buddhist Temples at the Yungang Grottoes. Huaxia Archaeol. 2025, 3, 142–150+160. [Google Scholar] [CrossRef]
- Wang, Y.K. Analysis of the Image Composition in Caves 5 and 6 at Datong Yungang. Dunhuang Res. 2019, 3, 17–31. [Google Scholar] [CrossRef]
- Liu, R.Z.; Zhang, B.J.; Zhang, H.; Shi, M.F. Deterioration of Yungang Grottoes: Diagnosis and research. J. Cult. Herit. 2011, 12, 494–499. [Google Scholar] [CrossRef]
- Sanmartín, P.; DeAraujo, A.; Vasanthakumar, A. Melding the old with the new: Trends in methods used to identify, monitor, and control microorganisms on cultural heritage materials. Microb. Ecol. 2018, 76, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Geweely, N.S. New frontiers review of some recent conservation techniques of organic and inorganic archaeological artefacts against microbial deterioration. Front. Microbiol. 2023, 14, 1146582. [Google Scholar] [CrossRef] [PubMed]
- Rosado, T.; Dias, L.; Lança, M.; Nogueira, C.; Santos, R.; Martins, M.R.; Candeias, A.; Mirão, J.; Caldeira, A.T. Assessment of microbiota present on a Portuguese historical stone convent using high-throughput sequencing approaches. MicrobiologyOpen 2020, 9, 1067–1084. [Google Scholar] [CrossRef]
- Perini, N.; Mercuri, F.; Orlanducci, S.; Thaller, M.C.; Migliore, L. The integration of metagenomics and chemical physical techniques biodecoded the buried traces of the biodeteriogens of parchment purple spots. Front. Microbiol. 2020, 11, 598945. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Shumskaya, M.; Lorusso, N.; Patel, U.; Leigh, M.; Somervuo, P.; Schigel, D. MycoPins: A metabarcoding-based method to monitor fungal colonization of fine woody debris. MycoKeys 2023, 96, 77. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, B.; He, Z.; Yang, X. Distribution and diversity of bacteria and fungi colonization in stone monuments analyzed by high-throughput sequencing. PLoS ONE 2016, 11, e0163287. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, R.N.; Felipe, M.T.C.; Silva, L.F.; Silva, E.A.; Silva, S.A.; Herculano, P.N.; Prazeres, J.F.S.A.; Lima, J.M.S.; Bezerra, J.D.P.; Moreira, K.A.; et al. A Review of the Biotechnological Potential of Cave Fungi: A Toolbox for the Future. J. Fungi 2025, 11, 145. [Google Scholar] [CrossRef]
- Wang, W.; Ma, X.; Ma, Y.; Mao, L.; Wu, F.; Ma, X.; An, L.; Feng, H. Molecular characterization of airborne fungi in caves of the Mogao Grottoes, Dunhuang, China. Int. Biodeterior. Biodegrad. 2011, 65, 726–731. [Google Scholar] [CrossRef]
- Zhu, X.; Jie, B. Analysis on the environment of cultural relic as tourist attraction—Take Yungang Grottoes as an example. IOP Conf. Ser. Earth Environ. Sci. 2018, 128, 012172. [Google Scholar]
- Sun, B. Archaeological and Craftsmanship Exploration of the Cave Wall Structure between Cave 5 and Cave 6 of Yungang Grottoes. Yungang Res. 2025, 5, 87–91. [Google Scholar] [CrossRef]
- Qian, H. Research on the Spatial Design of Datong Yungang Grottoes. Master’s Thesis, Taiyuan University of Technology, Taiyuan, China, 2014. [Google Scholar] [CrossRef]
- Vagelas, I.; Reizopoulou, A.; Exadactylos, A.; Madesis, P.; Karapetsi, L.; Michail, G. Stalactites core prospect as environmental “microbial ark”: The Actinomycetota diversity paradigm, first reported from a Greek cave. Pol. J. Microbiol. 2023, 72, 155. [Google Scholar] [CrossRef]
- Michail, G.; Karapetsi, L.; Madesis, P.; Reizopoulou, A.; Vagelas, I. Metataxonomic analysis of bacteria entrapped in a stalactite’s core and their possible environmental origins. Microorganisms 2021, 9, 2411. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Wickham, H. Data analysis. In ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Cham, Switzerland, 2016; pp. 189–201. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: http://www.R-project.org/ (accessed on 31 October 2024).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Koestler, R.J.; Warscheid, T.; Katayama, Y.; Gu, J.-D. Microbial deterioration and sustainable conservation of stone monuments and buildings. Nat. Sustain. 2020, 3, 991–1004. [Google Scholar] [CrossRef]
- Grbić, M.L.; Dimkić, I.; Savković, Ž.; Stupar, M.; Knežević, A.; Jelikić, A.; Unković, N. Mycobiome diversity of the cave church of sts. Peter and Paul in Serbia—Risk assessment implication for the conservation of rare cavern habitat housing a peculiar fresco painting. J. Fungi 2022, 8, 1263. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, H.; Du, Y.; Tian, T.; Xiang, T.; Liu, X.; Wu, F.; An, L.; Wang, W.; Gu, J.-D.; et al. The community distribution of bacteria and fungi on ancient wall paintings of the Mogao Grottoes. Sci. Rep. 2015, 5, 7752. [Google Scholar] [CrossRef]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 2, 47–55. [Google Scholar] [CrossRef]
- Chen, Y.; Kuang, J.; Wang, P.; Shu, W.; Barberán, A. Associations between human impacts and forest soil microbial communities. Elem. Sci. Anthr. 2020, 8, 5. [Google Scholar] [CrossRef]
- Sáiz-Jiménez, C. Deposition of airborne organic pollutants on historic buildings. Atmos. Environ. Part B Urban Atmos. 1993, 27, 77–85. [Google Scholar] [CrossRef]
- Mohammadipanah, F.; Wink, J. Actinobacteria from arid and desert habitats: Diversity and biological activity. Front. Microbiol. 2016, 6, 1541. [Google Scholar] [CrossRef]
- Martin-Pozas, T.; Fernandez-Cortes, A.; Cuezva, S.; Jurado, V.; Gonzalez-Pimentel, J.L.; Hermosin, B.; Ontañon, R.; Arias, P.; Cañaveras, J.C.; Sanchez-Moral, S.; et al. Microclimate, airborne particles, and microbiological monitoring protocol for conservation of rock-art caves: The case of the world-heritage site La Garma cave (Spain). J. Environ. Manag. 2024, 351, 119762. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.; Guseva, K.; Darcy, S.; Alteio, L.; Pjevac, P.; Schmidt, H.; Jenab, K.; Ranits, C.; Kaiser, C. Distinct microbial communities are linked to organic matter properties in millimetre-sized soil aggregates. ISME J. 2024, 18, Wrae156. [Google Scholar] [CrossRef]
- Li, Y.; Adams, J.; Shi, Y.; Wang, H.; He, J.-S.; Chu, H. Distinct Soil Microbial Communities in habitats of differing soil water balance on the Tibetan Plateau. Sci. Rep. 2017, 7, 46407. [Google Scholar] [CrossRef]
- Wang, W.; Ma, X.; Ma, Y.; Mao, L.; Wu, F.; Ma, X.; An, L.; Feng, H. Seasonal dynamics of airborne fungi in different caves of the Mogao Grottoes, Dunhuang, China. Int. Biodeterior. Biodegrad. 2010, 64, 461–466. [Google Scholar] [CrossRef]
- Kubera, Ł.; Kalwasińska, A.; Perliński, P. Assessment of the Structure and Activity of Bacterial Communities as a Tandem Tool for Estimating the Ecological Condition of Lakes in Poland: Assessment of the Structure and Activity of Bacterial Communities. Ecosystems 2025, 28, 32. [Google Scholar] [CrossRef]
- Mant, D.; Orevi, T.; Kashtan, N. Impact of micro-habitat fragmentation on microbial population growth dynamics. ISME J. 2025, 19, Wrae256. [Google Scholar] [CrossRef] [PubMed]
- Pyzik, A.; Ciuchcinski, K.; Dziurzynski, M.; Dziewit, L. The bad and the good—Microorganisms in cultural heritage environments—An update on biodeterioration and biotreatment approaches. Materials 2021, 14, 177. [Google Scholar] [CrossRef]
- Fidanza, M.R.; Caneva, G. Natural biocides for the conservation of stone cultural heritage: A review. J. Cult. Herit. 2019, 38, 271–286. [Google Scholar] [CrossRef]
- Ilieș, D.C.; Safarov, B.; Caciora, T.; Ilieș, A.; Grama, V.; Ilies, G.; Huniadi, A.; Zharas, B.; Hodor, N.; Sandor, M.; et al. Museal indoor air quality and public health: An integrated approach for exhibits preservation and ensuring human health. Sustainability 2022, 14, 2462. [Google Scholar] [CrossRef]
- Turrini, P.; Chebbi, A.; Riggio, F.P.; Visca, P. The geomicrobiology of limestone, sulfuric acid speleogenetic, and volcanic caves: Basic concepts and future perspectives. Front. Microbiol. 2024, 15, 1370520. [Google Scholar] [CrossRef] [PubMed]



| Group | Sampled Images | Sample | Sampling Location | Environmental Characteristics |
|---|---|---|---|---|
| Wall Junction/Hole Group |  | YGSK1, YGSK2 | (a): YGSK1 East wall, at the junction between the Ritual Buddha Layer and the North wall; (b): YGSK2 East wall, in a hole on the right side outside the niche of the first standing Buddha from north to south; | Low-disturbance deposition area |
| External Interaction Group |  | YGSK3, YGSK4 | (c): YGSK3 Mingchuang (window), right-side groove; (d): YGSK4 Mingchuang (window), lower wall surface, lower left corner; | Air circulation area |
| Pigment Group |  | YGSK5, YGSK6 | (e): YGSK5 East wall, red pigment layer of the third small seated Buddha from south to north; (f): YGSK6 South wall, red pigment layer of the fourth small seated Buddha from west to east; | Red pigment area |
| Buddha Statue Surface Group |  | YGSK7, YGSK8 | (g): YGSK7 West wall, leg portion of the first small seated Buddha from north to south; (h): YGSK8 North wall, circular hole on the abdomen of the first Vajrapani (guardian figure); | Buddha statue surface area |
| Inter-Buddha Area Group |  | YGSK9, YGSK10 | (i): YGSK9 West wall, between the first and second small seated Buddhas from south to north; (j): YGSK10 North wall, between the first and second small seated Buddhas from west to east. | Area between adjacent Buddha statues |
| Sample_Name | Chao1 | Dominance | Goods_Coverage | Observed_Features | Pielou_e | Shannon | Simpson |
|---|---|---|---|---|---|---|---|
| YGSK1 | 170 | 0.523 | 1 | 170 | 0.292 | 2.16 | 0.477 |
| YGSK2 | 198 | 0.237 | 1 | 198 | 0.422 | 3.218 | 0.763 |
| YGSK3 | 307.677 | 0.381 | 1 | 307 | 0.276 | 2.283 | 0.619 |
| YGSK4 | 780.444 | 0.034 | 1 | 774 | 0.687 | 6.596 | 0.966 |
| YGSK5 | 302.214 | 0.305 | 1 | 299 | 0.37 | 3.045 | 0.695 |
| YGSK6 | 36 | 0.095 | 1 | 36 | 0.761 | 3.932 | 0.905 |
| YGSK7 | 125.4 | 0.453 | 1 | 123 | 0.24 | 1.666 | 0.547 |
| YGSK8 | 197.462 | 0.296 | 1 | 194 | 0.37 | 2.815 | 0.704 |
| YGSK9 | 211.111 | 0.114 | 1 | 211 | 0.532 | 4.105 | 0.886 |
| YGSK10 | 203.118 | 0.283 | 1 | 201 | 0.361 | 2.76 | 0.717 |
| Sample_Name | Chao1 | Observed_Features | Dominance | Goods_Coverage | Pielou_e | Shannon | Simpson |
|---|---|---|---|---|---|---|---|
| YGSK1 | 326 | 323 | 0.072 | 1 | 0.644 | 5.365 | 0.928 |
| YGSK2 | 232.161 | 231 | 0.145 | 1 | 0.525 | 4.126 | 0.855 |
| YGSK3 | 1863.63 | 1857 | 0.115 | 0.999 | 0.594 | 6.449 | 0.885 |
| YGSK4 | 2151.58 | 2128 | 0.011 | 0.998 | 0.777 | 8.592 | 0.989 |
| YGSK5 | 1441.21 | 1433 | 0.039 | 0.999 | 0.757 | 7.939 | 0.961 |
| YGSK6 | 21.5 | 21 | 0.902 | 1 | 0.079 | 0.346 | 0.098 |
| YGSK7 | 926.774 | 918 | 0.124 | 0.999 | 0.587 | 5.777 | 0.876 |
| YGSK8 | 1129.65 | 1112 | 0.085 | 0.999 | 0.674 | 6.824 | 0.915 |
| YGSK9 | 632.203 | 612 | 0.125 | 0.999 | 0.487 | 4.505 | 0.875 |
| YGSK10 | 1123.76 | 1069 | 0.207 | 0.998 | 0.484 | 4.868 | 0.793 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, S.; Wang, Z.; Zhang, R.; Wang, Y.; Wang, C.; Gao, G.; Pan, J. Microbial Community Analysis and Environmental Association in Cave 6 of the Yungang Grottoes. Microorganisms 2025, 13, 2788. https://doi.org/10.3390/microorganisms13122788
Qiao S, Wang Z, Zhang R, Wang Y, Wang C, Gao G, Pan J. Microbial Community Analysis and Environmental Association in Cave 6 of the Yungang Grottoes. Microorganisms. 2025; 13(12):2788. https://doi.org/10.3390/microorganisms13122788
Chicago/Turabian StyleQiao, Shangxiao, Zeao Wang, Runping Zhang, Yu Wang, Cen Wang, Guoming Gao, and Jiao Pan. 2025. "Microbial Community Analysis and Environmental Association in Cave 6 of the Yungang Grottoes" Microorganisms 13, no. 12: 2788. https://doi.org/10.3390/microorganisms13122788
APA StyleQiao, S., Wang, Z., Zhang, R., Wang, Y., Wang, C., Gao, G., & Pan, J. (2025). Microbial Community Analysis and Environmental Association in Cave 6 of the Yungang Grottoes. Microorganisms, 13(12), 2788. https://doi.org/10.3390/microorganisms13122788







