Shared Gut Microbial and Functional Signatures Linking Parkinson’s Disease and Type 2 Diabetes Revealed by Function-Anchored Metagenomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Clinical and Lifestyle Data
2.3. Stool Collection
2.4. DNA Extraction and Shotgun Metagenomic Sequencing
2.5. Bioinformatic Processing and Taxonomic Profiling
2.6. Microbial Ecology Analyses
2.6.1. Alpha Diversity
2.6.2. Beta Diversity
2.6.3. Differential Abundance
2.6.4. Machine-Learning Validation
2.6.5. Functional Profiling and GO–KO–KEGG Integration
2.6.6. Species–Pathway Correlations
2.7. Statistical Analysis
3. Results
3.1. Cohort Characteristics
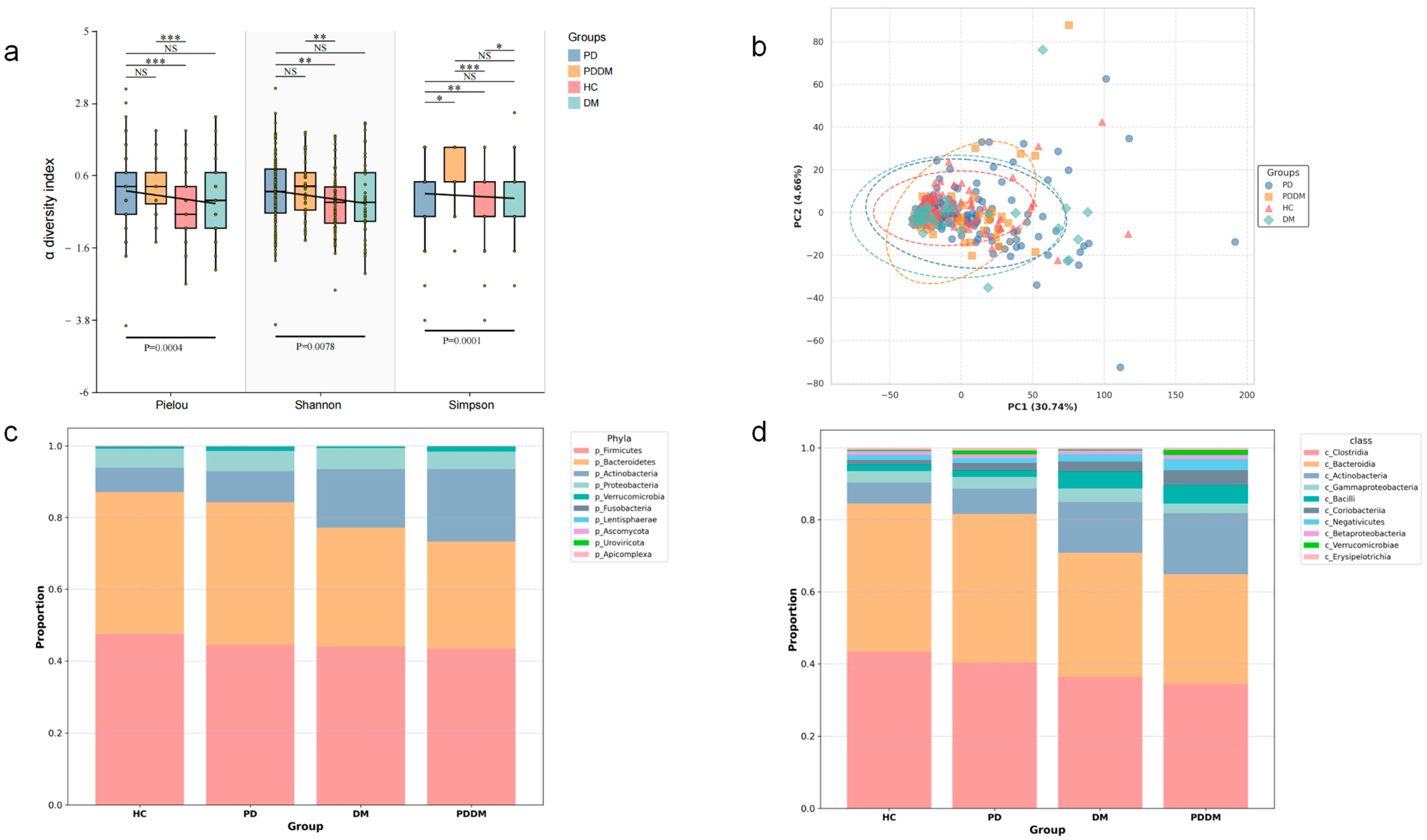
3.2. Overall Community Diversity: Convergent Trends Across Disease Groups
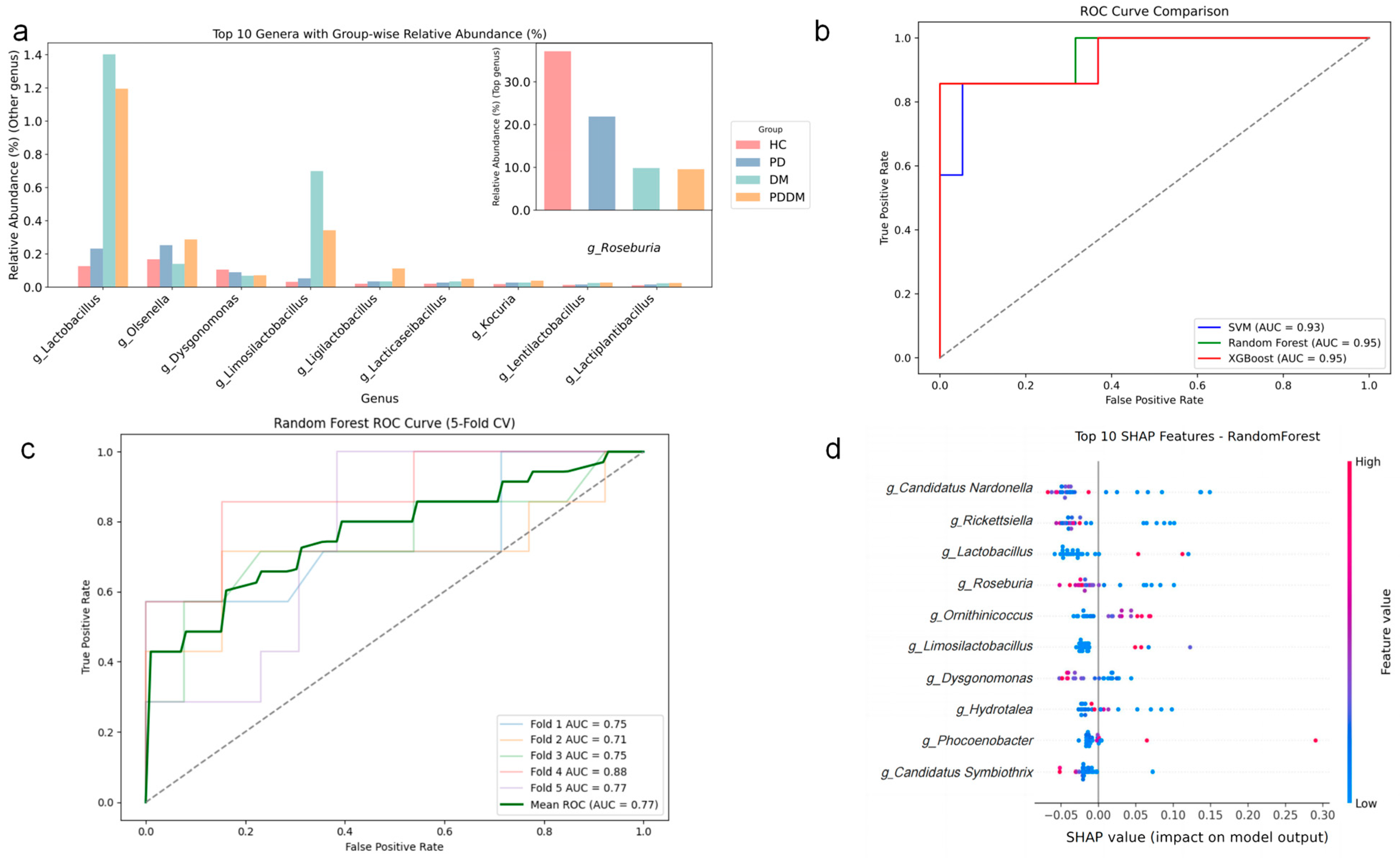
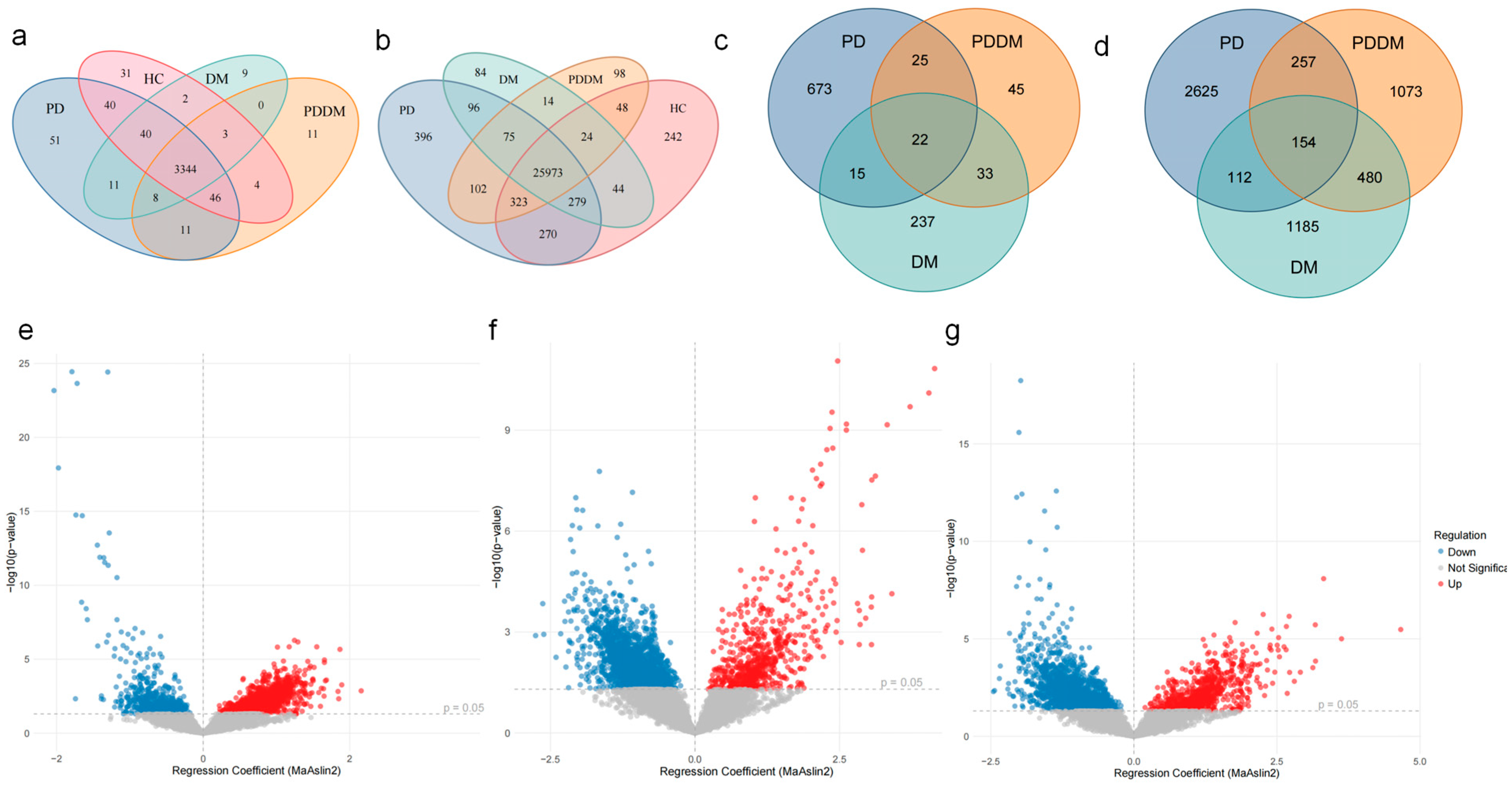
3.3. Shared Shifts in Gut Microbial Abundance at Genus and Species Levels Across Disease Groups
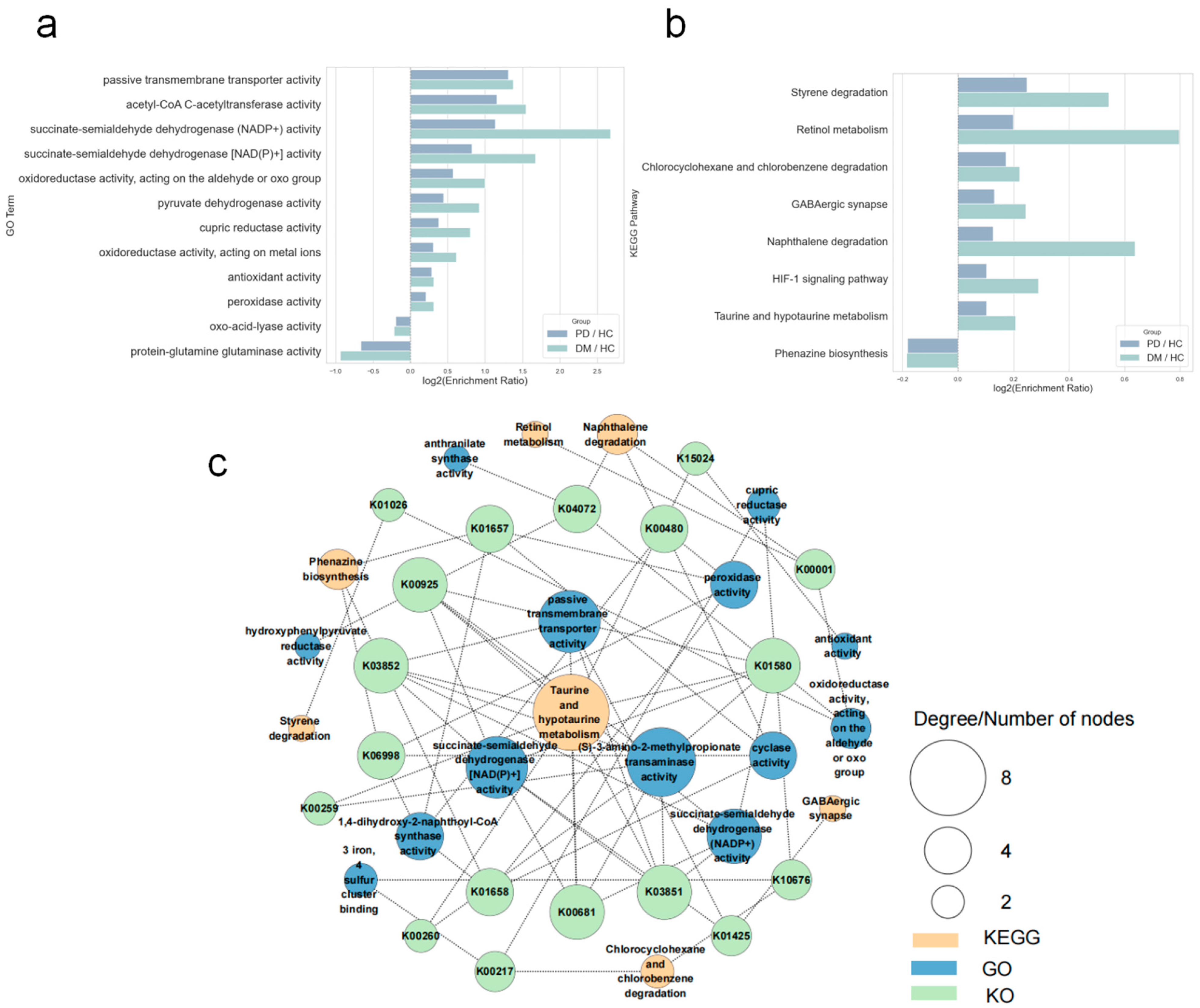
3.4. Concordant GO Terms and KEGG Pathways Between PD and DM
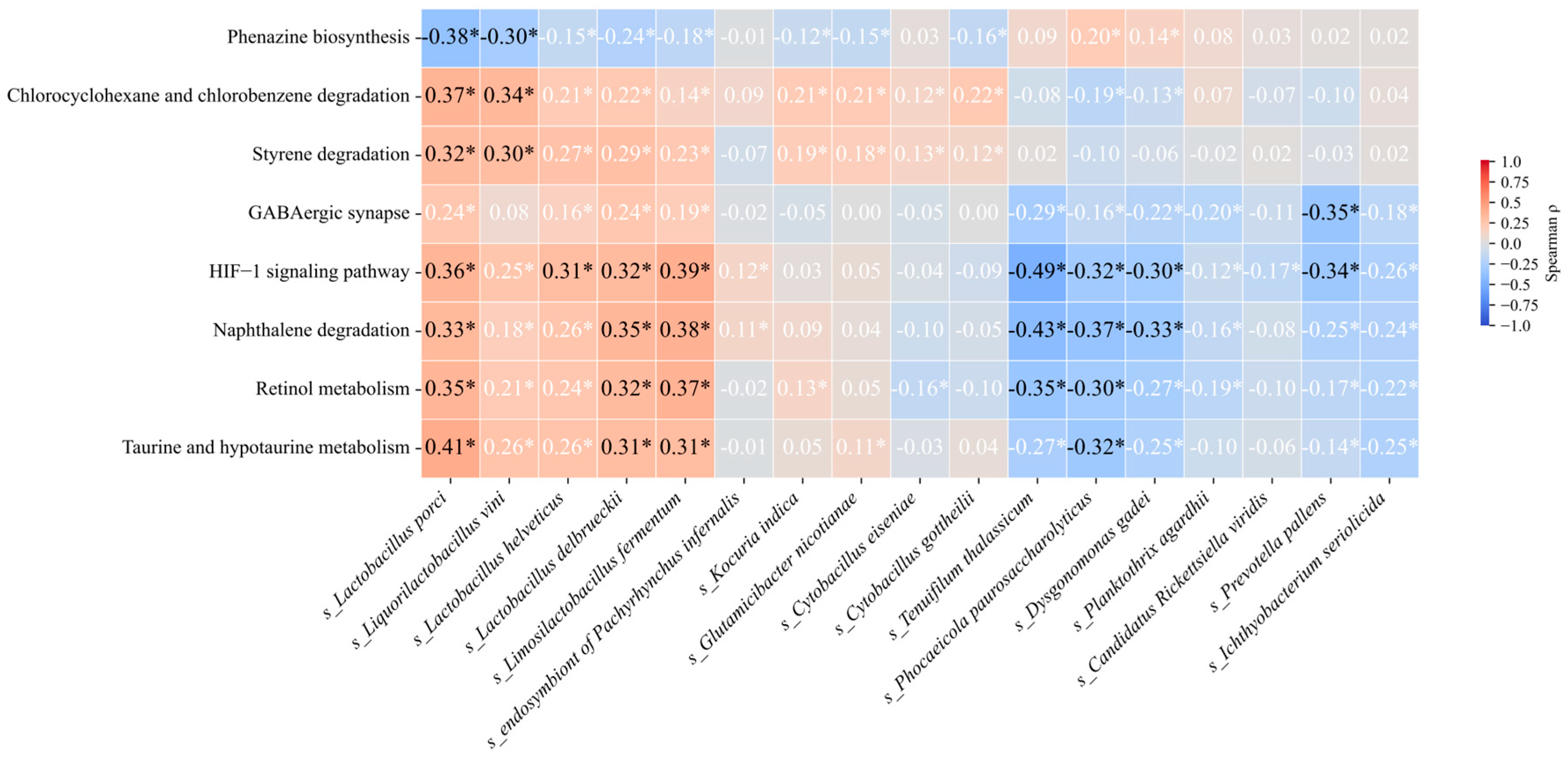
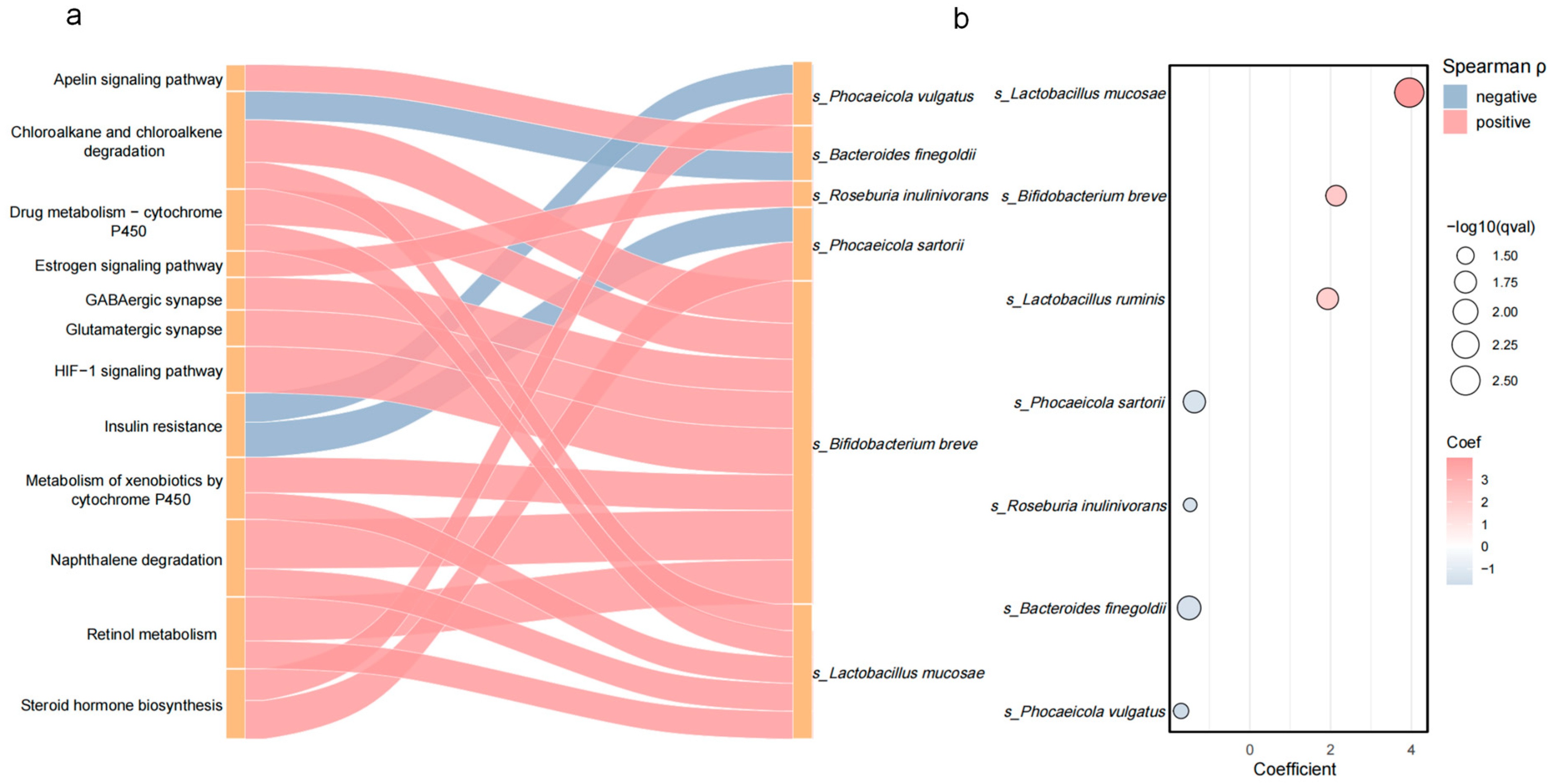
3.5. Correlation Between Differentially Abundant Species and Metabolic Pathways
3.6. Candidate Taxa Potentially Driving the Transition from PD to PDDM and Their Putative Pathways
4. Discussion
4.1. Concordant Gut Microbiota Dysbiosis in PD and DM
4.2. Common Mechanisms Revealed by Pathway Analysis
4.2.1. Taurine and Hypotaurine Metabolism as a Hub Pathway
4.2.2. Compensatory Activation of Detoxification Pathways
4.3. Key Genus Markers Indicated by Taxon–Function Correlations
4.3.1. Shared Species (PD and DM)
4.3.2. Conversion Species (PD → PDDM)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACE | Abundance-based Coverage Estimator |
| AUC-ROC | area under the receiver operating characteristic curve |
| BMI | body mass index |
| BWA | Burrows–Wheeler Aligner |
| D2R | dopamine D2 receptor |
| DAT | dopamine transporter |
| DM | diabetes mellitus |
| DNA | deoxyribonucleic acid |
| DPP-4 | dipeptidyl peptidase-4 |
| FASTQ | raw sequencing read file format |
| GABA | γ-aminobutyric acid |
| GLP-1 | glucagon-like peptide-1 |
| GO | Gene Ontology |
| HC | healthy controls |
| HIF-1 | hypoxia-inducible factor-1 |
| HR | hazard ratio |
| IAPP | islet amyloid polypeptide |
| InsR | insulin receptor |
| IRR | incidence rate ratio |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | KEGG Orthology |
| MaAsLin2 | Multivariate Association with Linear Models 2 |
| MAPK | mitogen-activated protein kinase |
| MDS | Movement Disorder Society |
| MOABI | monoamine oxidase type B inhibitors |
| OR | odds ratio |
| ORF | open reading frame |
| PCoA | principal coordinates analysis |
| PD | Parkinson’s disease |
| PDDM | PD with type 2 diabetes |
| PERMANOVA | Permutational Multivariate Analysis of Variance |
| PPARα | peroxisome proliferator-activated receptor alpha |
| RBF | radial basis function |
| SHAP | SHapley Additive exPlanations |
| SVM | support vector machine |
| T2DM | type 2 diabetes mellitus |
| TMAO | trimethylamine N-oxide |
| TPM | transcripts per million |
| UPDRS | Unified Parkinson’s Disease Rating Scale |
| XGBoost | eXtreme Gradient Boosting |
Appendix A
References
- Dorsey, E.R.; Elbaz, A.; Nichols, E.; Abbasi, N.; Abd-Allah, F.; Abdelalim, A.; Adsuar, J.C.; Ansha, M.G.; Brayne, C.; Choi, J.-Y.J.; et al. Global, regional, and national burden of Parkinson’s disease, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018, 17, 939–953. [Google Scholar] [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Han, K.; Kim, B.; Lee, S.H.; Kim, M.K. A nationwide cohort study on diabetes severity and risk of Parkinson disease. NPJ Park. Dis. 2023, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Pressley, J.C.; Louis, E.D.; Tang, M.X.; Cote, L.; Cohen, P.D.; Glied, S.; Mayeux, R. The impact of comorbid disease and injuries on resource use and expenditures in parkinsonism. Neurology 2003, 60, 87–93. [Google Scholar] [CrossRef]
- Brauer, R.; Wei, L.; Ma, T.; Athauda, D.; Girges, C.; Vijiaratnam, N.; Auld, G.; Whittlesea, C.; Wong, I.; Foltynie, T. Diabetes medications and risk of Parkinson’s disease: A cohort study of patients with diabetes. Brain 2020, 143, 3067–3076. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Brobert, G.P.; Johansson, S.; Jick, S.S.; Meier, C.R. Diabetes in patients with idiopathic Parkinson’s disease. Diabetes Care 2008, 31, 1808–1812. [Google Scholar] [CrossRef] [PubMed]
- D’Amelio, M.; Ragonese, P.; Callari, G.; Di Benedetto, N.; Palmeri, B.; Terruso, V.; Salemi, G.; Famoso, G.; Aridon, P.; Savettieri, G. Diabetes preceding Parkinson’s disease onset. A case–control study. Park. Relat. Disord. 2009, 15, 660–664. [Google Scholar] [CrossRef]
- Tomlinson, D.R.; Gardiner, N.J. Glucose neurotoxicity. Nat. Rev. Neurosci. 2008, 9, 36–45. [Google Scholar] [CrossRef]
- Barry, R.L.; Byun, N.E.; Williams, J.M.; Siuta, M.A.; Tantawy, M.N.; Speed, N.K.; Saunders, C.; Galli, A.; Niswender, K.D.; Avison, M.J. Brief exposure to obesogenic diet disrupts brain dopamine networks. PLoS ONE 2018, 13, e0191299. [Google Scholar] [CrossRef]
- Jones, K.T.; Woods, C.; Zhen, J.; Antonio, T.; Carr, K.D.; Reith, M.E. Effects of diet and insulin on dopamine transporter activity and expression in rat caudate-putamen, nucleus accumbens, and midbrain. J. Neurochem. 2017, 140, 728–740. [Google Scholar] [CrossRef]
- Cheng, H.; Gang, X.; Liu, Y.; Wang, G.; Zhao, X.; Wang, G. Mitochondrial dysfunction plays a key role in the development of neurodegenerative diseases in diabetes. Am. J. Physiol. -Endocrinol. Metab. 2020, 318, E750–E764. [Google Scholar] [CrossRef]
- Meng, L.; Li, Y.; Liu, C.; Zhang, G.; Chen, J.; Xiong, M.; Pan, L.; Zhang, X.; Chen, G.; Xiong, J. Islet amyloid polypeptide triggers α-synuclein pathology in Parkinson’s disease. Progress. Neurobiol. 2023, 226, 102462. [Google Scholar] [CrossRef]
- Alrouji, M.; Al-kuraishy, H.M.; Al-Gareeb, A.I.; Alexiou, A.; Papadakis, M.; Jabir, M.S.; Saad, H.M.; Batiha, G.E.S. NF-κB/NLRP3 inflammasome axis and risk of Parkinson’s disease in type 2 diabetes mellitus: A narrative review and new perspective. J. Cell. Mol. Med. 2023, 27, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Ghaisas, S.; Maher, J.; Kanthasamy, A. Gut microbiome in health and disease: Linking the microbiome–gut–brain axis and environmental factors in the pathogenesis of systemic and neurodegenerative diseases. Pharmacol. Ther. 2016, 158, 52–62. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, B.; Yu, D.; Zhu, C. Gut Microbiota: An Important Player in Type 2 Diabetes Mellitus. Front. Cell. Infect. Microbiol. 2022, 12, 834485. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wei, J.; Liu, P.; Zhang, Q.; Tian, Y.; Hou, G.; Meng, L.; Xin, Y.; Jiang, X. Role of the gut microbiota in type 2 diabetes and related diseases. Metabolism 2021, 117, 154712. [Google Scholar] [CrossRef]
- Elfil, M.; Kamel, S.; Kandil, M.; Koo, B.B.; Schaefer, S.M. Implications of the gut microbiome in Parkinson’s disease. Mov. Disord. 2020, 35, 921–933. [Google Scholar] [CrossRef]
- Gu, X.; Tang, J.; Chen, C. Efficacy of gut microbiota-targeted therapies in Parkinson’s disease: A systematic review and meta-analysis of randomized controlled trials. Front. Cell. Infect. Microbiol. 2025, 15, 1627406. [Google Scholar] [CrossRef]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef] [PubMed]
- Hensen, T.; Thiele, I. Metabolic modeling links gut microbiota to metabolic markers of Parkinson’s disease. Gut Microbes 2025, 17, 2554195. [Google Scholar] [CrossRef]
- Lotankar, M.; Houttu, N.; Mokkala, K.; Laitinen, K. Diet–Gut Microbiota Relations: Critical Appraisal of Evidence from Studies Using Metagenomics. Nutr. Rev. 2024, 83, e1917–e1938. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Li, N.; Zhang, R.; Tang, M.; Zhao, M.; Jiang, X.; Cai, X.; Ye, N.; Su, K.; Peng, J.; Zhang, X.; et al. Recent Progress and Prospects of Small Molecules for NLRP3 Inflammasome Inhibition. J. Med. Chem. 2023, 66, 14447–14473. [Google Scholar] [CrossRef] [PubMed]
- Pagano, G.; Polychronis, S.; Wilson, H.; Giordano, B.; Ferrara, N.; Niccolini, F.; Politis, M. Diabetes mellitus and Parkinson disease. Neurology 2018, 90, e1654–e1662. [Google Scholar] [CrossRef]
- Nam, G.E.; Kim, S.M.; Han, K.; Kim, N.H.; Chung, H.S.; Kim, J.W.; Han, B.; Cho, S.J.; Yu, J.H.; Park, Y.G. Metabolic syndrome and risk of Parkinson disease: A nationwide cohort study. PLoS Med. 2018, 15, e1002640. [Google Scholar] [CrossRef]
- Takagi, T.; Naito, Y.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Kamada, K.; Ishikawa, T.; Inoue, R.; Okuda, K.; Tsujimoto, Y.; et al. Changes in the Gut Microbiota are Associated with Hypertension, Hyperlipidemia, and Type 2 Diabetes Mellitus in Japanese Subjects. Nutrients 2020, 12, 2996. [Google Scholar] [CrossRef]
- Mao, L.; Zhang, Y.; Tian, J.; Sang, M.; Zhang, G.; Zhou, Y.; Wang, P. Cross-sectional study on the gut microbiome of Parkinson’s disease patients in central China. Front. Microbiol. 2021, 12, 728479. [Google Scholar] [CrossRef]
- Park, E.; Jia, J.; Quinn, M.R.; Schuller-Levis, G. Taurine Chloramine Inhibits Lymphocyte Proliferation and Decreases Cytokine Production in Activated Human Leukocytes. Clin. Immunol. 2002, 102, 179–184. [Google Scholar] [CrossRef]
- Rosa, F.T.; Freitas, E.C.; Deminice, R.; Jordão, A.A.; Marchini, J.S. Oxidative stress and inflammation in obesity after taurine supplementation: A double-blind, placebo-controlled study. Eur. J. Nutr. 2014, 53, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Wu, C.; Huang, Y.; Zheng, F.; Li, N.; Yan, W.; Li, C.; He, Q.; Hong, W.; Wu, S.; et al. Taurine as a supplement for treating type II diabetes mellitus with metformin and DPP4 inhibitor. Sci. Rep. 2025, 15, 18019. [Google Scholar] [CrossRef] [PubMed]
- Maleki, V.; Mahdavi, R.; Hajizadeh-Sharafabad, F.; Alizadeh, M. A Comprehensive Insight into Potential Roles of Taurine on Metabolic Variables in Type 2 Diabetes: A Systematic Review. Pharm. Sci. 2020, 26, 225–238. [Google Scholar] [CrossRef]
- Santos-Silva, J.C.; Ribeiro, R.A.; Vettorazzi, J.F.; Irles, E.; Rickli, S.; Borck, P.C.; Porciuncula, P.M.; Quesada, I.; Nadal, A.; Boschero, A.C.; et al. Taurine supplementation ameliorates glucose homeostasis, prevents insulin and glucagon hypersecretion, and controls β, α, and δ-cell masses in genetic obese mice. Amino Acids 2015, 47, 1533–1548. [Google Scholar] [CrossRef]
- Cui, C.; Song, H.; Han, Y.; Yu, H.; Li, H.; Yang, Y.; Zhang, B. Gut microbiota-associated taurine metabolism dysregulation in a mouse model of Parkinson’s disease. mSphere 2023, 8, e0043123. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Bargues-Carot, A.; Riaz, Z.; Wickham, H.; Zenitsky, G.; Jin, H.; Anantharam, V.; Kanthasamy, A.; Kanthasamy, A.G. Impact of environmental risk factors on mitochondrial dysfunction, neuroinflammation, protein misfolding, and oxidative stress in the etiopathogenesis of Parkinson’s disease. Int. J. Mol. Sci. 2022, 23, 10808. [Google Scholar] [CrossRef]
- Liang, B.; Huang, Y.; Zhong, Y.; Li, Z.; Ye, R.; Wang, B.; Zhang, B.; Meng, H.; Lin, X.; Du, J.; et al. Brain single-nucleus transcriptomics highlights that polystyrene nanoplastics potentially induce Parkinson’s disease-like neurodegeneration by causing energy metabolism disorders in mice. J. Hazard. Mater. 2022, 430, 128459. [Google Scholar] [CrossRef]
- Chen, Y.; Nan, Y.; Xu, L.; Dai, A.; Orteg, R.M.M.; Ma, M.; Zeng, Y.; Li, J. Polystyrene nanoplastics exposure induces cognitive impairment in mice via induction of oxidative stress and ERK/MAPK-mediated neuronal cuproptosis. Part. Fibre Toxicol. 2025, 22, 13. [Google Scholar] [CrossRef]
- Liang, X.; Zeng, Y.; Zhang, P.; Zhu, B.; Feng, J.; Deng, T.; Fu, Z.; Liu, C.; Chen, C.; Zhang, Y. Polystyrene nanoplastics trigger pyroptosis in dopaminergic neurons through TSC2/TFEB-mediated disruption of autophagosome-lysosome fusion in Parkinson’s disease. J. Transl. Med. 2025, 23, 631. [Google Scholar] [CrossRef]
- Shah, A.; Ong, C.E.; Pan, Y. Unveiling the role of cytochrome P450 (2E1) in human brain specifically in Parkinson’s Disease-Literature Review. Curr. Drug Metab. 2021, 22, 698–708. [Google Scholar] [CrossRef]
- Isse, F.A.; El-Sherbeni, A.A.; El-Kadi, A.O. The multifaceted role of cytochrome P450-Derived arachidonic acid metabolites in diabetes and diabetic cardiomyopathy. Drug Metab. Rev. 2022, 54, 141–160. [Google Scholar] [CrossRef]
- Marie, A.; Darricau, M.; Touyarot, K.; Parr-Brownlie, L.C.; Bosch-Bouju, C. Role and Mechanism of Vitamin A Metabolism in the Pathophysiology of Parkinson’s Disease. J. Park. Dis. 2021, 11, 949–970. [Google Scholar] [CrossRef]
- Van, Y.-H.; Lee, W.-H.; Ortiz, S.; Lee, M.-H.; Qin, H.-J.; Liu, C.-P. All-trans Retinoic Acid Inhibits Type 1 Diabetes by T Regulatory (Treg)-Dependent Suppression of Interferon-γ–Producing T-cells Without Affecting Th17 Cells. Diabetes 2009, 58, 146–155. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef]
- Al Ebrahim, R.N.; Alekseeva, M.G.; Bazhenov, S.V.; Fomin, V.V.; Mavletova, D.A.; Nesterov, A.A.; Poluektova, E.U.; Danilenko, V.N.; Manukhov, I.V. ClpL Chaperone as a Possible Component of the Disaggregase Activity of Limosilactobacillus fermentum U-21. Biology 2024, 13, 592. [Google Scholar] [CrossRef]
- Marsova, M.; Poluektova, E.; Odorskaya, M.; Ambaryan, A.; Revishchin, A.; Pavlova, G.; Danilenko, V. Protective effects of Lactobacillus fermentum U-21 against paraquat-induced oxidative stress in Caenorhabditis elegans and mouse models. World J. Microbiol. Biotechnol. 2020, 36, 104. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Z.; Zhao, L.; Zhao, Y.; Yang, G.; Wang, C.; Gao, L.; Niu, C.; Li, S. Lactobacillus plantarum DP189 Reduces α-SYN Aggravation in MPTP-Induced Parkinson’s Disease Mice via Regulating Oxidative Damage, Inflammation, and Gut Microbiota Disorder. J. Agric. Food Chem. 2022, 70, 1163–1173. [Google Scholar] [CrossRef]
- Wang, C.H.; Yen, H.R.; Lu, W.L.; Ho, H.H.; Lin, W.Y.; Kuo, Y.W.; Huang, Y.Y.; Tsai, S.Y.; Lin, H.C. Adjuvant Probiotics of Lactobacillus salivarius subsp. salicinius AP-32, L. johnsonii MH-68, and Bifidobacterium animalis subsp. lactis CP-9 Attenuate Glycemic Levels and Inflammatory Cytokines in Patients With Type 1 Diabetes Mellitus. Front. Endocrinol. 2022, 13, 754401. [Google Scholar] [CrossRef]
- Montgomery, T.L.; Toppen, L.C.; Eckstrom, K.; Heney, E.R.; Kennedy, J.J.; Scarborough, M.J.; Krementsov, D.N. Lactobacillaceae differentially impact butyrate-producing gut microbiota to drive CNS autoimmunity. Gut Microbes 2024, 16, 2418415. [Google Scholar] [CrossRef]
- Wang, Q.W.; Jia, D.J.C.; He, J.M.; Sun, Y.; Qian, Y.; Ge, Q.W.; Qi, Y.D.; Wang, Q.Y.; Hu, Y.Y.; Wang, L. Lactobacillus intestinalis primes epithelial cells to suppress colitis-related Th17 response by host-microbe retinoic acid biosynthesis. Adv. Sci. 2023, 10, 2303457. [Google Scholar] [CrossRef]
- Shao, X.; Xu, B.; Chen, C.; Li, P.; Luo, H. The function and mechanism of lactic acid bacteria in the reduction of toxic substances in food: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 5950–5963. [Google Scholar] [CrossRef]
- Saban Güler, M.; Arslan, S.; Ağagündüz, D.; Cerqua, I.; Pagano, E.; Berni Canani, R.; Capasso, R. Butyrate: A potential mediator of obesity and microbiome via different mechanisms of actions. Food Res. Int. 2025, 199, 115420. [Google Scholar] [CrossRef]
- Wang, S.; Ren, H.; Zhong, H.; Zhao, X.; Li, C.; Ma, J.; Gu, X.; Xue, Y.; Huang, S.; Yang, J.; et al. Combined berberine and probiotic treatment as an effective regimen for improving postprandial hyperlipidemia in type 2 diabetes patients: A double blinded placebo controlled randomized study. Gut Microbes 2022, 14, 2003176. [Google Scholar] [CrossRef]
- Qian, X.; Li, Q.; Zhu, H.; Chen, Y.; Lin, G.; Zhang, H.; Chen, W.; Wang, G.; Tian, P. Bifidobacteria with indole-3-lactic acid-producing capacity exhibit psychobiotic potential via reducing neuroinflammation. Cell Rep. Med. 2024, 5, 101798. [Google Scholar] [CrossRef]
- Jin, S.; Chen, P.; Yang, J.; Li, D.; Liu, X.; Zhang, Y.; Xia, Q.; Li, Y.; Chen, G.; Li, Y.; et al. Phocaeicola vulgatus alleviates diet-induced metabolic dysfunction-associated steatotic liver disease progression by downregulating histone acetylation level via 3-HPAA. Gut Microbes 2024, 16, 2309683. [Google Scholar] [CrossRef]
- Ryan, P.M.; Stolte, E.H.; London, L.E.E.; Wells, J.M.; Long, S.L.; Joyce, S.A.; Gahan, C.G.M.; Fitzgerald, G.F.; Ross, R.P.; Caplice, N.M.; et al. Lactobacillus mucosae DPC 6426 as a bile-modifying and immunomodulatory microbe. BMC Microbiol. 2019, 19, 33. [Google Scholar] [CrossRef]
- London, L.E.; Kumar, A.H.; Wall, R.; Casey, P.G.; O’Sullivan, O.; Shanahan, F.; Hill, C.; Cotter, P.D.; Fitzgerald, G.F.; Ross, R.P.; et al. Exopolysaccharide-producing probiotic Lactobacilli reduce serum cholesterol and modify enteric microbiota in ApoE-deficient mice. J. Nutr. 2014, 144, 1956–1962. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Mörbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Mølgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Yang, J.Y.; Lee, Y.S.; Kim, Y.; Lee, S.H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.S.; Lim, H.S.; Kim, M.S.; et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef]
- Bae, H.J.; Lee, D.H.; Choi, H.S.; Lim, H.J.; Chung, E.C.; Oh, N.S. Lacticaseibacillus rhamnosus BELR47 Mitigates Inflammatory Response in a Gardnerella vaginalis-Induced Bacterial Vaginosis Mouse Model via Gut Microbiome Modulation. Probiotics Antimicrob. Proteins 2025. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Wang, S.; Zhao, W.; Du, Y.; Wang, L.; Han, B.; Zhang, M.; Xu, X.; Wang, S.; Ma, X.; et al. Shared Gut Microbial and Functional Signatures Linking Parkinson’s Disease and Type 2 Diabetes Revealed by Function-Anchored Metagenomics. Microorganisms 2025, 13, 2705. https://doi.org/10.3390/microorganisms13122705
Cui Y, Wang S, Zhao W, Du Y, Wang L, Han B, Zhang M, Xu X, Wang S, Ma X, et al. Shared Gut Microbial and Functional Signatures Linking Parkinson’s Disease and Type 2 Diabetes Revealed by Function-Anchored Metagenomics. Microorganisms. 2025; 13(12):2705. https://doi.org/10.3390/microorganisms13122705
Chicago/Turabian StyleCui, Ying, Shiya Wang, Wenlu Zhao, Yitong Du, Lin Wang, Bingyu Han, Mingkai Zhang, Xiaojiao Xu, Sichen Wang, Xiaolong Ma, and et al. 2025. "Shared Gut Microbial and Functional Signatures Linking Parkinson’s Disease and Type 2 Diabetes Revealed by Function-Anchored Metagenomics" Microorganisms 13, no. 12: 2705. https://doi.org/10.3390/microorganisms13122705
APA StyleCui, Y., Wang, S., Zhao, W., Du, Y., Wang, L., Han, B., Zhang, M., Xu, X., Wang, S., Ma, X., Xu, X., Zhao, Y., Liu, S., Wang, Y., & Tuo, H. (2025). Shared Gut Microbial and Functional Signatures Linking Parkinson’s Disease and Type 2 Diabetes Revealed by Function-Anchored Metagenomics. Microorganisms, 13(12), 2705. https://doi.org/10.3390/microorganisms13122705







