Diversity and Functional Potential of Yeasts Inhabiting Honey Bee Drones
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Honey Bee Drones and Ethics Statement
2.2. Cultivable Microorganisms Isolation
2.3. Identification of Isolated Yeasts
2.4. Phylogenetic Analysis
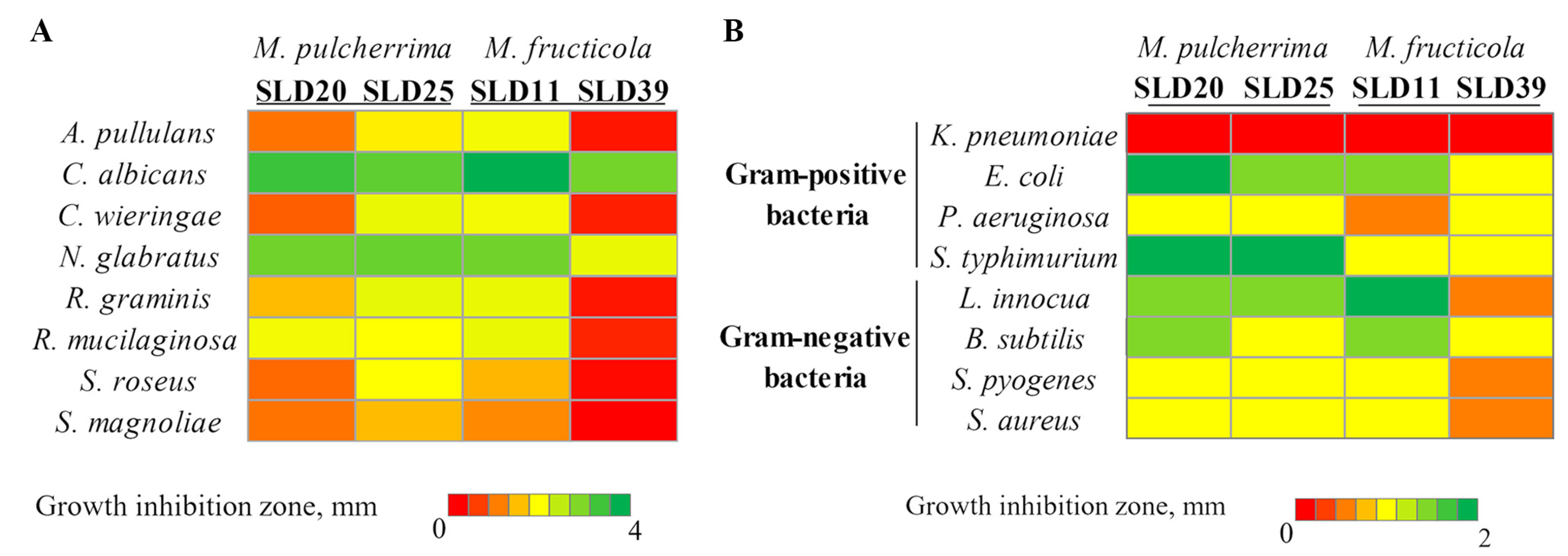
2.5. In Vitro Evaluation of Antifungal Activity
2.6. Determination of Yeast Antibacterial Activity
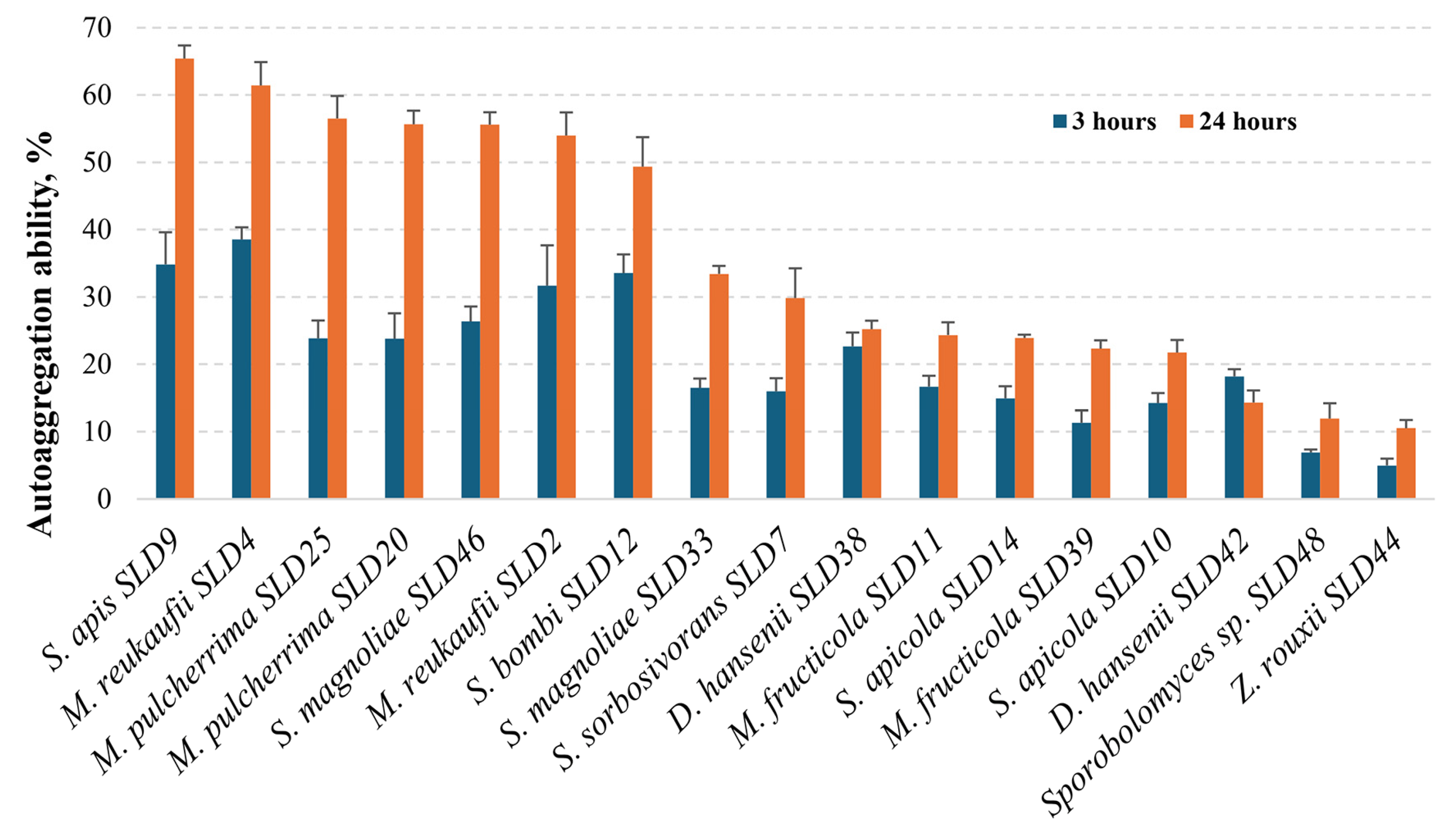
2.7. Determination of Yeast Autoaggregation Properties
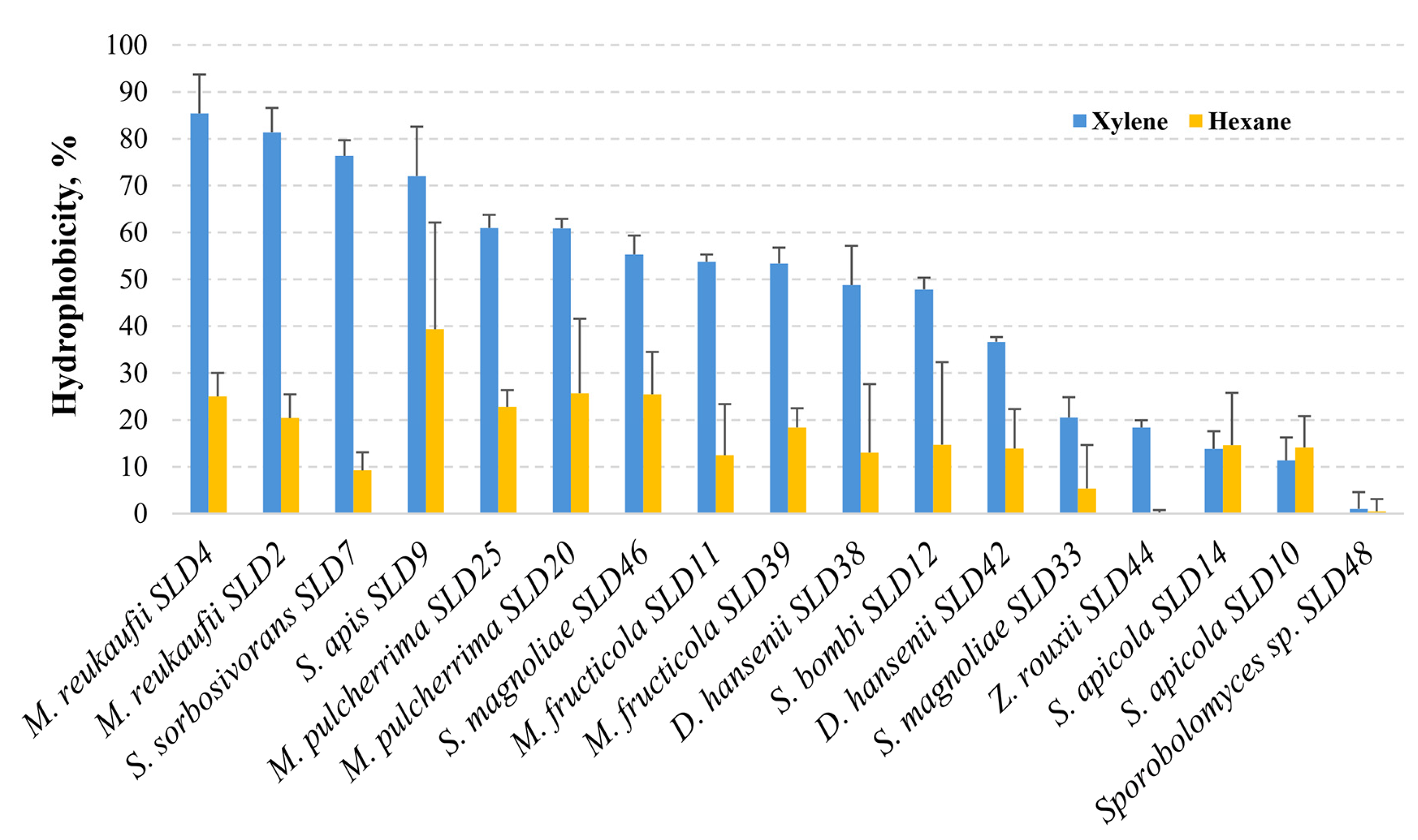
2.8. Yeast Adhesion to Hydrocarbons
2.9. Biofilm Formation
2.10. Statistical Analysis
3. Results and Discussion
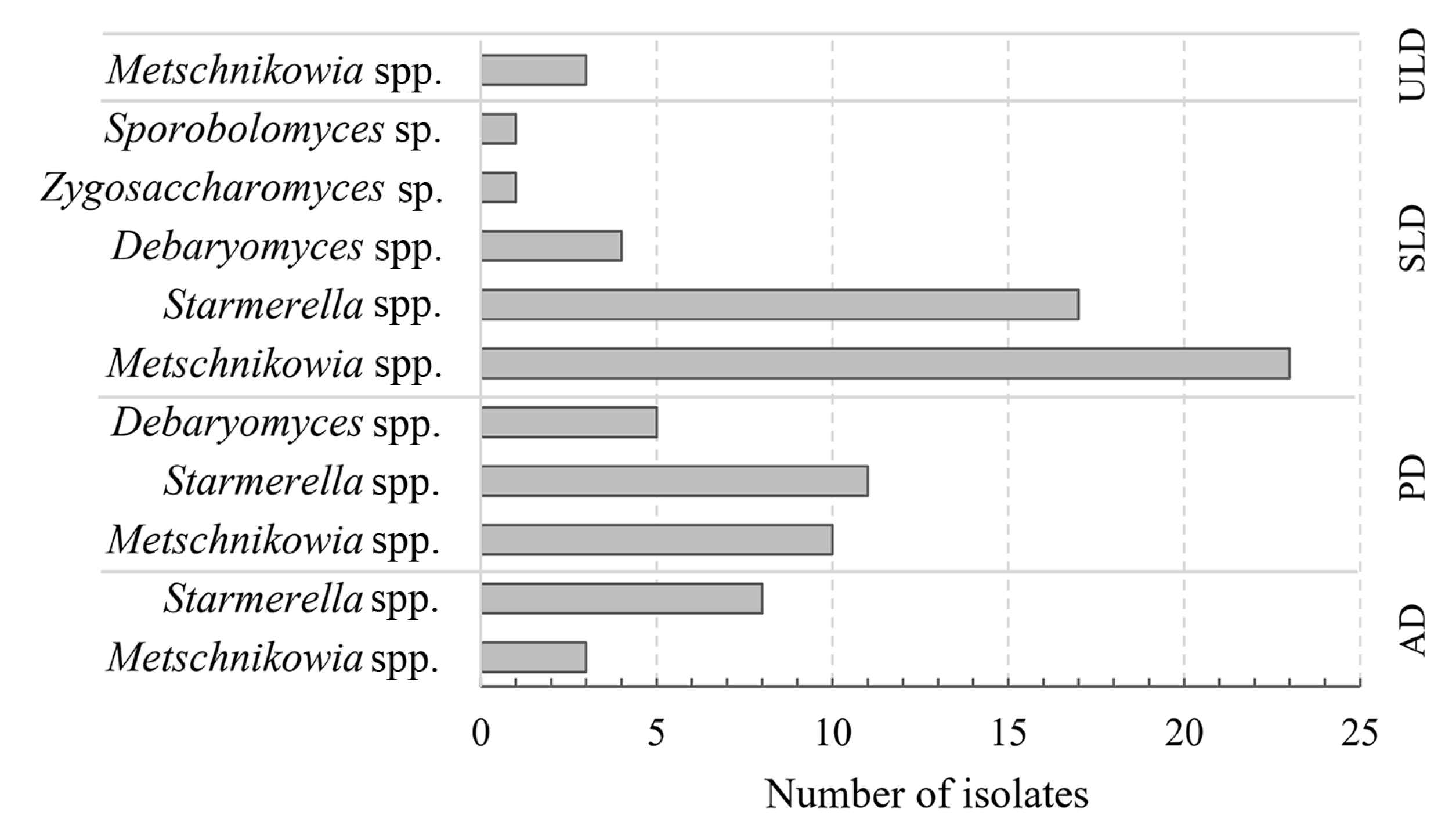
3.1. Cultivable Yeasts Associated with Honey Bee Drones at Different Development Stages and Phylogenetic Characterization
3.2. Biocontrol Features of Honey Bee Drones-Inhabiting Yeasts
3.3. Evaluation of Probiotic Traits
3.3.1. Variation in Autoaggregation Capacity Across Strains
3.3.2. Hydrophobic Interaction with Hydrocarbons
3.3.3. Determination of Biofilm Formation Potential
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fikadu, Z. The Contribution of Managed Honey Bees to Crop Pollination, Food Security, and Economic Stability: Case of Ethiopia. Open Agric. J. 2020, 13, 175–181. [Google Scholar] [CrossRef]
- Tsadila, C.; Amoroso, C.; Mossialos, D. Microbial Diversity in Bee Species and Bee Products: Pseudomonads Contribution to Bee Well-Being and the Biological Activity Exerted by Honey Bee Products: A Narrative Review. Diversity 2023, 15, 1088. [Google Scholar] [CrossRef]
- Khalifa, S.A.M.; Elshafiey, E.H.; Shetaia, A.A.; El-Wahed, A.A.A.; Algethami, A.F.; Musharraf, S.G.; Alajmi, M.F.; Zhao, C.; Masry, S.H.D.; Abdel-Daim, M.M.; et al. Overview of Bee Pollination and Its Economic Value for Crop Production. Insects 2021, 12, 688. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Dong, Y.; Gu, C.; Zhang, X.; Ma, H. Processing Technologies for Bee Products: An Overview of Recent Developments and Perspectives. Front. Nutr. 2021, 8, 727181. [Google Scholar] [CrossRef] [PubMed]
- Palma-Morales, M.; Huertas, J.R.; Rodríguez-Pérez, C. A Comprehensive Review of the Effect of Honey on Human Health. Nutrients 2023, 15, 3056. [Google Scholar] [CrossRef] [PubMed]
- Nong, Y.; Maloh, J.; Natarelli, N.; Gunt, H.B.; Tristani, E.; Sivamani, R.K. A Review of the Use of Beeswax in Skincare. J. Cosmet. Dermatol. 2023, 22, 2166–2173. [Google Scholar] [CrossRef]
- Ilie, C.I.; Spoiala, A.; Geana, E.I.; Chircov, C.; Ficai, A.; Ditu, L.M.; Oprea, E. Bee Bread: A Promising Source of Bioactive Compounds with Antioxidant Properties—First Report on Some Antimicrobial Features. Antioxidants 2024, 13, 353. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Baer, B.; Niño, E.L. Putative Drone Copulation Factors Regulating Honey Bee (Apis mellifera) Queen Reproduction and Health: A Review. Insects 2019, 10, 8. [Google Scholar] [CrossRef]
- Gätschenberger, H.; Azzami, K.; Tautz, J.; Beier, H. Antibacterial Immune Competence of Honey Bees (Apis mellifera) Is Adapted to Different Life Stages and Environmental Risks. PLoS ONE 2013, 8, e66415. [Google Scholar] [CrossRef]
- Kachhwaha, N. Apiculture; Fedorov, S., Ed.; Weser Books: Zittau, Germany, 2019; Volume 1, ISBN 9783964921192. [Google Scholar]
- Chen, J.; DeGrandi-Hoffman, G.; Ratti, V.; Kang, Y.; Chen, J.; DeGrandi-Hoffman, G.; Ratti, V.; Kang, Y. Review on Mathematical Modeling of Honeybee Population Dynamics. Math. Biosci. Eng. 2021, 18, 9606–9650. [Google Scholar] [CrossRef]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional Diversity within the Simple Gut Microbiota of the Honey Bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The Gut Microbiota of Insects—Diversity in Structure and Function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut Microbial Communities of Social Bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Agarbati, A.; Gattucci, S.; Canonico, L.; Ciani, M.; Comitini, F. Yeast Communities Related to Honeybees: Occurrence and Distribution in Flowers, Gut Mycobiota, and Bee Products. Appl. Microbiol. Biotechnol. 2024, 108, 175. [Google Scholar] [CrossRef]
- Callegari, M.; Crotti, E.; Fusi, M.; Marasco, R.; Gonella, E.; De Noni, I.; Romano, D.; Borin, S.; Tsiamis, G.; Cherif, A.; et al. Compartmentalization of Bacterial and Fungal Microbiomes in the Gut of Adult Honeybees. npj Biofilms Microbiomes 2021, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Januário da Costa Neto, D.; Benevides de Morais, P. The Vectoring of Starmerella Species and Other Yeasts by Stingless Bees in a Neotropical Savanna. Fungal Ecol. 2020, 47, 100973. [Google Scholar] [CrossRef]
- Türkel, S.; Korukluoğlu, M.; Yavuz, M. Biocontrol Activity of the Local Strain of Metschnikowia pulcherrima on Different Postharvest Pathogens. Biotechnol. Res. Int. 2014, 2014, 397167. [Google Scholar] [CrossRef]
- Di Canito, A.; Mateo-Vargas, M.A.; Mazzieri, M.; Cantoral, J.; Foschino, R.; Cordero-Bueso, G.; Vigentini, I. The Role of Yeasts as Biocontrol Agents for Pathogenic Fungi on Postharvest Grapes: A Review. Foods 2021, 10, 1650. [Google Scholar] [CrossRef]
- Iorizzo, M.; Coppola, F.; Pannella, G.; Ganassi, S.; Matarazzo, C.; Albanese, G.; Tedino, C.; Di Donato, L.M.; Iacovino, V.P.; Cozzolino, R.; et al. First Report on Antifungal Activity of Metschnikowia pulcherrima Against Ascosphaera apis, the Causative Agent of Chalkbrood Disease in Honeybee (Apis Mellifera L.) Colonies. J. Fungi 2025, 11, 336. [Google Scholar] [CrossRef]
- Dzialo, M.C.; Park, R.; Steensels, J.; Lievens, B.; Verstrepen, K.J. Physiology, Ecology and Industrial Applications of Aroma Formation in Yeast. FEMS Microbiol. Rev. 2017, 41, S95–S128. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Gattucci, S.; Ciani, M.; Comitini, F. From Pollen to Bee Bread: A Reservoir of Functional Yeasts. Fermentation 2025, 11, 290. [Google Scholar] [CrossRef]
- Echeverrigaray, S.; Scariot, F.J.; Foresti, L.; Schwarz, L.V.; Rocha, R.K.M.; da Silva, G.P.; Moreira, J.P.; Delamare, A.P.L. Yeast Biodiversity in Honey Produced by Stingless Bees Raised in the Highlands of Southern Brazil. Int. J. Food Microbiol. 2021, 347, 109200. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Moran, N.A. The Honeybee Microbiota and Its Impact on Health and Disease. Nat. Rev. Microbiol. 2023, 22, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Todorov, S.D.; Alves, M.V.; Bueno, G.C.A.; Alves, V.F.; Ivanova, I.V. Bee-Associated Beneficial Microbes—Importance for Bees and for Humans. Insects 2024, 15, 430. [Google Scholar] [CrossRef] [PubMed]
- Matuszewska-Mach, E.; Packi, K.; Rzetecka, N.; Wieliński, W.; Kokot, Z.J.; Kowalczyk, D.; Matysiak, J. Insights into the Nutritional Value of Honeybee Drone Larvae (Apis mellifera) through Proteomic Profiling. Sci. Rep. 2024, 14, 28562. [Google Scholar] [CrossRef] [PubMed]
- Burks, A.; Gallagher, P.; Raymann, K. Discovery of Reproductive Tissue-Associated Bacteria and the Modes of Microbiota Acquisition in Male Honey Bees (Drones). mSphere 2025, 10, e00705-24. [Google Scholar] [CrossRef]
- Ayup, M.M.; Gärtner, P.; Agosto-Rivera, J.L.; Marendy, P.; de Souza, P.; Galindo-Cardona, A. Analysis of Honeybee Drone Activity during the Mating Season in Northwestern Argentina. Insects 2021, 12, 566. [Google Scholar] [CrossRef]
- Santorelli, L.A.; Wilkinson, T.; Abdulmalik, R.; Rai, Y.; Creevey, C.J.; Huws, S.; Gutierrez-Merino, J. Beehives Possess Their Own Distinct Microbiomes. Environ. Microbiomes 2023, 18, 1. [Google Scholar] [CrossRef]
- Kashchenko, G.; Taldaev, A.; Adonin, L.; Smutin, D. Investigating Aerobic Hive Microflora: Role of Surface Microbiome of Apis mellifera. Biology 2025, 14, 88. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Rao, V.D.; Yeoman, C.J.; Wilson, B.A.; White, B.A.; Goldenfeld, N.; Robinson, G.E. Caste-Specific Differences in Hindgut Microbial Communities of Honey Bees (Apis mellifera). PLoS ONE 2015, 10, e0123911. [Google Scholar] [CrossRef] [PubMed]
- Smutin, D.; Lebedev, E.; Selitskiy, M.; Panyushev, N.; Adonin, L. Micro”bee”ota: Honey Bee Normal Microbiota as a Part of Superorganism. Microorganisms 2022, 10, 2359. [Google Scholar] [CrossRef] [PubMed]
- Lanh, P.T.; Duong, B.T.T.; Thu, H.T.; Hoa, N.T.; Yoo, M.S.; Cho, Y.S.; Quyen, D.V. The Gut Microbiota at Different Developmental Stages of Apis Cerana Reveals Potential Probiotic Bacteria for Improving Honeybee Health. Microorganisms 2022, 10, 1938. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Letizia, F.; Ganassi, S.; Testa, B.; Petrarca, S.; Albanese, G.; Di Criscio, D.; De Cristofaro, A. Functional Properties and Antimicrobial Activity from Lactic Acid Bacteria as Resources to Improve the Health and Welfare of Honey Bees. Insects 2022, 13, 308. [Google Scholar] [CrossRef]
- Leska, A.; Nowak, A.; Szulc, J.; Motyl, I.; Czarnecka-Chrebelska, K.H. Antagonistic Activity of Potentially Probiotic Lactic Acid Bacteria against Honeybee (Apis mellifera L.) Pathogens. Pathogens 2022, 11, 1367. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Bonatsou, S.; Karamouza, M.; Zoumpopoulou, G.; Mavrogonatou, E.; Kletsas, D.; Papadimitriou, K.; Tsakalidou, E.; Nychas, G.J.E.; Panagou, E. Evaluating the Probiotic Potential and Technological Characteristics of Yeasts Implicated in Cv. Kalamata Natural Black Olive Fermentation. Int. J. Food Microbiol. 2018, 271, 48–59. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, W.; Gao, Y.; Zha, M.; Zhang, D.; Peng, X.; Zhang, H.; Wang, C.; Xu, C.; Zhou, T.; et al. Engineering Saccharomyces cerevisiae for Improved Biofilm Formation and Ethanol Production in Continuous Fermentation. Biotechnol. Biofuels Bioprod. 2023, 16, 119. [Google Scholar] [CrossRef]
- Wang, D.; Zeng, N.; Li, C.; Li, Z.; Zhang, N.; Li, B. Fungal Biofilm Formation and Its Regulatory Mechanism. Heliyon 2024, 10, e32766. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- George, T.; Sivam, V.; Vaiyapuri, M.; Anandan, R.; Sivaraman, G.K.; Joseph, T.C. Standardizing Biofilm Quantification: Harmonizing Crystal Violet Absorbance Measurements through Extinction Coefficient Ratio Adjustment. Arch. Microbiol. 2025, 207, 59. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical 689 Computing: Vienna, Austria, 2025; Available online: https://www.R-project.org/ (accessed on 15 September 2025).
- Ogle, D.H.; Wheeler, P.; Dinno, A. FSA: Fisheries Stock Analysis. R Package Version 0.10.0. 2024. Available online: https://CRAN.R-project.org/package=FSA (accessed on 15 September 2025).
- Dinno, A. dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums. R Package Version 1.3.6. 693. 2024. Available online: https://CRAN.R-project.org/package=dunn.test (accessed on 15 September 2025).
- Rutkowski, D.; Weston, M.; Vannette, R.L. Bees Just Wanna Have Fungi: A Review of Bee Associations with Nonpathogenic Fungi. FEMS Microbiol. Ecol. 2023, 99, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Inglis, G.D.; Sigler, L.; Goette, M.S. Aerobic Microorganisms Associated with Alfalfa Leafcutter Bees (megachile rotundata). Microb. Ecol. 1993, 26, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Detry, R.; Simon-Delso, N.; Bruneau, E.; Daniel, H.M. Specialisation of Yeast Genera in Different Phases of Bee Bread Maturation. Microorganisms 2020, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
- Wirth, F.; Goldani, L.Z. Epidemiology of Rhodotorula: An Emerging Pathogen. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 465717. [Google Scholar] [CrossRef]
- Liu, D.; Ma, L.; Shi, Y.; Wang, A.; Liu, C. Molecular Diagnosis and Source Tracing of an Infection of Aureobasidium pullulans. J. Infect. Dev. Ctries. 2019, 13, 1174–1179. [Google Scholar] [CrossRef]
- McNicholas, S.; McDermott, H.; Power, L.; Johnson, E.M.; Moroney, J.; Humphreys, H.; Smyth, E.G. Sporobolomyces roseus in the Cerebrospinal Fluid of an Immunocompetent Patient—To Treat or Not to Treat? J. Med. Microbiol. 2012, 61, 295–296. [Google Scholar] [CrossRef]
- Bernal-Martinez, L.; Gomez-Lopez, A.; Castelli, M.V.; Mesa-Arango, A.C.; Zaragoza, O.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. Susceptibility Profile of Clinical Isolates of Non-Cryptococcus neoformans/Non-Cryptococcus gattii Cryptococcus Species and Literature Review. Med. Mycol. 2010, 48, 90–96. [Google Scholar] [CrossRef]
- Martorell, P.; Fernández-Espinar, M.T.; Querol, A. Molecular Monitoring of Spoilage Yeasts during the Production of Candied Fruit Nougats to Determine Food Contamination Sources. Int. J. Food Microbiol. 2005, 101, 293–302. [Google Scholar] [CrossRef]
- Di Francesco, A.; Zajc, J.; Stenberg, J.A. Aureobasidium spp.: Diversity, Versatility, and Agricultural Utility. Horticulturae 2023, 9, 59. [Google Scholar] [CrossRef]
- Hallas-Møller, M.; Burow, M.; Henrissat, B.; Johansen, K.S. Cryptococcus neoformans: Plant–Microbe Interactions and Ecology. Trends Microbiol. 2024, 32, 984–995. [Google Scholar] [CrossRef] [PubMed]
- El Dana, F.; David, V.; Hallal, M.A.; Tourdot-Maréchal, R.; Hayar, S.; Colosio, M.C.; Alexandre, H. Metschnikowia pulcherrima and Lachancea thermotolerans Killer Toxins: Contribution to Must Bioprotection. Foods 2025, 14, 1462. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, P.K.; Kaur, M.; Sharma, S.C.; Mahmood, A. Harnessing Killer Yeast System: From Molecular Insight to Real World Biocontrol Solution. Arch. Microbiol. 2025, 207, 116. [Google Scholar] [CrossRef] [PubMed]
- Vepštaitė-Monstavičė, I.; Ravoitytė, B.; Būdienė, J.; Valys, A.; Lukša, J.; Servienė, E. Essential Oils of Mentha arvensis and Cinnamomum cassia Exhibit Distinct Antibacterial Activity at Different Temperatures In Vitro and on Chicken Skin. Foods 2023, 12, 3938. [Google Scholar] [CrossRef]
- Büyüksırıt-Bedir, T.; Kuleaşan, H. Purification and Characterization of a Metschnikowia pulcherrima Killer Toxin with Antagonistic Activity against Pathogenic Microorganisms. Arch. Microbiol. 2022, 204, 337. [Google Scholar] [CrossRef]
- Sipiczki, M.; Pfliegler, W.P.; Holb, I.J. Metschnikowia Species Share a Pool of Diverse RRNA Genes Differing in Regions That Determine Hairpin-Loop Structures and Evolve by Reticulation. PLoS ONE 2013, 8, e67384. [Google Scholar] [CrossRef]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol Yeasts: Mechanisms and Applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef]
- Stanevičienė, R.; Lukša, J.; Strazdaitė-žielienė, Ž.; Ravoitytė, B.; Losinska-sičiūnienė, R.; Mozūraitis, R.; Servienė, E. Mycobiota in the Carposphere of Sour and Sweet Cherries and Antagonistic Features of Potential Biocontrol Yeasts. Microorganisms 2021, 9, 1423. [Google Scholar] [CrossRef]
- Hicks, R.H.; Moreno-Beltrán, M.; Gore-Lloyd, D.; Chuck, C.J.; Henk, D.A. The Oleaginous Yeast Metschnikowia pulcherrima Displays Killer Activity against Avian-Derived Pathogenic Bacteria. Biology 2021, 10, 1227. [Google Scholar] [CrossRef]
- Gomaa, E.Z. Antimicrobial and Anti-Adhesive Properties of Biosurfactant Produced by Lactobacilli Isolates, Biofilm Formation and Aggregation Ability. J. Gen. Appl. Microbiol. 2013, 59, 425–436. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion Mechanisms Mediated by Probiotics and Prebiotics and Their Potential Impact on Human Health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef]
- Saito, K.; Tomita, S.; Nakamura, T. Aggregation of Lactobacillus brevis Associated with Decrease in PH by Glucose Fermentation. Biosci. Biotechnol. Biochem. 2019, 83, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Staniszewski, A.; Kordowska-Wiater, M. Probiotic Yeasts and How to Find Them—Polish Wines of Spontaneous Fermentation as Source for Potentially Probiotic Yeasts. Foods 2023, 12, 3392. [Google Scholar] [CrossRef]
- Tomičić, R.; Tomičić, Z.; Raspor, P. Influence of Culture Conditions on Co-Aggregation of Probiotic Yeast Saccharomyces boulardii with Candida spp. and Their Auto-Aggregation. Folia Microbiol. 2022, 67, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
- Ashwini Hariwal, C.J.D. Autoaggregation, Coaggregation and Hydrophobicity—A Mechanism to Explain Oral Probiotic Function by Isolates from Dairy Source. J. Chem. Health Risks 2024, 14, 1735–1743. [Google Scholar]
- Doyle, R.J. Contribution of the Hydrophobic Effect to Microbial Infection. Microbes Infect. 2000, 2, 391–400. [Google Scholar] [CrossRef]
- Kelesidis, T.; Pothoulakis, C. Efficacy and Safety of the Probiotic Saccharomyces boulardii for the Prevention and Therapy of Gastrointestinal Disorders. Therap Adv. Gastroenterol. 2012, 5, 111. [Google Scholar] [CrossRef]
- Coimbra-Gomes, J.; Reis, P.J.M.; Tavares, T.G.; Faria, M.A.; Malcata, F.X.; Macedo, A.C. Evaluating the Probiotic Potential of Lactic Acid Bacteria Implicated in Natural Fermentation of Table Olives, Cv. Cobrançosa. Molecules 2023, 28, 3285. [Google Scholar] [CrossRef]
- De Graeve, M.; De Maeseneire, S.L.; Roelants, S.L.K.W.; Soetaert, W. Starmerella bombicola, an Industrially Relevant, yet Fundamentally Underexplored Yeast. FEMS Yeast Res. 2018, 18, foy072. [Google Scholar] [CrossRef]
- Kalia, V.C.; Patel, S.K.S.; Lee, J.K. Bacterial Biofilm Inhibitors: An Overview. Ecotoxicol. Environ. Saf. 2023, 264, 115389. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, L.; Johansen, P.G.; Petersen, M.A.; Arneborg, N.; Jespersen, L. Debaryomyces hansenii Strains Isolated from Danish Cheese Brines Act as Biocontrol Agents to Inhibit Germination and Growth of Contaminating Molds. Front. Microbiol. 2021, 12, 662785. [Google Scholar] [CrossRef]

| OD | Result |
|---|---|
| OD ≤ ODc | non-biofilm |
| ODc < OD < 2 × ODc | weak biofilm |
| 2 × ODc < OD < 4 × ODc | moderate biofilm |
| OD ≥ 4 × ODc | strong biofilm |
| Sample | Total Aerobic Counts (TAC), Log (CFU/g) | |
|---|---|---|
| PCA | YPD | |
| Unsealed larvae | 4.13 ± 0.54 | 1.52 ± 0.79 |
| Sealed larvae | 2.11 ± 0.37 | 2.83 ± 0.95 |
| Pupae | 3.81 ± 0.84 | 2.26 ± 0.26 |
| Adults | 5.10 ± 0.46 | 4.60 ± 0.35 |
| Yeast Species | Strain | Accession No. | Reference Accession No. | Identity (%) |
|---|---|---|---|---|
| Metschnikowia pulcherrima | SLD25 | PX353702 | OQ305009.1 | 100 |
| SLD20 | PX353703 | OQ304824.1 | 100 | |
| PD2 | PX353704 | OR475100.1 | 99.18 | |
| PD5 | PX353705 | OR475100.1 | 100 | |
| AD1 | PX353706 | OR475100.1 | 99.79 | |
| AD6 | PX353707 | MT821086.1 | 100 | |
| ULD4 | PX353708 | ON428299.1 | 99.79 | |
| ULD5 | PX353709 | ON428298.1 | 99.58 | |
| Metschnikowia fructicola | SLD11 | PX353710 | MZ185370.1 | 98.56 |
| SLD39 | PX353711 | MZ185363.1 | 99.59 | |
| Starmerella magnoliae | SLD46 | PX353712 | NG_060814.1 | 98.88 |
| SLD33 | PX353713 | MH910971.1 | 99.1 | |
| PD13 | PX353714 | NG_060814.1 | 99.55 | |
| PD14 | PX353715 | NG_060814.1 | 99.33 | |
| AD3 | PX353716 | NG_060814.1 | 98.88 | |
| AD8 | PX353717 | MH910969.1 | 99.54 | |
| Metschnikowia reukaufii | SLD4 | PX353718 | KM281731.1 | 100 |
| SLD2 | PX353719 | MT472086.1 | 100 | |
| Starmerella apis | SLD9 | PX353720 | PP235714.1 | 99.77 |
| Starmerella apicola | SLD10 | PX353721 | JN004197.1 | 99.79 |
| SLD14 | PX353722 | KT718105.1 | 99.17 | |
| PD1 | PX353723 | KT718105.1 | 99.38 | |
| Starmerella sorbosivorans | SLD7 | PX353724 | MW587726.1 | 99.88 |
| Starmerella bombi | SLD12 | PX353726 | KY106366.1 | 99.38 |
| PD7 | PX353727 | KY106366.1 | 99.38 | |
| PD6 | PX353728 | KY106366.1 | 100 | |
| Debaryomyces hansenii | SLD38 | PX353729 | KY511956.1 | 100 |
| SLD42 | PX353730 | KY511819.1 | 100 | |
| PD16 | PX353731 | HM988696.1 | 100 | |
| PD17 | PX353732 | KY512083.1 | 100 | |
| Zygosaccharomyces rouxii | SLD44 | PX353733 | KJ739844.1 | 100 |
| Sporobolomyces sp. | SLD48 | PX353734 | MG588966.1 | 98.58 |
| No. | Strain | OD Average Values ± ST | Biofilm-Forming Ability |
|---|---|---|---|
| 1 | YPD medium | 0.28 ± 0.07 | control, non-biofilm |
| 2 | S. cerevisiae flo11 | 0.68 ± 0.09 | weak biofilm |
| 3 | D. hansenii SLD38 | 0.56 ± 0.03 | weak biofilm |
| 4 | Z. rouxii SLD44 | 0.88 ± 0.13 | weak biofilm |
| 5 | M. fructicola SLD39 | 1.13 ± 0.22 | moderate biofilm |
| 6 | M. reukaufii SLD2 | 1.13 ± 0.24 | moderate biofilm |
| 7 | M. fructicola SLD11 | 1.21 ± 0.05 | moderate biofilm |
| 8 | M. pulcherrima SLD25 | 1.31 ± 0.70 | moderate biofilm |
| 9 | Sporobolomyces sp. SLD48 | 1.34 ± 0.12 | moderate biofilm |
| 10 | M. pulcherrima SLD20 | 1.61 ± 0.18 | moderate biofilm |
| 11 | M. reukaufii SLD4 | 1.70 ± 0.28 | moderate biofilm |
| 12 | D. hansenii SLD42 | 2.00 ± 0.30 | strong biofilm |
| 13 | S. sorbosivorans SLD7 | 2.41 ± 0.07 | strong biofilm |
| 14 | S. magnoliae SLD46 | 2.58 ± 0.59 | strong biofilm |
| 15 | S. apicola SLD10 | 2.68 ± 0.48 | strong biofilm |
| 16 | S. bombi SLD12 | 2.99 ± 0.29 | strong biofilm |
| 17 | S. apis SLD9 | 3.21 ± 0.38 | strong biofilm |
| 18 | S. magnoliae SLD33 | 3.56 ± 0.08 | strong biofilm |
| 19 | S. apicola SLD14 | 3.63 ± 0.01 | strong biofilm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lapinskaitė, V.; Bartkutė, P.; Lukša-Žebelovič, J.; Strazdaitė-Žielienė, Ž.; Servienė, E. Diversity and Functional Potential of Yeasts Inhabiting Honey Bee Drones. Microorganisms 2025, 13, 2614. https://doi.org/10.3390/microorganisms13112614
Lapinskaitė V, Bartkutė P, Lukša-Žebelovič J, Strazdaitė-Žielienė Ž, Servienė E. Diversity and Functional Potential of Yeasts Inhabiting Honey Bee Drones. Microorganisms. 2025; 13(11):2614. https://doi.org/10.3390/microorganisms13112614
Chicago/Turabian StyleLapinskaitė, Vilija, Paulina Bartkutė, Juliana Lukša-Žebelovič, Živilė Strazdaitė-Žielienė, and Elena Servienė. 2025. "Diversity and Functional Potential of Yeasts Inhabiting Honey Bee Drones" Microorganisms 13, no. 11: 2614. https://doi.org/10.3390/microorganisms13112614
APA StyleLapinskaitė, V., Bartkutė, P., Lukša-Žebelovič, J., Strazdaitė-Žielienė, Ž., & Servienė, E. (2025). Diversity and Functional Potential of Yeasts Inhabiting Honey Bee Drones. Microorganisms, 13(11), 2614. https://doi.org/10.3390/microorganisms13112614







