Crystal Digital PCR™ Enables Precise Quantification of Species Abundance in Microbial Mixtures
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media and Growth Conditions
2.2. DNA Extraction from Bacterial Culture
2.3. Identification of Species-Specific Genes and Primer Design
2.4. PCR Experiment
3. Results
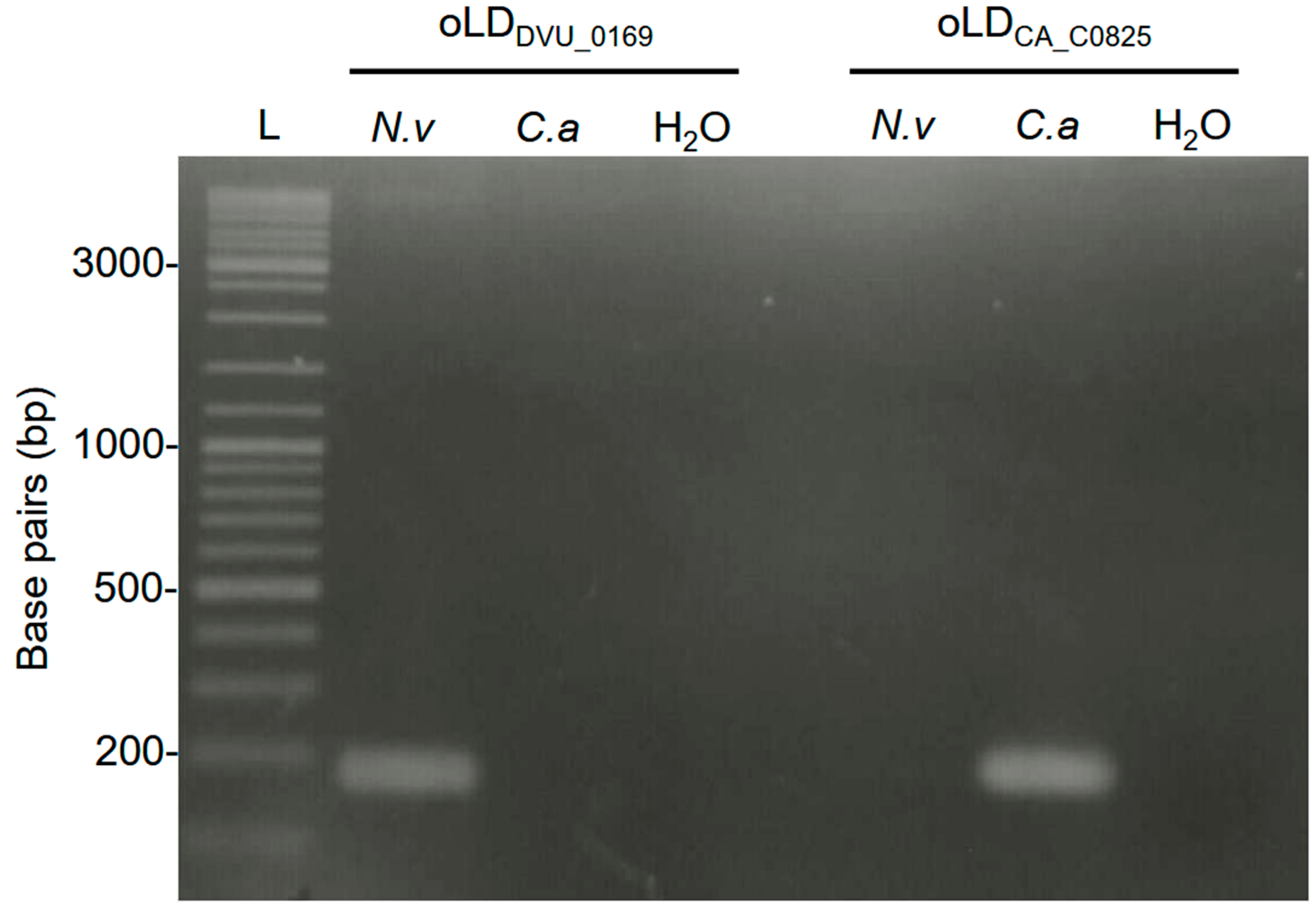
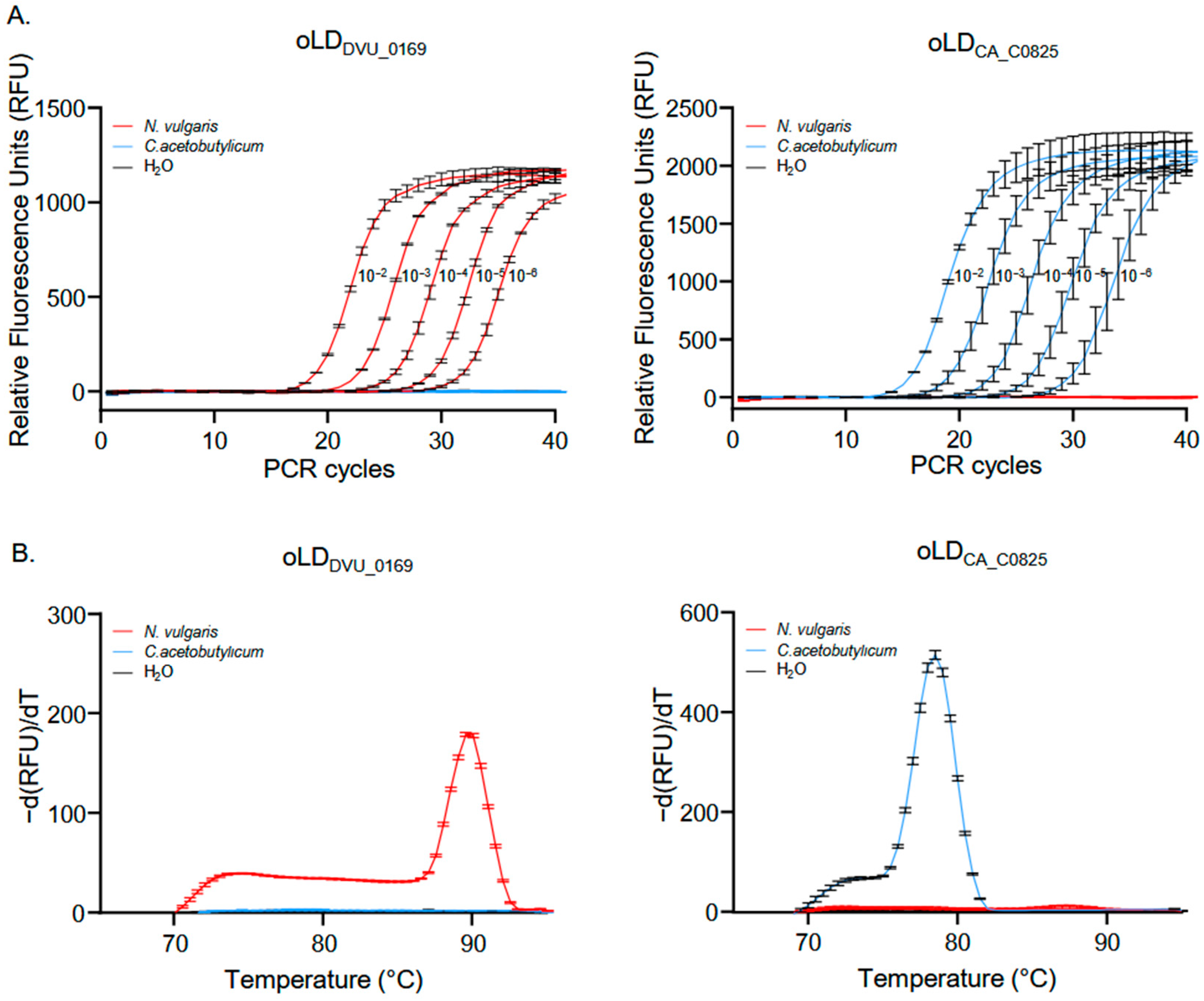
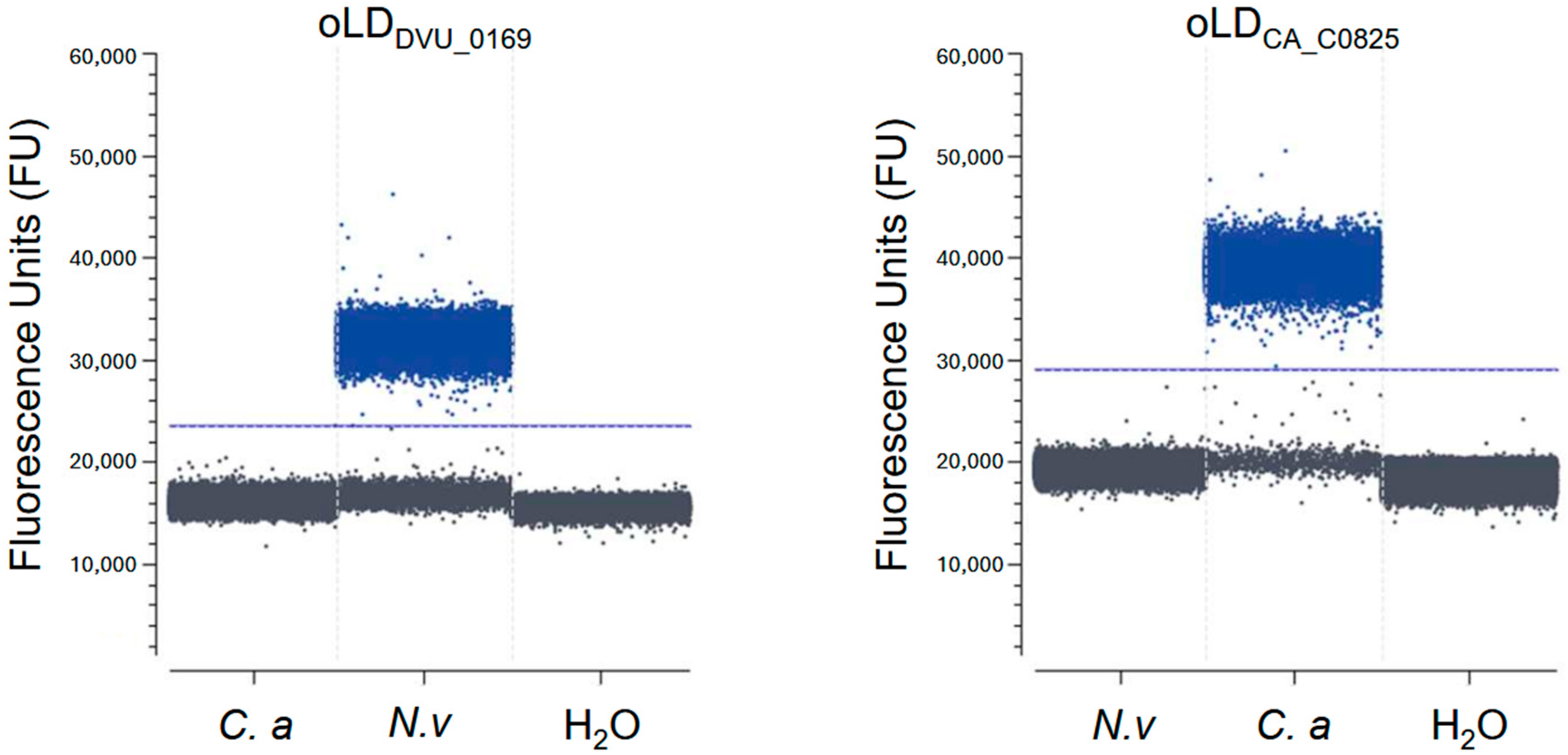
3.1. Primer Selection and Specificity Testing for Crystal Digital PCRTM
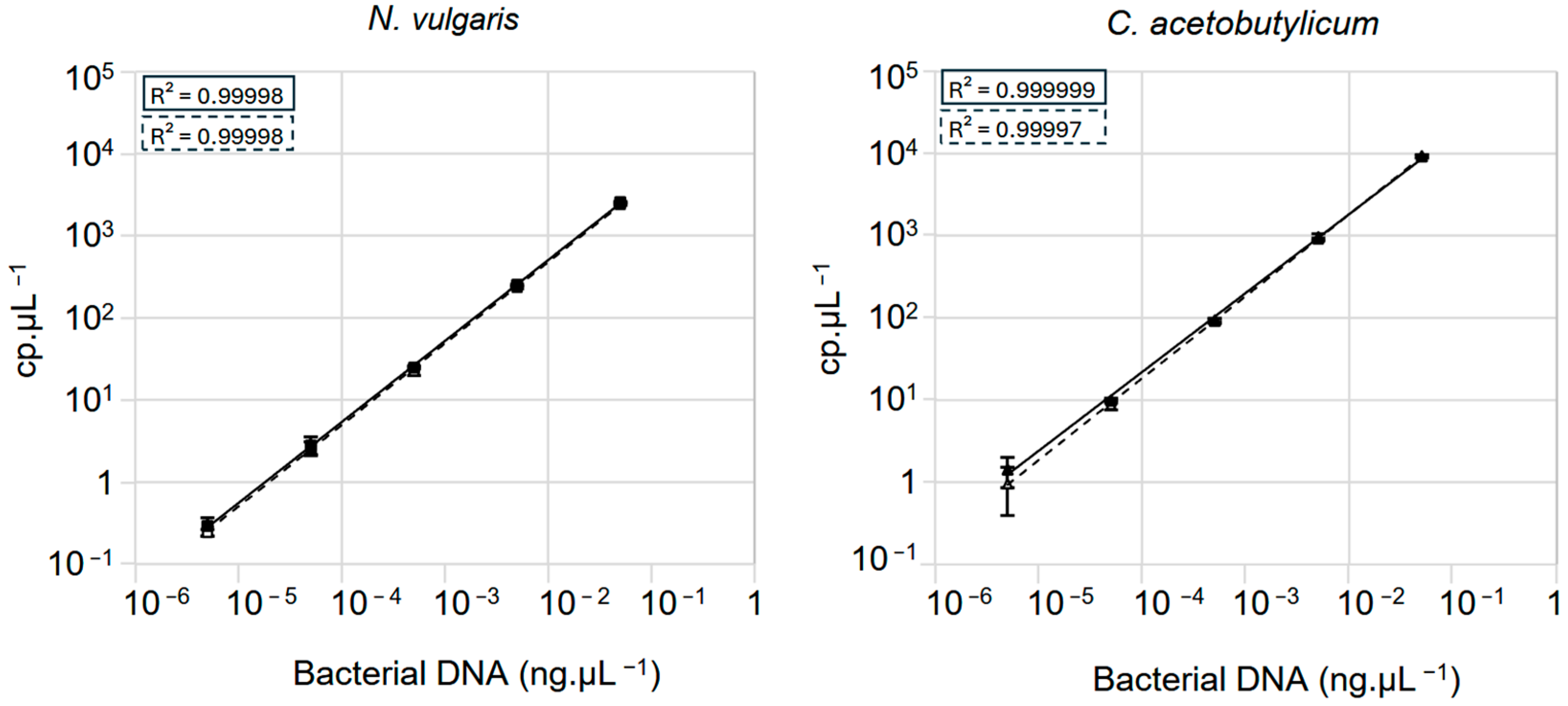
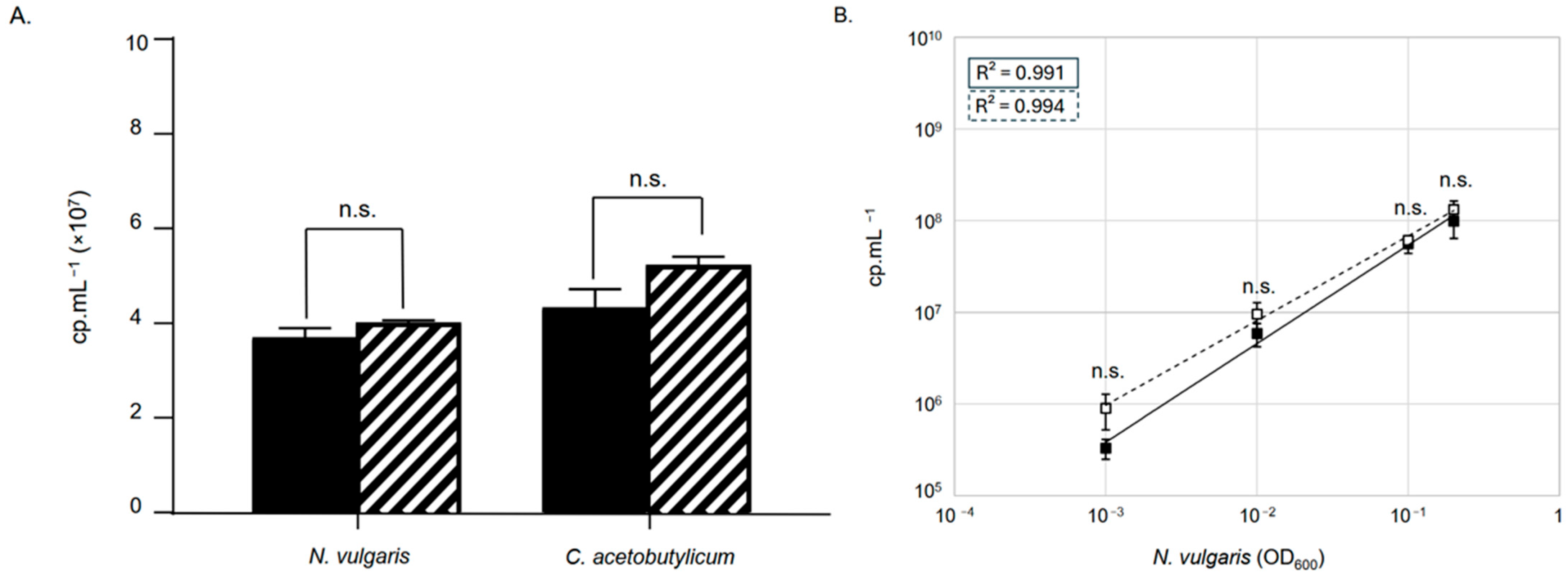
3.2. Assessment of Crystal Digital PCRTM Sensitivity for Quantifying Mixed Genomes in Samples
3.3. Bacterial Quantification in a Controlled Mixed Community
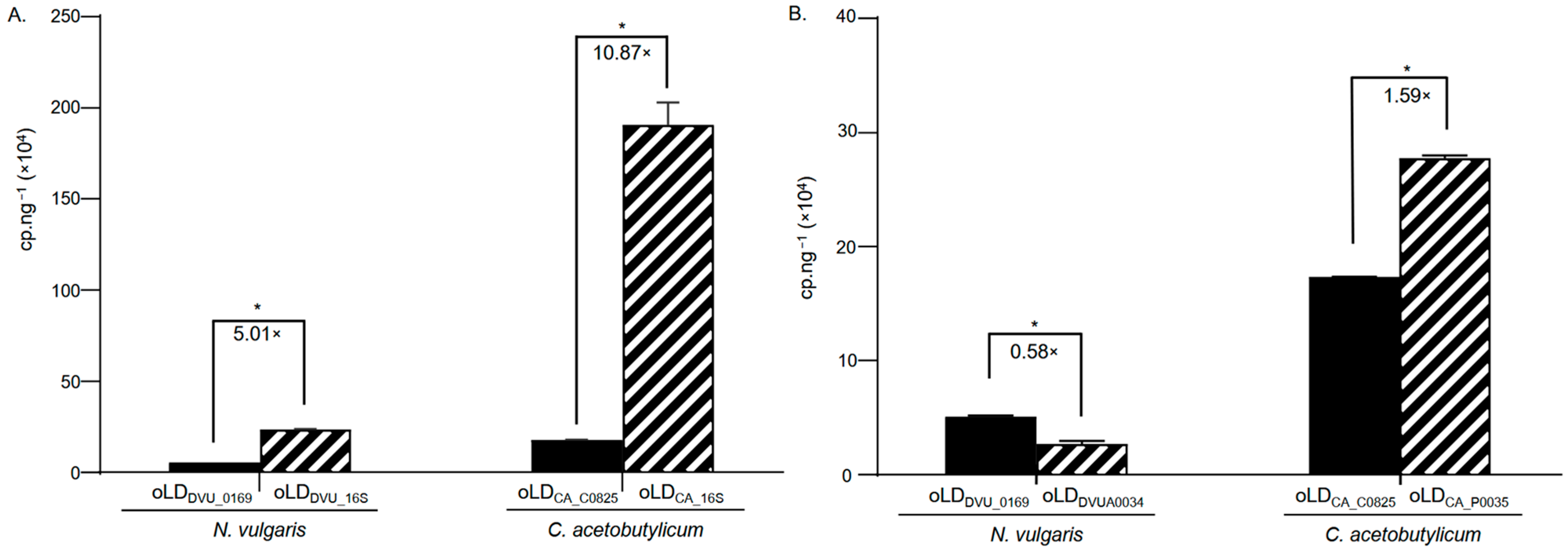
3.4. Plasmid Quantification in N. vulgaris and C. acetobutylicum Cultures
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bramucci, M.; Nagarajan, V. Bacterial Communities in Industrial Wastewater Bioreactors. Curr. Opin. Microbiol. 2006, 9, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.-L.; Zhou, J.-J.; Quan, C.-S.; Xiu, Z.-L. Advances in Industrial Microbiome Based on Microbial Consortium for Biorefinery. Bioresour. Bioprocess. 2017, 4, 11. [Google Scholar] [CrossRef]
- Wang, D.; Hunt, K.A.; Candry, P.; Tao, X.; Wofford, N.Q.; Zhou, J.; McInerney, M.J.; Stahl, D.A.; Tanner, R.S.; Zhou, A.; et al. Cross-Feedings, Competition, and Positive and Negative Synergies in a Four-Species Synthetic Community for Anaerobic Degradation of Cellulose to Methane. mBio 2023, 14, e03189-22. [Google Scholar] [CrossRef]
- Contreras-Salgado, E.A.; Sánchez-Morán, A.G.; Rodríguez-Preciado, S.Y.; Sifuentes-Franco, S.; Rodríguez-Rodríguez, R.; Macías-Barragán, J.; Díaz-Zaragoza, M. Multifaceted Applications of Synthetic Microbial Communities: Advances in Biomedicine, Bioremediation, and Industry. Microbiol. Res. 2024, 15, 1709–1727. [Google Scholar] [CrossRef]
- Mittermeier, F.; Bäumler, M.; Arulrajah, P.; García Lima, J.d.J.; Hauke, S.; Stock, A.; Weuster-Botz, D. Artificial Microbial Consortia for Bioproduction Processes. Eng. Life Sci. 2023, 23, e2100152. [Google Scholar] [CrossRef]
- Sgobba, E.; Wendisch, V.F. Synthetic Microbial Consortia for Small Molecule Production. Curr. Opin. Biotechnol. 2020, 62, 72–79. [Google Scholar] [CrossRef]
- Benomar, S.; Ranava, D.; Cárdenas, M.L.; Trably, E.; Rafrafi, Y.; Ducret, A.; Hamelin, J.; Lojou, E.; Steyer, J.-P.; Giudici-Orticoni, M.-T. Nutritional Stress Induces Exchange of Cell Material and Energetic Coupling between Bacterial Species. Nat. Commun. 2015, 6, 6283. [Google Scholar] [CrossRef] [PubMed]
- Marbehan, X.; Roger, M.; Fournier, F.; Infossi, P.; Guedon, E.; Delecourt, L.; Lebrun, R.; Giudici-Orticoni, M.T.; Delaunay, S. Combining Metabolic Flux Analysis with Proteomics to Shed Light on the Metabolic Flexibility: The Case of Desulfovibrio vulgaris Hildenborough. Front. Microbiol. 2024, 15, 1336360. [Google Scholar] [CrossRef] [PubMed]
- Ranava, D.; Backes, C.; Karthikeyan, G.; Ouari, O.; Soric, A.; Guiral, M.; Cárdenas, M.L.; Giudici-Orticoni, M.T. Metabolic Exchange and Energetic Coupling between Nutritionally Stressed Bacterial Species: Role of Quorum-Sensing Molecules. mBio 2021, 12, e02758-20. [Google Scholar] [CrossRef]
- Daly, A.J.; Baetens, J.M.; De Baets, B. The Impact of Initial Evenness on Biodiversity Maintenance for a Four-Species in Silico Bacterial Community. J. Theor. Biol. 2015, 387, 189–205. [Google Scholar] [CrossRef]
- Ehsani, E.; Hernandez-Sanabria, E.; Kerckhof, F.-M.; Props, R.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Boon, N. Initial Evenness Determines Diversity and Cell Density Dynamics in Synthetic Microbial Ecosystems. Sci. Rep. 2018, 8, 340. [Google Scholar] [CrossRef]
- Ferrari, A.; Lombardi, S.; Signoroni, A. Bacterial Colony Counting by Convolutional Neural Networks. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 7458–7461. [Google Scholar] [CrossRef]
- Kerstens, M.; Boulet, G.; Tritsmans, C.; Horemans, T.; Hellings, M.; Delputte, P.; Maes, L.; Cos, P. Flow Cytometric Enumeration of Bacteria Using TO-PRO®-3 Iodide as a Single-Stain Viability Dye. J. Lab. Autom. 2014, 19, 555–561. [Google Scholar] [CrossRef]
- Inoue, Y.; Nakaho, K. Sensitive Quantitative Detection of Ralstonia Solanacearum in Soil by the Most Probable Number-Polymerase Chain Reaction (MPN-PCR) Method. Appl. Microbiol. Biotechnol. 2014, 98, 4169–4177. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.S.; Jo, H.Y.; Kwon, M.J. Revisiting Soil Bacterial Counting Methods: Optimal Soil Storage and Pretreatment Methods and Comparison of Culture-Dependent and -Independent Methods. PLoS ONE 2021, 16, e0246142. [Google Scholar] [CrossRef]
- Madigan, M.T.; Bender, K.S.; Buckley, D.H.; Sattley, W.M.; Stahl, D.A. Brock Biology of Microorganisms, 16th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2020. [Google Scholar]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to Culturing the Uncultured Microbial Majority. Nat. Rev. Microbiol. 2021, 19, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for Culture of “unculturable” Bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef]
- Smith, C.J.; Osborn, A.M. Advantages and Limitations of Quantitative PCR (Q-PCR)-Based Approaches in Microbial Ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Analysis and Normalization of Real-Time Polymerase Chain Reaction (PCR) Experimental Data. Cold Spring Harb. Protoc. 2018, 2018, pdb-top095000. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; Moorman, A.F.M. Amplification Efficiency: Linking Baseline and Bias in the Analysis of Quantitative PCR Data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef]
- Nocker, A.; Camper, A.K. Selective Removal of DNA from Dead Cells of Mixed Bacterial Communities by Use of Ethidium Monoazide. Appl. Environ. Microbiol. 2006, 72, 1997–2004. [Google Scholar] [CrossRef]
- Müller, S.; Nebe-von-Caron, G. Functional Single-Cell Analyses: Flow Cytometry and Cell Sorting of Microbial Populations and Communities. FEMS Microbiol. Rev. 2010, 34, 554–587. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hammes, F.; Boon, N.; Chami, M.; Egli, T. Isolation and Characterization of Low Nucleic Acid (LNA)-Content Bacteria. ISME J. 2009, 3, 889–902. [Google Scholar] [CrossRef]
- Veal, D.A.; Deere, D.; Ferrari, B.; Piper, J.; Attfield, P.V. Fluorescence Staining and Flow Cytometry for Monitoring Microbial Cells. J. Immunol. Methods 2000, 243, 191–210. [Google Scholar] [CrossRef]
- Power, A.L.; Barber, D.G.; Groenhof, S.R.M.; Wagley, S.; Liu, P.; Parker, D.A.; Love, J. The Application of Imaging Flow Cytometry for Characterisation and Quantification of Bacterial Phenotypes. Front. Cell. Infect. Microbiol. 2021, 11, 716592. [Google Scholar] [CrossRef]
- Hammes, F.; Berney, M.; Wang, Y.; Vital, M.; Köster, O.; Egli, T. Flow-Cytometric Total Bacterial Cell Counts as a Descriptive Microbiological Parameter for Drinking Water Treatment Processes. Water Res. 2008, 42, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Props, R.; Monsieurs, P.; Mysara, M.; Clement, L.; Boon, N. Measuring the Biodiversity of Microbial Communities by Flow Cytometry. Methods Ecol. Evol. 2016, 7, 1376–1385. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun Metagenomics, from Sampling to Analysis. Nat. Biotechnol. 2017, 35, 833–844, Erratum in Nat. Biotechnol. 2017, 35, 1211. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s Microbes: Tools for Distinguishing the Living from the Dead in Microbial Ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Fang, W.; Liu, X.; Maiga, M.; Cao, W.; Mu, Y.; Yan, Q.; Zhu, Q. Digital PCR for Single-Cell Analysis. Biosensors 2024, 14, 64. [Google Scholar] [CrossRef]
- Zhang, L.; Parvin, R.; Fan, Q.; Ye, F. Emerging Digital PCR Technology in Precision Medicine. Biosens. Bioelectron. 2022, 211, 114344. [Google Scholar] [CrossRef]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Starkey, R.L. Characteristics and Cultivation of Sulfate-Reducing Bacteria. J. Am. Water Works Assoc. 1948, 40, 1291–1298. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Poggio, P.; Songia, P.; Vavassori, C.; Ricci, V.; Banfi, C.; Barbieri, S.S.; Garoffolo, G.; Myasoedova, V.A.; Piacentini, L.; Raucci, A.; et al. Digital PCR for High Sensitivity Viral Detection in False-Negative SARS-CoV-2 Patients. Sci. Rep. 2021, 11, 4310. [Google Scholar] [CrossRef] [PubMed]
- Heidelberg, J.; Seshadri, R.; Haveman, S.; Hemme, C.; Paulsen, I.; Kolonay, J.; Eisen, J.; Ward, N.; Methe, B.; Brinkac, L.; et al. The Genome Sequence of the Anaerobic, Sulfate-Reducing Bacterium Desulfovibrio Vulgaris Hildenborough. Nat. Biotechnol. 2004, 22, 554–559. [Google Scholar] [CrossRef]
- Mo, X.; Pei, J.; Guo, Y.; Lin, L.; Peng, L.; Kou, C.; Fan, D.; Pang, H. Genome Sequence of Clostridium Acetobutylicum GXAS18-1, a Novel Biobutanol Production Strain. Genome Announc. 2015, 3, e00033-15. [Google Scholar] [CrossRef]
- Cavaliere, M.; Feng, S.; Soyer, O.S.; Jiménez, J.I. Cooperation in Microbial Communities and Their Biotechnological Applications. Environ. Microbiol. 2017, 19, 2949–2963. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR Inhibition in qPCR, dPCR and MPS—Mechanisms and Solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Dingle, T.C.; Sedlak, R.H.; Cook, L.; Jerome, K.R. Tolerance of Droplet-Digital PCR vs Real-Time Quantitative PCR to Inhibitory Substances. Clin. Chem. 2013, 59, 1670–1672. [Google Scholar] [CrossRef]
- Liu, H.; Shi, K.; Feng, S.; Yin, Y.; Long, F.; Si, H. Development of a Crystal Digital RT-PCR for the Detection of Atypical Porcine Pestivirus. Vet. Sci. 2023, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- del Solar, G.; Giraldo, R.; Ruiz-Echevarría, M.J.; Espinosa, M.; Díaz-Orejas, R. Replication and Control of Circular Bacterial Plasmids. Microbiol. Mol. Biol. Rev. MMBR 1998, 62, 434–464. [Google Scholar] [CrossRef] [PubMed]
- Nordström, K.; Dasgupta, S. Copy-Number Control of the Escherichia Coli Chromosome: A Plasmidologist’s view. EMBO Rep. 2006, 7, 484–489. [Google Scholar] [CrossRef]
- Postgate, J.R. The Sulfate-Reducing Bacteria, 2nd ed.; Cambridge University Press: Cambridge, UK; Scientific Research Publishing: Wuhan, China, 1984; Available online: https://www.scirp.org/reference/referencespapers?referenceid=1617902 (accessed on 31 October 2025).
| N. vulgaris | Primer Name | Sequence |
|---|---|---|
| Genes | ||
| DVU_16S | oLDDVU_16S_Fwd | TCTGGCTCAGATTGAACGCT |
| oLDDVU_16S_Rev | GCAAGCAGAGGCCACCTTTC | |
| DVU_0169 | oLDDVU_0169_Fwd | AAGAAGTTCCCCCAGTTCGC |
| oLDDVU_0169_Rev | TTGTCGAGATTGTAGCGGGG | |
| DVUA0034 | oLDDVUA0034_Fwd | GGGTTGGTCGAGAAGTGGTT |
| oLDDVUA0034_Rev | AGTTGCAGGAGAAGTACGGC | |
| C. acetobutylicum | Primer Name | Sequence |
| Genes | ||
| CA_16S | oLDCA_16S_Fwd | CAGGATGACAGGTGGTGCAT |
| oLDCA_16S_Rev | AGCCCTAGACATAAGGGGCA | |
| CA_C0825 | oLDCA_C0825_Fwd | AGAGACACCGGTGCAAAGAA |
| oLDCA_C0825_Rev | CTTTGCGCTTCCCCAAGATG | |
| CA_P0035 | oLDCA_P0035_Fwd | AGTGCCGCATCCAAGAGTAA |
| oLDCA_P0035_Rev | ATGCCTTCTTCACAGGGAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delecourt, L.; Reset, M.; Bertaux, L.; Sturgis, J.; Denis, Y.; Giudici-Orticoni, M.-T.; Roger, M.; Bordi, C. Crystal Digital PCR™ Enables Precise Quantification of Species Abundance in Microbial Mixtures. Microorganisms 2025, 13, 2592. https://doi.org/10.3390/microorganisms13112592
Delecourt L, Reset M, Bertaux L, Sturgis J, Denis Y, Giudici-Orticoni M-T, Roger M, Bordi C. Crystal Digital PCR™ Enables Precise Quantification of Species Abundance in Microbial Mixtures. Microorganisms. 2025; 13(11):2592. https://doi.org/10.3390/microorganisms13112592
Chicago/Turabian StyleDelecourt, Louis, Mila Reset, Lionel Bertaux, James Sturgis, Yann Denis, Marie-Thérèse Giudici-Orticoni, Magali Roger, and Christophe Bordi. 2025. "Crystal Digital PCR™ Enables Precise Quantification of Species Abundance in Microbial Mixtures" Microorganisms 13, no. 11: 2592. https://doi.org/10.3390/microorganisms13112592
APA StyleDelecourt, L., Reset, M., Bertaux, L., Sturgis, J., Denis, Y., Giudici-Orticoni, M.-T., Roger, M., & Bordi, C. (2025). Crystal Digital PCR™ Enables Precise Quantification of Species Abundance in Microbial Mixtures. Microorganisms, 13(11), 2592. https://doi.org/10.3390/microorganisms13112592






