Establishment of Specific Multiplex PCR Detection Methods for the Predominant tet(X)-Positive Acinetobacter Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Collection and Quality Evaluation
2.2. Identification of Acinetobacter Species, Antibiotic Resistance Genes, and Sequence Types (STs)
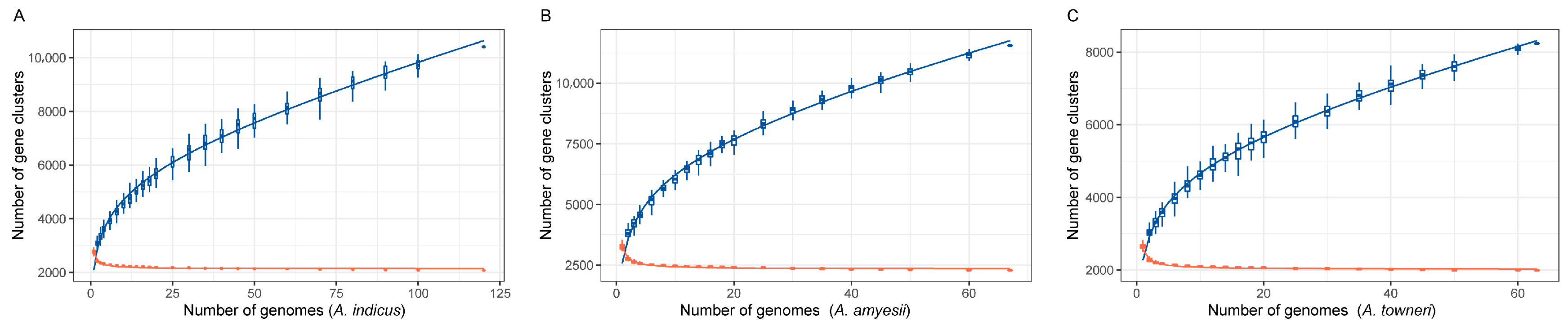
2.3. Primer Design Based on Pan-Genome Analyses
2.4. Primer Confirmation
2.5. Determination of the Optimal Annealing Temperature
2.6. Determination of the Optimal Primer Ratio
2.7. Determination of the Minimum Detection Limit
2.8. Primer Stability Under Different Storage Temperatures
2.9. Determination of the Multiplex PCR Detection Accuracy
3. Results
3.1. Distribution of tet(X) and Associated Genes in Acinetobacter Species
3.2. Optimized Primers of Three Predominan tet(X)-Positive Acinetobacter Species
3.3. Evaluation of Specific Detection Primers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CRAb | Carbapenem-resistant Acinetobacter baumannii |
| MDR | Multidrug-resistant |
| qPCR | Quantitative real-time PCR |
| ANI | Average Nucleotide Identity |
| MALDI-TOF MS | Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| NCBI | National Center for Biotechnology Information |
| STs | Sequence Types |
| CARD | Comprehensive Antibiotic Resistance Database |
| RPA | Recombinase polymerase amplification |
| LC-MS/MS | Liquid chromatography-tandem mass spectrometry |
References
- Chen, C.; Wu, T.; Liu, J.; Gao, J. Threat and Control of tet(X)-Mediated Tigecycline-Resistant Acinetobacter sp. Bacteria. Foods 2025, 14, 3374. [Google Scholar] [CrossRef]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2017, 16, 91–102. [Google Scholar] [CrossRef]
- Qin, J.; Feng, Y.; Lu, X.; Zong, Z. Precise Species Identification for Acinetobacter: A Genome-Based Study with Description of Two Novel Acinetobacter Species. mSystems 2021, 6, e0023721. [Google Scholar] [CrossRef]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, Y.; Wang, Y.; Tang, K.; Wang, D.; Hong, J.; Wang, P.; Ye, S.; Yan, J.; Li, S.; et al. Genetic landscape and evolution of Acinetobacter pittii, an underestimated emerging nosocomial pathogen. Commun. Biol. 2025, 8, 738. [Google Scholar] [CrossRef]
- Li, S.; Jiang, G.; Wang, S.; Wang, M.; Wu, Y.; Zhang, J.; Liu, X.; Zhong, L.; Zhou, M.; Xie, S.; et al. Emergence and global spread of a dominant multidrug-resistant clade within Acinetobacter baumannii. Nat. Commun. 2025, 16, 2787. [Google Scholar] [CrossRef]
- Aguilar-Vera, A.; Bello-López, E.; Pantoja-Nuñez, G.I.; Rodríguez-López, G.M.; Morales-Erasto, V.; Castillo-Ramírez, S.; Tringe, S.G. Acinetobacter junii: An emerging One Health pathogen. mSphere 2024, 9, e0016224. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-T.; Sun, J.-R.; Li, L.-H.; Yang, Y.-S.; Chang, H.-M.; Lin, P.-Y.; Liao, P.-H.; Kang, F.-Y.; Chang, Y.-Y.; Yang, Y.-S.; et al. Comparison of clinical manifestations, antimicrobial susceptibility patterns, and carbapenem resistance determinants between Acinetobacter seifertii and Acinetobacter nosocomialis isolated in Taiwan. J. Microbiol. Immunol. Infect. 2025, Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655, Correction in Lancet 2022, 400, 1102. [Google Scholar] [CrossRef] [PubMed]
- Talaat, M.; Zayed, B.; Tolba, S.; Abdou, E.; Gomaa, M.; Itani, D.; Hutin, Y.; Hajjeh, R. Increasing Antimicrobial Resistance in World Health Organization Eastern Mediterranean Region, 2017–2019. Emerg. Infect. Dis. 2022, 28, 717–724. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 6 November 2025).
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.J.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.B.; Ji, Q.J.; Wei, R.C.; Liu, Z.H.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cui, C.Y.; Yu, J.J.; He, Q.; Wu, X.T.; He, Y.Z.; Cui, Z.H.; Li, C.; Jia, Q.L.; Shen, X.G.; et al. Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome Med. 2020, 12, 111. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.Y.; Chen, C.; Liu, B.T.; He, Q.; Wu, X.T.; Sun, R.Y.; Zhang, Y.; Cui, Z.H.; Guo, W.Y.; Jia, Q.L.; et al. Co-occurrence of Plasmid-Mediated Tigecycline and Carbapenem Resistance in Acinetobacter spp. from Waterfowls and Their Neighboring Environment. Antimicrob. Agents Chemother. 2020, 64, e02502-19. [Google Scholar] [CrossRef]
- Zheng, X.R.; Zhu, J.H.; Zhang, J.; Cai, P.; Sun, Y.H.; Chang, M.X.; Fang, L.X.; Sun, J.; Jiang, H.X. A novel plasmid-borne tet(X6) variant co-existing with blaNDM-1 and blaOXA-58 in a chicken Acinetobacter baumannii isolate. J. Antimicrob. Chemoth 2020, 75, 3397–3399. [Google Scholar] [CrossRef]
- Mallonga, Z.; Tokuda, M.; Yamazaki, R.; Tsuruga, S.; Nogami, I.; Sato, Y.; Tarrayo, A.G.; Fuentes, R.; Parilla, R.; Kimbara, K.; et al. Emergence of Acinetobacter towneri harbouring a novel plasmid with blaNDM-1 and tet(X7) from hospital wastewater in the Philippines. J. Glob. Antimicrob. Resist. 2025, 41, 287–289. [Google Scholar] [CrossRef]
- Lee, M.J.; Jang, S.J.; Li, X.M.; Park, G.; Kook, J.K.; Kim, M.J.; Chang, Y.H.; Shin, J.H.; Kim, S.H.; Kim, D.M.; et al. Comparison of rpoB gene sequencing, 16S rRNA gene sequencing, gyrB multiplex PCR, and the VITEK2 system for identification of Acinetobacter clinical isolates. Diagn. Microbiol. Infect. Dis. 2014, 78, 29–34. [Google Scholar] [CrossRef]
- Sun, J.; Chen, C.; Cui, C.Y.; Zhang, Y.; Liu, X.; Cui, Z.H.; Ma, X.Y.; Feng, Y.; Fang, L.X.; Lian, X.L.; et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef]
- Wei, S.; Zhao, H.; Xian, Y.; Hussain, M.A.; Wu, X. Multiplex PCR assays for the detection of Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae with an internal amplification control. Diagn. Microbiol. Infect. Dis. 2014, 79, 115–118. [Google Scholar] [CrossRef]
- Hammad, M.I.; Conrads, G.; Abdelbary, M.M.H. Isolation, identification, and significance of salivary Veillonella spp., Prevotella spp., and Prevotella salivae in patients with inflammatory bowel disease. Front. Cell Infect. Microbiol. 2023, 13, 1278582. [Google Scholar] [CrossRef]
- Gao, D.; Yu, J.; Dai, X.; Tian, Y.; Sun, J.; Xu, X.; Cai, X. Development and evaluation of an indirect enzyme-linked immunosorbent assay based on a recombinant SifA protein to detect Salmonella infection in poultry. Poult. Sci. 2023, 102, 102513. [Google Scholar] [CrossRef]
- Poveda, A.; Krell, J.; Heinz, V.; Terjung, N.; Tomasevic, I.; Gibis, M. The Use of MALDI--TOF MS for Microbial Identification of Discolored Vacuum--Packaged Beef. J. Food Sci. 2025, 90, e70591. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Chatzimichail, S.; Turner, P.; Feehily, C.; Farrar, A.; Crook, D.; Andersson, M.; Oakley, S.; Barrett, L.; El Sayyed, H.; Kyropoulos, J.; et al. Rapid identification of bacterial isolates using microfluidic adaptive channels and multiplexed fluorescence microscopy. Lab Chip 2024, 24, 4843–4858. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chen, C.; Wu, T.; Liu, J.; Lv, Y. Genomic Diversity of the tet(X)-Positive Myroides Species. Microorganisms 2025, 13, 1180. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, Y.; Fan, G.; Sun, D.; Zhang, X.; Yu, Z.; Wang, J.; Wu, L.; Shi, W.; Ma, J. IPGA: A handy integrated prokaryotes genome and pan--genome analysis web service. iMeta 2022, 1, e55. [Google Scholar] [CrossRef]
- Xu, Q.; Mu, X.; He, J.; Liu, H.; Liu, X.; Wang, Y.; Hua, X.; Yu, Y.; Manning, S.D. Phenotypic and genotypic characterization of clinical carbapenem-resistant Acinetobacter species harboring the metallo-beta-lactamases IMP-8 or NDM-1 in China. Microbiol. Spectr. 2025, 13, e0115824. [Google Scholar] [CrossRef]
- Malhotra, J.; Anand, S.; Jindal, S.; Rajagopal, R.; Lal, R. Acinetobacter indicus sp. nov., isolated from a hexachlorocyclohexane dump site. Int. J. Syst. Evol. Microbiol. 2012, 62, 2883–2890. [Google Scholar] [CrossRef]
- Bello-Lopez, E.; Rocha-Gracia, R.D.C.; Castro-Jaimes, S.; Cevallos, M.A.; Vargas-Cruz, M.; Verdugo-Yocupicio, R.; Saenz, Y.; Torres, C.; Gutierrez-Cazarez, Z.; Arenas-Hernandez, M.M.P.; et al. Antibiotic resistance mechanisms in Acinetobacter spp. strains isolated from patients in a paediatric hospital in Mexico. J. Glob. Antimicrob. Resist. 2020, 23, 120–129. [Google Scholar] [CrossRef]
- Rui, Y.; Qiu, G. Analysis of Antibiotic Resistance Genes in Water Reservoirs and Related Wastewater from Animal Farms in Central China. Microorganisms 2024, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Carr, E.L.; Kämpfer, P.; Patel, B.K.C.; Gürtler, V.; Seviour, R.J. Seven novel species of Acinetobacter isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2003, 53, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Huang, Y.; Liu, W.; Yang, Z.; Dong, D.; Huang, S.; He, X.; Ao, D.; Liu, N.; Wang, S.; et al. Complete sequences of two novel blaNDM-1-harbouring plasmids from two Acinetobacter towneri isolates in China associated with the acquisition of Tn125. Sci. Rep. 2017, 7, 9405. [Google Scholar] [CrossRef] [PubMed]
- Tobin, L.A.; Jarocki, V.M.; Kenyon, J.; Drigo, B.; Donner, E.; Djordjevic, S.P.; Hamidian, M.; Elkins, C.A. Genomic analysis of diverse environmental Acinetobacter isolates identifies plasmids, antibiotic resistance genes, and capsular polysaccharides shared with clinical strains. Appl. Environ. Microbiol. 2024, 90, e0165423. [Google Scholar] [CrossRef]
- Ma, J.; Wang, J.; Feng, J.; Liu, Y.; Yang, B.; Li, R.; Bai, L.; He, T.; Wang, X.; Yang, Z. Characterization of Three Porcine Acinetobacter towneri Strains Co-Harboring tet(X3) and blaOXA-58. Front. Cell Infect. Microbiol. 2020, 10, 586507. [Google Scholar] [CrossRef]
- Maehana, S.; Kitasato, H.; Suzuki, M.; Stewart, F.J. Genome Sequence of Acinetobacter towneri Strain DSM 16313, Previously Known as the Proposed Type Strain of Acinetobacter seohaensis. Microbiol. Resour. Announc. 2021, 10, e0069021. [Google Scholar] [CrossRef]
- Nemec, A.; Radolfová-Křížová, L.; Maixnerová, M.; Nemec, M.; Shestivska, V.; Španělová, P.; Kyselková, M.; Wilharm, G.; Higgins, P.G. Acinetobacter amyesii sp. nov., widespread in the soil and water environment and animals. Int. J. Syst. Evol. Microbiol. 2022, 72, 005642. [Google Scholar] [CrossRef]
- Dede, A.; Pérez-Valera, E.; Elhottová, D. Genome analysis of manure and soil-dwelling Acinetobacter strains indicates potential health risks associated with antibiotic resistance and virulence factors. Microb. Pathog. 2025, 205, 107610. [Google Scholar] [CrossRef]
- Wang, L.; Liu, D.; Lv, Y.; Cui, L.; Li, Y.; Li, T.; Song, H.; Hao, Y.; Shen, J.; Wang, Y.; et al. Novel Plasmid-Mediated tet(X5) Gene Conferring Resistance to Tigecycline, Eravacycline, and Omadacycline in a Clinical Acinetobacter baumannii Isolate. Antimicrob. Agents Chemother. 2019, 64, e01326-19. [Google Scholar] [CrossRef]
- Tang, B.; Yang, H.; Jia, X.; Feng, Y. Coexistence and characterization of Tet(X5) and NDM-3 in the MDR-Acinetobacter indicus of duck origin. Microb. Pathog. 2021, 150, 104697. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.; Xu, Y.; Sun, J.; Huang, M.; Jia, X.; Jiang, C.; Feng, Y. Harnessing efficient multiplex PCR methods to detect the expanding Tet(X) family of tigecycline resistance genes. Virulence 2019, 11, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, D.; Song, H.; Liu, Z.; Jiang, H.; Wang, Y. Development of a Multiplex Real-Time PCR Assay for Rapid Detection of Tigecycline Resistance Gene tet(X) Variants from Bacterial, Fecal, and Environmental Samples. Antimicrob. Agents Chemother. 2020, 64, e02292-19. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shen, Z.; Ding, S.; Wang, S. A TaqMan-based multiplex real-time PCR assay for the rapid detection of tigecycline resistance genes from bacteria, faeces and environmental samples. BMC Microbiol. 2020, 20, 174. [Google Scholar] [CrossRef]
- Chen, G.; Chen, L.; Lin, S.; Yang, C.; Liang, H.; Huang, K.; Guo, Z.; Lv, F. Sensitive and rapid detection of tet(X2) ~ tet(X5) by loop-mediated isothermal amplification based on visual OTG dye. BMC Microbiol. 2023, 23, 329. [Google Scholar] [CrossRef]
- Cui, Z.H.; Ni, W.N.; Tang, T.; He, B.; Zhong, Z.X.; Fang, L.X.; Chen, L.; Chen, C.; Cui, C.Y.; Liu, Y.H.; et al. Rapid detection of plasmid-mediated high-level tigecycline resistance in Escherichia coli and Acinetobacter spp. J. Antimicrob. Chemother. 2020, 75, 1479–1483. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Pan, Q.; Wang, J.; Jiao, X.a.; Zhang, Y. Development and evaluation of rapid and accurate one-tube RPA-CRISPR-Cas12b-based detection of mcr-1 and tet(X4). Appl. Microbiol. Biotechnol. 2024, 108, 345. [Google Scholar] [CrossRef]
- Cui, Z.-H.; Zheng, Z.-J.; Tang, T.; Zhong, Z.-X.; Cui, C.-Y.; Lian, X.-L.; Fang, L.-X.; He, Q.; Wang, X.-R.; Chen, C.; et al. Rapid Detection of High-Level Tigecycline Resistance in Tet(X)-Producing Escherichia coli and Acinetobacter spp. Based on MALDI-TOF MS. Front. Cell Infect. Microbiol. 2020, 10, 583341. [Google Scholar] [CrossRef]
- Zheng, Z.J.; Cui, Z.H.; Diao, Q.Y.; Ye, X.Q.; Zhong, Z.X.; Tang, T.; Wu, S.B.; He, H.L.; Lian, X.L.; Fang, L.X.; et al. MALDI-TOF MS for rapid detection and differentiation between Tet(X)-producers and non-Tet(X)-producing tetracycline-resistant Gram-negative bacteria. Virulence 2022, 13, 77–88. [Google Scholar] [CrossRef]
- Zhang, L.; Xie, J.; Qu, Z.; Duan, D.; Liu, C.; Zhang, D.; Jiang, H.; Dai, X.; Jiang, Y.; Fang, X.; et al. A rapid liquid chromatography-tandem mass spectrometry based method for the detection of Tet(X) resistance gene in Enterobacteriaceae. Front. Microbiol. 2024, 15, 1477740. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.; Ruan, Z.; Feng, Y.; Fu, Y.; Jiang, Y.; Wang, H.; Yu, Y. Species Distribution of Clinical Acinetobacter Isolates Revealed by Different Identification Techniques. PLoS ONE 2014, 9, e104882. [Google Scholar] [CrossRef]
- Marinova-Bulgaranova, T.V.; Hitkova, H.Y.; Balgaranov, N.K. Current Methods for Reliable Identification of Species in the Acinetobacter calcoaceticus–Acinetobacter baumannii Complex. Microorganisms 2025, 13, 1819. [Google Scholar] [CrossRef]
- Rassoulian Barrett, S.; Hoffman, N.G.; Rosenthal, C.; Bryan, A.; Marshall, D.A.; Lieberman, J.; Greninger, A.L.; Peddu, V.; Cookson, B.T.; Salipante, S.J.; et al. Sensitive Identification of Bacterial DNA in Clinical Specimens by Broad-Range 16S rRNA Gene Enrichment. J. Clin. Microbiol. 2020, 58, e01605-20. [Google Scholar] [CrossRef]
- Liu, Y.; Raymond, J.J.; Wu, X.; Chua, P.W.L.; Ling, S.Y.H.; Chan, C.C.; Chan, C.; Loh, J.X.Y.; Song, M.X.Y.; Ong, M.Y.Y.; et al. Electrostatic microfiltration (EM) enriches and recovers viable microorganisms at low-abundance in large-volume samples and enhances downstream detection. Lab Chip 2024, 24, 4275–4287. [Google Scholar] [CrossRef]
- Jansriphibul, K.; Krohn, C.; Ball, A.S. Sources of variability for viability PCR using propidium monoazide. Microbiol. Res. 2025, 298, 128224. [Google Scholar] [CrossRef]
- Chen, C.; Cui, C.Y.; Wu, X.T.; Fang, L.X.; He, Q.; He, B.; Long, T.F.; Liao, X.P.; Chen, L.; Liu, Y.H.; et al. Spread of tet(X5) and tet(X6) genes in multidrug-resistant Acinetobacter baumannii strains of animal origin. Vet. Microbiol. 2021, 253, 108954. [Google Scholar] [CrossRef]
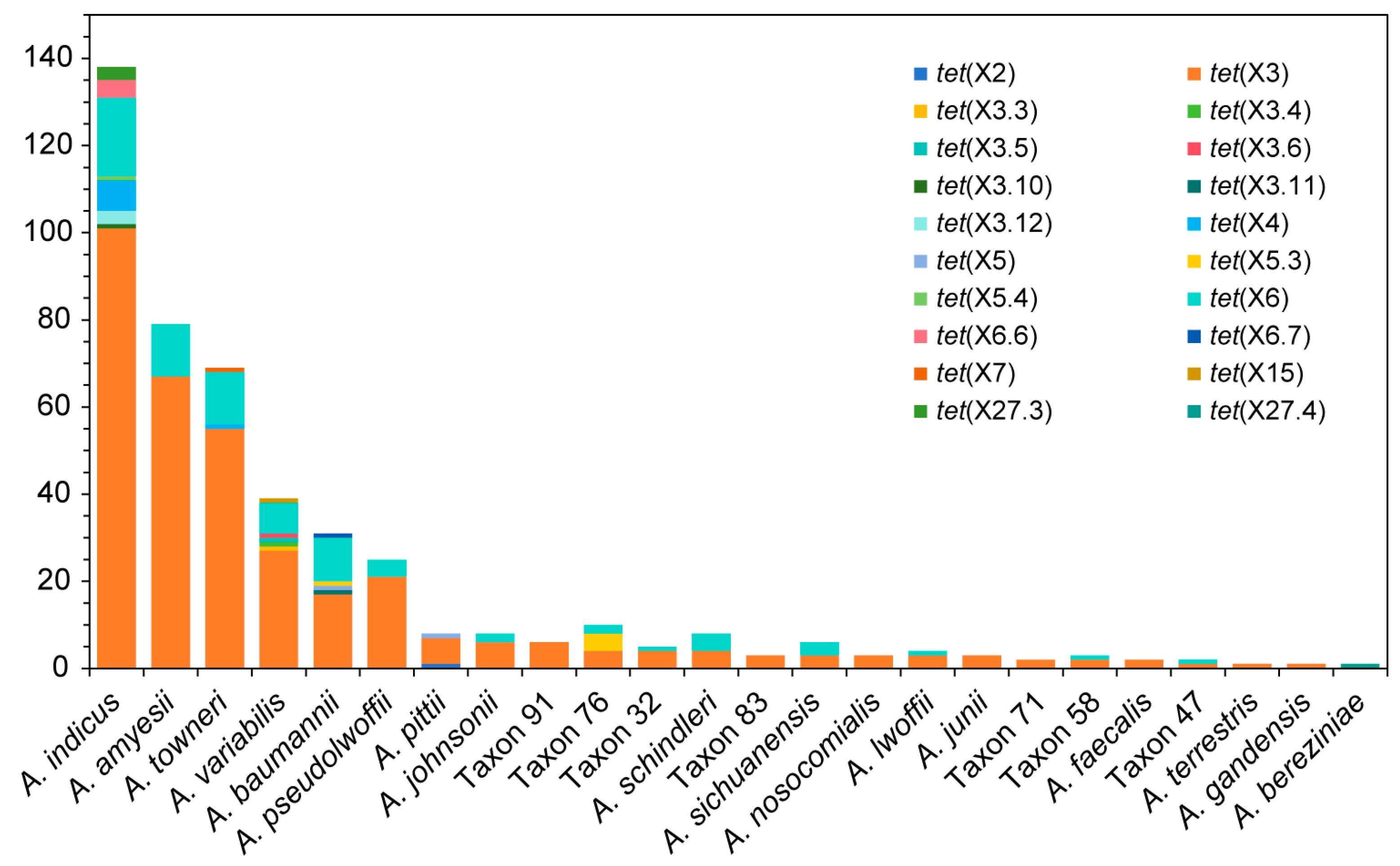
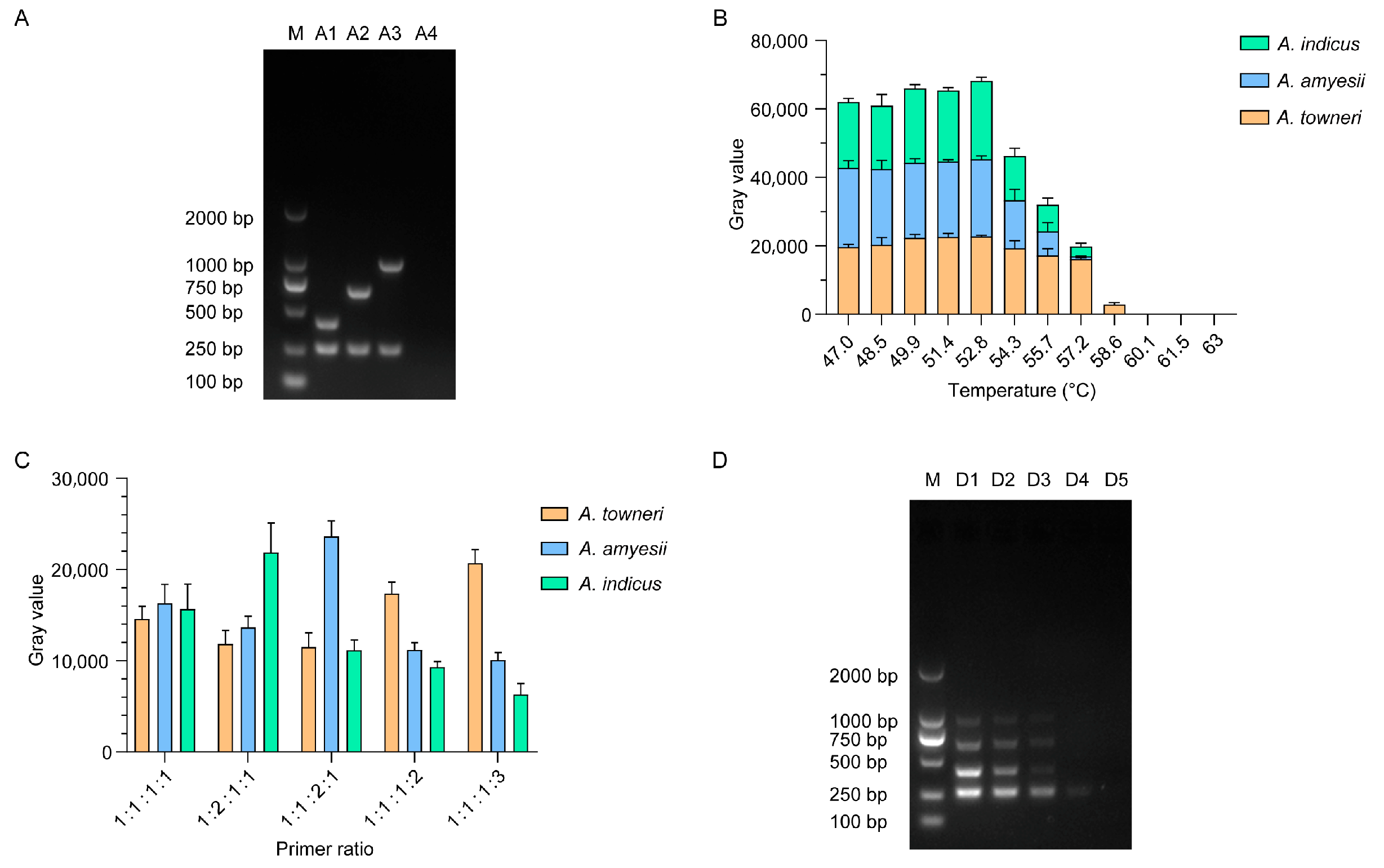

| Primer | Sequence (5′-3′) | Size (bp) | Target |
|---|---|---|---|
| tetX-F | GCGGGATTGTTACAAACTTA | 267 | tet(X) |
| tetX-R | ATCTGCTGTTTCACTCG | ||
| indicus-F | ATGCAATTAACCGATTATCCAG | 424 | A. indicus |
| indicus-R | CCAGATAATGCCCCACACT | ||
| amyesii-F | GCCTATGTTTTTGACCCAAT | 690 | A. amyesii |
| amyesii-R | GCACCATAAAACCAATACC | ||
| towneri-F | TGGGTAGATGTGTCACAGG | 990 | A. towneri |
| towneri-R | GGTATTCAAACCAATGACTGC |
| Species | Number | Percentage (Positive Strains/Samples) | |
|---|---|---|---|
| Bacterial Suspensions | Genomic DNA | ||
| tet(X)-positive strains | 145 | 47.6% (69/145) | 49% (71/145) |
| A. indicus | 46 | 93.5% (43/46) | 97.8% (45/46) |
| A. amyesii | 17 | 100% (17/17) | 100% (17/17) |
| A. towneri | 8 | 100% (8/8) | 100% (8/8) |
| A. variabilis | 28 | 3.6% (1/28) | 3.6% (1/28) |
| A. schindleri | 5 | 0% (0/5) | 0% (0/5) |
| A. pseudolwoffii | 5 | 0% (0/5) | 0% (0/5) |
| A. sichuanensis | 2 | 0% (0/2) | 0% (0/2) |
| A. lwoffii | 1 | 0% (0/1) | 0% (0/1) |
| A. defluvii | 1 | 0% (0/1) | 0% (0/1) |
| E. coli | 9 | 0% (0/9) | 0% (0/9) |
| A. caviae | 1 | 0% (0/1) | 0% (0/1) |
| E. stercoris | 13 | 0% (0/13) | 0% (0/13) |
| M. tengzhouensis | 1 | 0% (0/1) | 0% (0/1) |
| M. odoratimimus | 5 | 0% (0/5) | 0% (0/5) |
| M. zaozhuangensis | 1 | 0% (0/1) | 0% (0/1) |
| M. faecalis | 2 | 0% (0/2) | 0% (0/2) |
| tet(X)-negative strains | 6 | 0% (0/6) | 0% (0/6) |
| E. coli | 2 | 0% (0/2) | 0% (0/2) |
| A. baumannii | 1 | 0% (0/1) | 0% (0/1) |
| A. baylyi | 1 | 0% (0/1) | 0% (0/1) |
| S. enterica | 1 | 0% (0/1) | 0% (0/1) |
| K. pneumoniae | 1 | 0% (0/1) | 0% (0/1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Liu, J.; Gao, J.; Wu, T.; Huang, J. Establishment of Specific Multiplex PCR Detection Methods for the Predominant tet(X)-Positive Acinetobacter Species. Microorganisms 2025, 13, 2584. https://doi.org/10.3390/microorganisms13112584
Chen C, Liu J, Gao J, Wu T, Huang J. Establishment of Specific Multiplex PCR Detection Methods for the Predominant tet(X)-Positive Acinetobacter Species. Microorganisms. 2025; 13(11):2584. https://doi.org/10.3390/microorganisms13112584
Chicago/Turabian StyleChen, Chong, Jing Liu, Jie Gao, Taotao Wu, and Jinlin Huang. 2025. "Establishment of Specific Multiplex PCR Detection Methods for the Predominant tet(X)-Positive Acinetobacter Species" Microorganisms 13, no. 11: 2584. https://doi.org/10.3390/microorganisms13112584
APA StyleChen, C., Liu, J., Gao, J., Wu, T., & Huang, J. (2025). Establishment of Specific Multiplex PCR Detection Methods for the Predominant tet(X)-Positive Acinetobacter Species. Microorganisms, 13(11), 2584. https://doi.org/10.3390/microorganisms13112584






