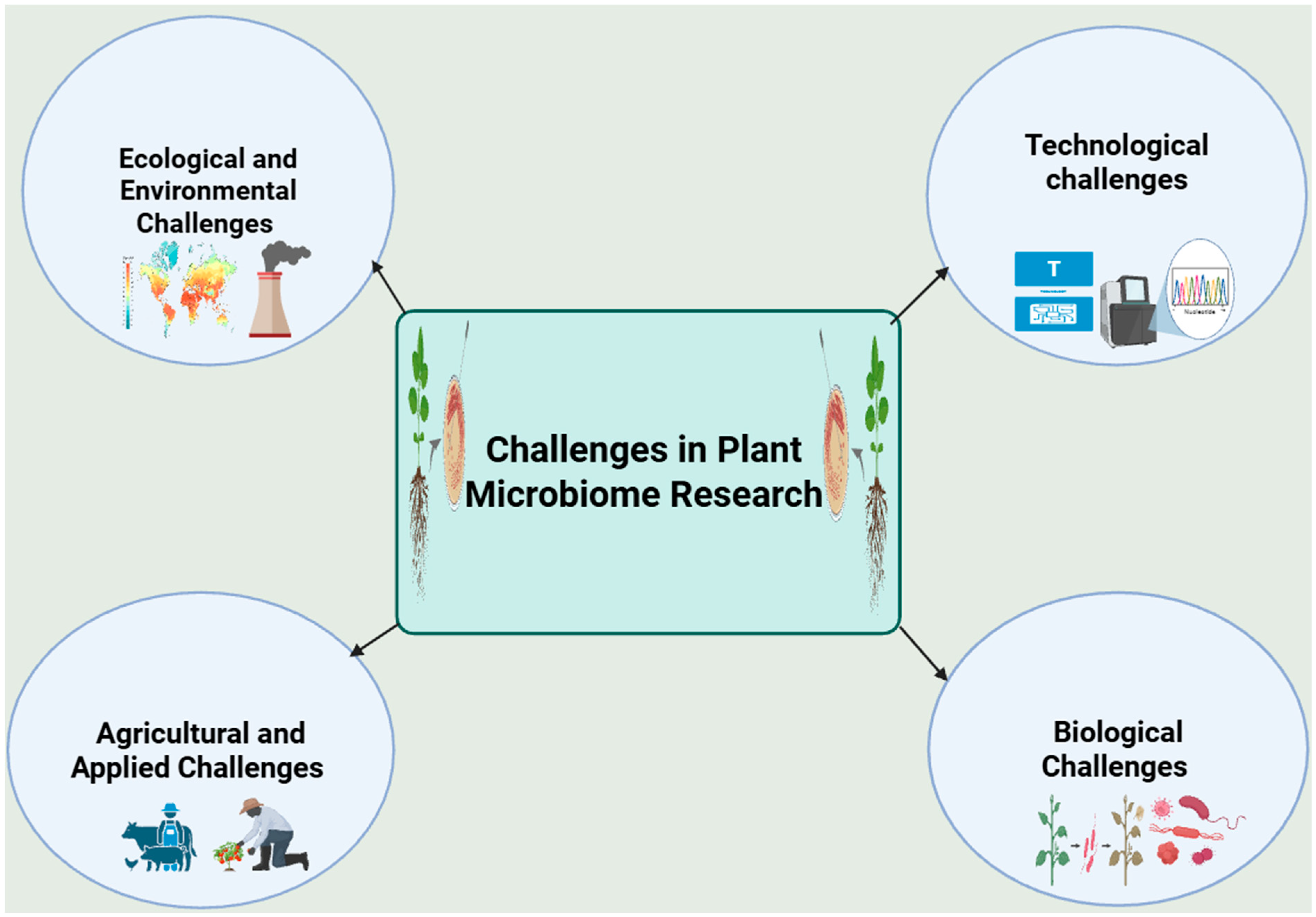
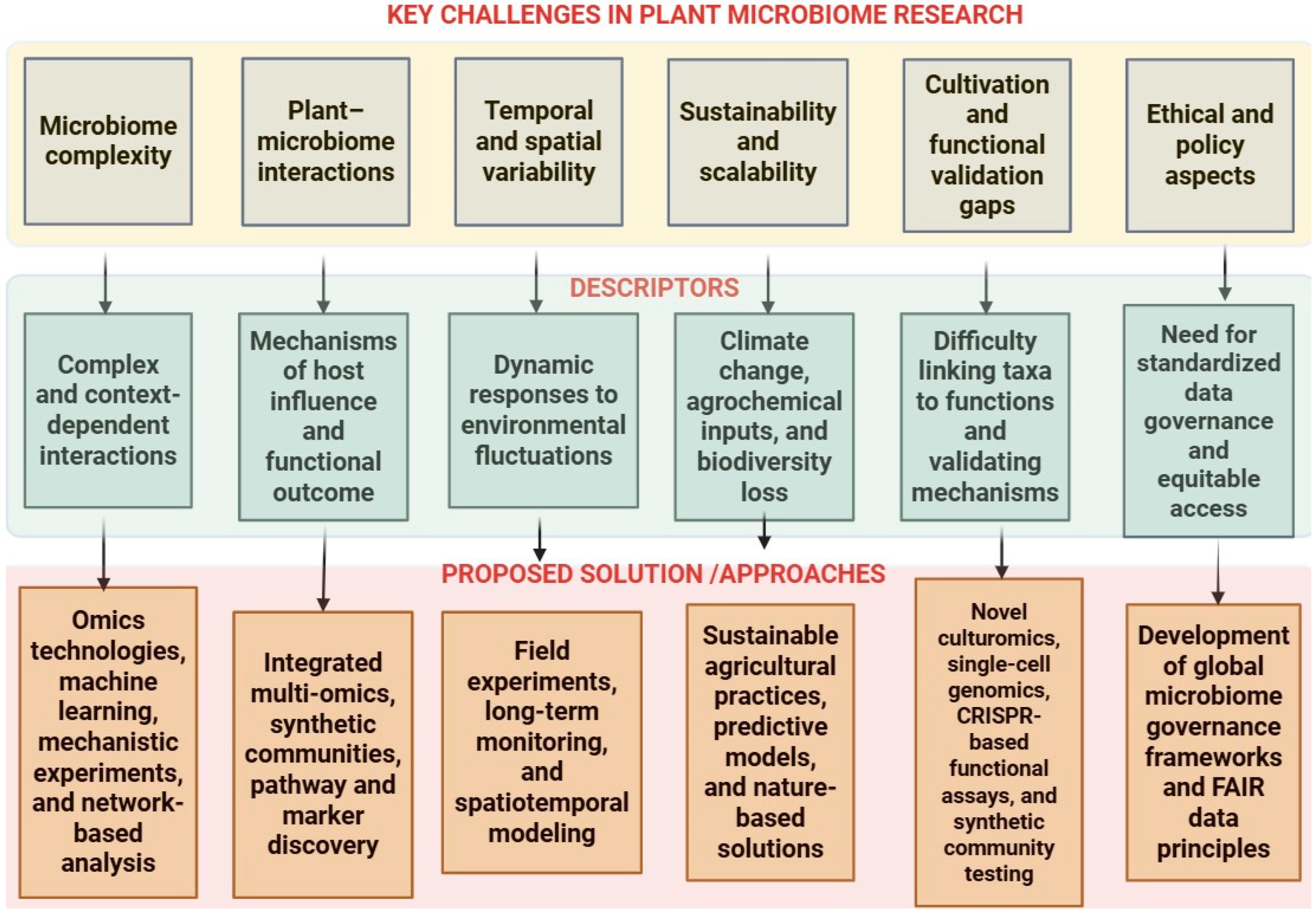
Key Challenges in Plant Microbiome Research in the Next Decade
Abstract
1. Introduction
2. Technological Challenges in Plant Microbiome Research
2.1. Sequencing Technologies: Limitations, Standardization and Reproducibility
2.2. Data Integration and Analysis
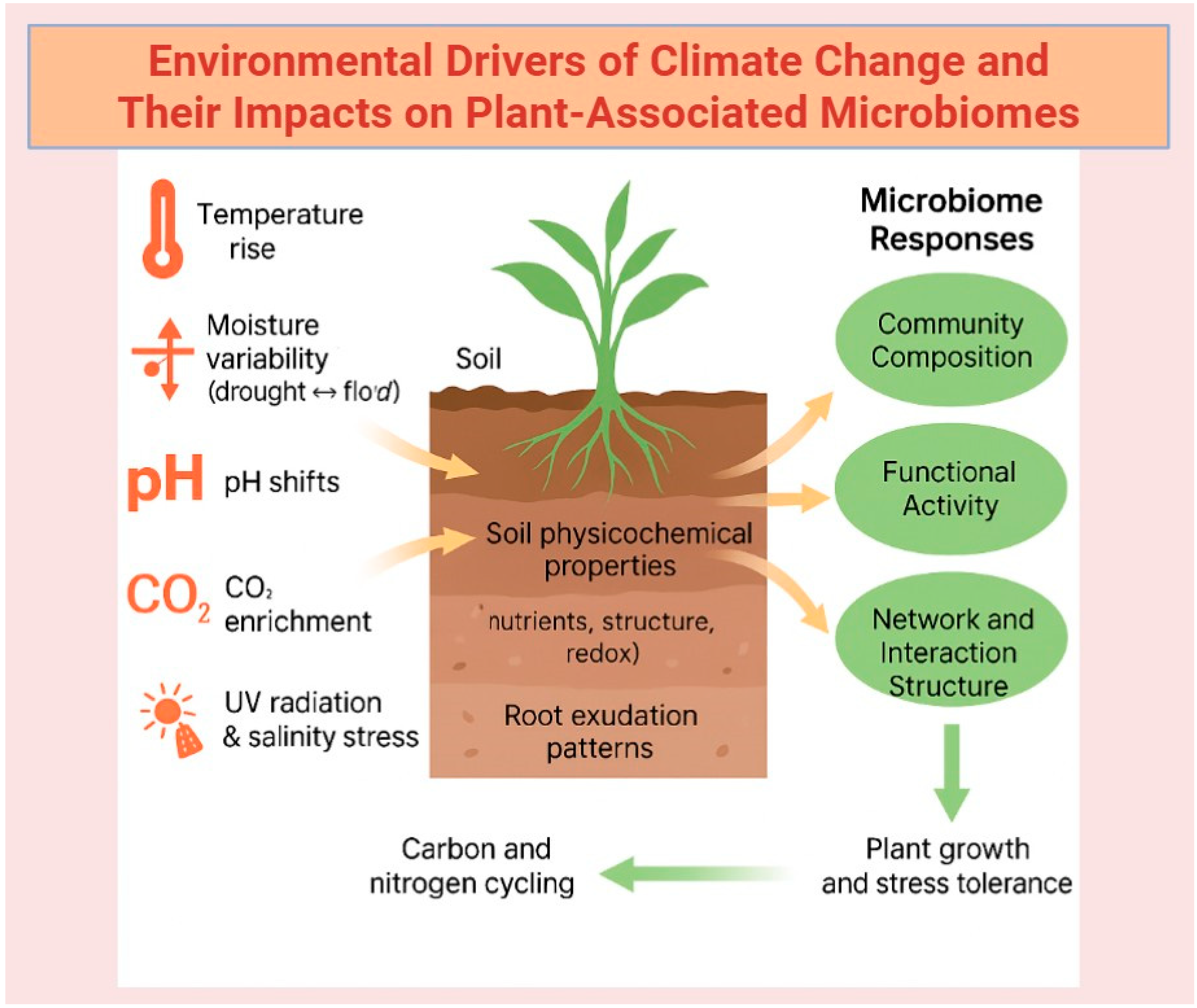
3. Ecological and Environmental Challenges
3.1. Understanding the Dynamic Nature of Plant–Microbiome Interactions
3.2. Influence of Abiotic Factors on Plant Microbiome Composition and Function
3.2.1. Soil pH
3.2.2. Soil Temperature
3.2.3. Soil Aeration
3.2.4. Soil Moisture
3.3. Managing the Impact of Climate Change on Plant Microbiomes
3.3.1. Elevated Carbon Dioxide (CO2)
3.3.2. Increased Temperature
3.3.3. Permafrost Thaw
3.3.4. Drought
3.3.5. Increased Precipitation and Flooding
3.4. The Role of Horizontal Gene Transfer and Microbial Adaptation in Shaping Plant–Microbe Interactions
4. Biological Challenges
4.1. Limited Functional Characterization of Microbiome Members
4.2. Host-Specificity and Environmental Context Dependency of Microbiome Assemblies
4.3. Achieving Stability, Resilience and Predictability of Engineered Microbiomes
4.4. Exploring the Co-Evolution and Domestication Effects on Plant–Microbe Interactions
5. Agricultural and Applied Challenges
5.1. Translating Research to Field Applications
5.2. Regulatory and Ethical Considerations in Microbiome-Based Products
5.3. Economic Viability of Microbiome-Based Interventions in Agriculture
6. Future Directions and Emerging Opportunities
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, M.; Verma, S.; Choudhary, P.; Chakdar, H. Plant Microbiome: Trends and Prospects for Sustainable Agriculture. In Plant Microbe Symbiosis; Varma, A., Tripathi, S., Prasad, R., Eds.; Springer: Cham, Switzerland, 2020; pp. 129–151. [Google Scholar] [CrossRef]
- French, E.; Kaplan, I.; Iyer-Pascuzzi, A.; Nakatsu, C.H.; Enders, L. Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants 2021, 7, 256–267. [Google Scholar] [CrossRef]
- Berruto, C.A.; Demirer, G.S. Engineering agricultural soil microbiomes and predicting plant phenotypes. Trends Microbiol. 2024, 32, 858–873. [Google Scholar] [CrossRef]
- Rocca, J.D.; Simonin, M.; Blaszczak, J.R.; Ernakovich, J.G.; Gibbons, S.M.; Midani, F.S.; Washburne, A.D. The microbiome stress project: Toward a global meta-analysis of environmental stressors and their effects on microbial communities. Front. Microbiol. 2019, 9, 3272. [Google Scholar] [CrossRef]
- Finkel, O.M.; Castrillo, G.; Paredes, S.H.; González, I.S.; Dangl, J.L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 2017, 38, 155–163. [Google Scholar] [CrossRef]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; De Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.; Gomez-Exposito, R.; Elsayed, S.S. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Ridout, M.; Newcombe, G. Fungal endophytes: Modifiers of plant disease. Plant Mol. Biol. 2016, 90, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O. Exploring the potentialities of beneficial endophytes for improved plant growth. Saudi J. Biol. Sci. 2020, 27, 3622–3633. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Lange, L.; Berg, G.; Cernava, T.; Champomier-Vergès, M.-C.; Charles, T.; Cocolin, L.; Cotter, P.; D’hondt, K.; Kostic, T.; Maguin, E. Microbiome ethics, guiding principles for microbiome research, use and knowledge management. Environ. Microbiome 2022, 17, 50. [Google Scholar] [CrossRef]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil. Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Yadav, A.N.; Santoyo, G.; Babalola, O.O. Understanding the plant-microbe interactions in environments exposed to abiotic stresses: An overview. Microbiol. Res. 2023, 271, 127368. [Google Scholar] [CrossRef] [PubMed]
- Adeniji, A.; Fadiji, A.E.; Li, S.; Guo, R. From lab bench to farmers’ fields: Co-creating microbial inoculants with farmers input. Rhizosphere 2024, 31, 100920. [Google Scholar] [CrossRef]
- Babalola, O.O.; Fadiji, A.E.; Enagbonma, B.J.; Alori, E.T.; Ayilara, M.S.; Ayangbenro, A.S. The nexus between plant and plant microbiome: Revelation of the networking strategies. Front. Microbiol. 2020, 11, 548037. [Google Scholar] [CrossRef] [PubMed]
- Nath, M.; Bhatt, D.; Bhargava, P.; Choudhary, D.K. Microbial Metatranscriptomics Below Ground; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Wen, T.; Yuan, J.; He, X.; Lin, Y.; Huang, Q.; Shen, Q. Enrichment of beneficial cucumber rhizosphere microbes mediated by organic acid secretion. Hortic. Res. 2020, 7, 154. [Google Scholar] [CrossRef]
- Zhang, F.; Xu, N.; Zhang, Z.; Zhang, Q.; Yang, Y.; Yu, Z.; Sun, L.; Lu, T.; Qian, H. Shaping effects of rice, wheat, maize, and soybean seedlings on their rhizosphere microbial community. Environ. Sci. Pollut. Res. 2023, 30, 35972–35984. [Google Scholar] [CrossRef]
- Bansal, A.K. Bioinformatics in microbial biotechnology–a mini review. Microb. Cell Factories 2005, 4, 19. [Google Scholar]
- Odom, A.R.; Faits, T.; Castro-Nallar, E.; Crandall, K.A.; Johnson, W.E. Metagenomic profiling pipelines improve taxonomic classification for 16S amplicon sequencing data. Sci. Rep. 2023, 13, 13957. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O. Metagenomics methods for the study of plant-associated microbial communities: A review. J. Microbiol. Methods 2020, 170, 105860. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Qureshi, A.I.; Winther-Larsen, H.C.; Rounge, T.B. Challenges in capturing the mycobiome from shotgun metagenome data: Lack of software and databases. Microbiome 2025, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Cheng, L.; Liu, Q.; Wang, S.; Zhou, Y.; Zhong, H.; Tang, M.; Nian, H.; Lian, T. Effects of waterlogging on soybean rhizosphere bacterial community using V4, LoopSeq, and PacBio 16S rRNA sequence. Microbiol. Spectr. 2022, 10, e02011–e02021. [Google Scholar] [CrossRef]
- Meslier, V.; Quinquis, B.; Da Silva, K.; Plaza Oñate, F.; Pons, N.; Roume, H.; Podar, M.; Almeida, M. Benchmarking second and third-generation sequencing platforms for microbial metagenomics. Sci. Data 2022, 9, 694. [Google Scholar] [CrossRef]
- Jeong, J.; Yun, K.; Mun, S.; Chung, W.-H.; Choi, S.-Y.; Nam, Y.-d.; Lim, M.Y.; Hong, C.P.; Park, C.; Ahn, Y.J. The effect of taxonomic classification by full-length 16S rRNA sequencing with a synthetic long-read technology. Sci. Rep. 2021, 11, 1727. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G. Next-generation sequencing technology: Current trends and advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Markusková, B.; Minarovičová, J.; Véghová, A.; Drahovská, H.; Kaclíková, E. Impact of DNA extraction methods on 16S rRNA-based profiling of bacterial communities in cheese. J. Microbiol. Methods 2021, 184, 106210. [Google Scholar] [CrossRef]
- Sare, A.R.; Stouvenakers, G.; Eck, M.; Lampens, A.; Goormachtig, S.; Jijakli, M.H.; Massart, S. Standardization of plant microbiome studies: Which proportion of the microbiota is really harvested? Microorganisms 2020, 8, 342. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef]
- Greenlon, A.; Sieradzki, E.; Zablocki, O.; Koch, B.J.; Foley, M.M.; Kimbrel, J.A.; Hungate, B.A.; Blazewicz, S.J.; Nuccio, E.E.; Sun, C.L. Quantitative stable-isotope probing (qSIP) with metagenomics links microbial physiology and activity to soil moisture in Mediterranean-climate grassland ecosystems. Msystems 2022, 7, e00417–e00422. [Google Scholar] [CrossRef]
- Berg, G.; Köberl, M.; Rybakova, D.; Müller, H.; Grosch, R.; Smalla, K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 2017, 93, fix050. [Google Scholar] [CrossRef]
- Zancarini, A.; Westerhuis, J.A.; Smilde, A.K.; Bouwmeester, H.J. Integration of omics data to unravel root microbiome recruitment. Curr. Opin. Biotechnol. 2021, 70, 255–261. [Google Scholar] [CrossRef]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 16, 2533–2544. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Almeida, A.; Beracochea, M.; Boland, M.; Burgin, J.; Cochrane, G.; Crusoe, M.R.; Kale, V.; Potter, S.C.; Richardson, L.J. MGnify: The microbiome analysis resource in 2020. Nucleic Acids Res. 2020, 48, D570–D578. [Google Scholar] [CrossRef] [PubMed]
- Eloe-Fadrosh, E.A.; Ahmed, F.; Babinski, M.; Baumes, J.; Borkum, M.; Bramer, L.; Canon, S.; Christianson, D.S.; Corilo, Y.E.; Davenport, K.W. The National Microbiome Data Collaborative Data Portal: An integrated multi-omics microbiome data resource. Nucleic Acids Res. 2022, 50, D828–D836. [Google Scholar]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R. The IMG/M data management and analysis system v. 6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S. KBase: The United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking plant secondary metabolites and plant microbiomes: A review. Front. Plant Sci. 2021, 12, 621276. [Google Scholar] [CrossRef]
- Hutchison, W.J.; Keyes, T.J.; Consortium, T.; Crowell, H.L.; Serizay, J.; Soneson, C.; Davis, E.S.; Sato, N.; Moses, L.; Tarlinton, B. The tidyomics ecosystem: Enhancing omic data analyses. Nat. Methods 2024, 21, 1166–1170. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Dillon, M.R.; Bolyen, E.; Kaehler, B.D.; Huttley, G.A.; Caporaso, J.G. q2-sample-classifier: Machine-learning tools for microbiome classification and regression. J. Open Res. Softw. 2018, 3, 934. [Google Scholar] [CrossRef]
- Wirbel, J.; Zych, K.; Essex, M.; Karcher, N.; Kartal, E.; Salazar, G.; Bork, P.; Sunagawa, S.; Zeller, G. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol. 2021, 22, 93. [Google Scholar] [CrossRef]
- Morton, J.T.; Marotz, C.; Washburne, A.; Silverman, J.; Zaramela, L.S.; Edlund, A.; Zengler, K.; Knight, R. Establishing microbial composition measurement standards with reference frames. Nat. Commun. 2019, 10, 2719. [Google Scholar] [CrossRef]
- Morton, J.T.; Aksenov, A.A.; Nothias, L.F.; Foulds, J.R.; Quinn, R.A.; Badri, M.H.; Swenson, T.L.; Van Goethem, M.W.; Northen, T.R.; Vazquez-Baeza, Y. Learning representations of microbe–metabolite interactions. Nat. Methods 2019, 16, 1306–1314. [Google Scholar] [CrossRef]
- Noecker, C.; Eng, A.; Muller, E.; Borenstein, E. MIMOSA2: A metabolic network-based tool for inferring mechanism-supported relationships in microbiome-metabolome data. Bioinformatics 2022, 38, 1615–1623. [Google Scholar] [CrossRef]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef]
- Haney, C.H.; Samuel, B.S.; Bush, J.; Ausubel, F.M. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 2015, 1, 15051. [Google Scholar] [CrossRef]
- Pieterse, C.M.; de Jonge, R.; Berendsen, R.L. The soil-borne supremacy. Trends Plant Sci. 2016, 21, 171–173. [Google Scholar] [CrossRef]
- Davenport, E.R.; Sanders, J.G.; Song, S.J.; Amato, K.R.; Clark, A.G.; Knight, R. The human microbiome in evolution. BMC Biol. 2017, 15, 127. [Google Scholar] [CrossRef]
- Sherwin, E.; Bordenstein, S.R.; Quinn, J.L.; Dinan, T.G.; Cryan, J.F. Microbiota and the social brain. Science 2019, 366, eaar2016. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef]
- Aziz, S.; Waqas, M.; Mohanta, T.K.; Halim, S.A.; Iqbal, A.; Ali, A.; Khalid, A.; Abdalla, A.N.; Khan, A.; Al-Harrasi, A. Identifying non-nucleoside inhibitors of RNA-dependent RNA-polymerase of SARS-CoV-2 through per-residue energy decomposition-based pharmacophore modeling, molecular docking, and molecular dynamics simulation. J. Infect. Public Health 2023, 16, 501–519. [Google Scholar] [CrossRef]
- Nguyen, J.; Lara-Gutiérrez, J.; Stocker, R. Environmental fluctuations and their effects on microbial communities, populations and individuals. FEMS Microbiol. Rev. 2021, 45, fuaa068. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A.H.M. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Fu, Y.; Shao, J.; Liu, Y.; Xuan, W.; Xu, G.; Zhang, R. Signal communication during microbial modulation of root system architecture. J. Exp. Bot. 2024, 75, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef]
- Paine, R.T. Food web complexity and species diversity. Am. Nat. 1966, 100, 65–75. [Google Scholar] [CrossRef]
- Walker, B.H. Biodiversity and ecological redundancy. Conserv. Biol. 1992, 6, 18–23. [Google Scholar] [CrossRef]
- Cardinale, B.J.; Matulich, K.L.; Hooper, D.U.; Byrnes, J.E.; Duffy, E.; Gamfeldt, L.; Balvanera, P.; O’connor, M.I.; Gonzalez, A. The functional role of producer diversity in ecosystems. Am. J. Bot. 2011, 98, 572–592. [Google Scholar] [CrossRef]
- Moya, A.; Ferrer, M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016, 24, 402–413. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Carlström, C.I.; Field, C.M.; Bortfeld-Miller, M.; Müller, B.; Sunagawa, S.; Vorholt, J.A. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 2019, 3, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Liu, W.; Qiu, X.; Zhang, J.; Zhang, J.; Bai, Y. The root microbiome: Community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 2022, 64, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Pacheco, A.R.; Künzler, R.; Bortfeld-Miller, M.; Field, C.M.; Vayena, E.; Hatzimanikatis, V.; Vorholt, J.A. Metabolic interaction models recapitulate leaf microbiota ecology. Science 2023, 381, eadf5121. [Google Scholar] [CrossRef]
- Schlechter, R.O.; Kear, E.J.; Bernach, M.; Remus, D.M.; Remus-Emsermann, M.N. Metabolic resource overlap impacts competition among phyllosphere bacteria. ISME J. 2023, 17, 1445–1454. [Google Scholar] [CrossRef]
- Getzke, F.; Hassani, M.A.; Crüsemann, M.; Malisic, M.; Zhang, P.; Ishigaki, Y.; Böhringer, N.; Jiménez Fernández, A.; Wang, L.; Ordon, J. Cofunctioning of bacterial exometabolites drives root microbiota establishment. Proc. Natl. Acad. Sci. USA 2023, 120, e2221508120. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Edwards, J.A.; Santos-Medellín, C.M.; Liechty, Z.S.; Nguyen, B.; Lurie, E.; Eason, S.; Phillips, G.; Sundaresan, V. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 2018, 16, e2003862. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef]
- Beilsmith, K.; Perisin, M.; Bergelson, J. Natural bacterial assemblages in Arabidopsis thaliana tissues become more distinguishable and diverse during host development. MBio 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- Smets, W.; Chock, M.K.; Walsh, C.M.; Vanderburgh, C.Q.; Kau, E.; Lindow, S.E.; Fierer, N.; Koskella, B. Leaf side determines the relative importance of dispersal versus host filtering in the phyllosphere microbiome. MBio 2023, 14, e01111–e01123. [Google Scholar] [CrossRef]
- Aziz, U.; Rehmani, M.S.; Wang, L.; Luo, X.; Xian, B.; Wei, S.; Wang, G.; Shu, K. Toward a Molecular Understanding of Rhizosphere, Phyllosphere, and Spermosphere Interactions in Plant Growth and Stress Response. Crit. Rev. Plant Sci. 2021, 40, 479–500. [Google Scholar] [CrossRef]
- Sullivan, T.; Barth, V.; Lewis, R. Soil Acidity Impacts Beneficial Soil; WSU Press: Washington, DC, USA, 2016. [Google Scholar]
- Fernández-Calviño, D.; Bååth, E. Growth response of the bacterial community to pH in soils differing in pH. FEMS Microbiol. Ecol. 2010, 73, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.A.; Puissant, J.; Buckeridge, K.M.; Goodall, T.; Jehmlich, N.; Chowdhury, S.; Gweon, H.S.; Peyton, J.M.; Mason, K.E.; van Agtmaal, M. Land use driven change in soil pH affects microbial carbon cycling processes. Nat. Commun. 2018, 9, 3591. [Google Scholar] [CrossRef]
- Smercina, D.N.; Evans, S.E.; Friesen, M.L.; Tiemann, L.K. To fix or not to fix: Controls on free-living nitrogen fixation in the rhizosphere. Appl. Environ. Microbiol. 2019, 85, e02546-18. [Google Scholar] [CrossRef]
- Jones, R.T.; Robeson, M.S.; Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 2009, 3, 442–453. [Google Scholar] [CrossRef]
- Mayerhofer, J.; Wächter, D.; Calanca, P.; Kohli, L.; Roth, T.; Meuli, R.G.; Widmer, F. Environmental and anthropogenic factors shape major bacterial community types across the complex mountain landscape of Switzerland. Front. Microbiol. 2021, 12, 581430. [Google Scholar] [CrossRef]
- Dubey, A.; Malla, M.A.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.L. Soil microbiome: A key player for conservation of soil health under changing climate. Biodivers. Conserv. 2019, 28, 2405–2429. [Google Scholar] [CrossRef]
- Goicoechea, N. Mycorrhizal fungi as bioprotectors of crops against verticillium wilt—A hypothetical scenario under changing environmental conditions. Plants 2020, 9, 1468. [Google Scholar] [CrossRef] [PubMed]
- Frater, P.N.; Borer, E.T.; Fay, P.A.; Jin, V.; Knaeble, B.; Seabloom, E.; Sullivan, L.; Wedin, D.A.; Harpole, W.S. Nutrients and environment influence arbuscular mycorrhizal colonization both independently and interactively in Schizachyrium scoparium. Plant Soil 2018, 425, 493–506. [Google Scholar] [CrossRef]
- Jerbi, M.; Labidi, S.; Lounes-Hadj Sahraoui, A.; Chaar, H.; Ben Jeddi, F. Higher temperatures and lower annual rainfall do not restrict, directly or indirectly, the mycorrhizal colonization of barley (Hordeum vulgare L.) under rainfed conditions. PLoS ONE 2020, 15, e0241794. [Google Scholar] [CrossRef]
- Loreti, E.; Perata, P. The many facets of hypoxia in plants. Plants 2020, 9, 745. [Google Scholar] [CrossRef]
- Li, Y.; Niu, W.; Zhang, M.; Wang, J.; Zhang, Z. Artificial soil aeration increases soil bacterial diversity and tomato root performance under greenhouse conditions. Land. Degrad. Dev. 2020, 31, 1443–1461. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Siebielec, S.; Siebielec, G.; Klimkowicz-Pawlas, A.; Gałązka, A.; Grządziel, J.; Stuczyński, T. Impact of water stress on microbial community and activity in sandy and loamy soils. Agronomy 2020, 10, 1429. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Martin, D.D.; Ciulla, R.A.; Roberts, M.F. Osmoadaptation in archaea. Appl. Environ. Microbiol. 1999, 65, 1815–1825. [Google Scholar] [CrossRef]
- Cohen, S.P.; Leach, J.E. High temperature-induced plant disease susceptibility: More than the sum of its parts. Curr. Opin. Plant Biol. 2020, 56, 235–241. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Guerra, C.A.; Cano-Díaz, C.; Egidi, E.; Wang, J.-T.; Eisenhauer, N.; Singh, B.K.; Maestre, F.T. The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Change 2020, 10, 550–554. [Google Scholar] [CrossRef]
- Burdon, J.J.; Zhan, J. Climate change and disease in plant communities. PLoS Biol. 2020, 18, e3000949. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.T.; Zhang, L.; He, S.Y. Plant-microbe interactions facing environmental challenge. Cell Host Microbe 2019, 26, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, C.; Frachon, L.; Barret, M.; Rigal, M.; Huard-Chauveau, C.; Mayjonade, B.; Zanchetta, C.; Bouchez, O.; Roby, D.; Carrère, S. In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 2018, 12, 2024–2038. [Google Scholar] [CrossRef] [PubMed]
- Carol Adair, E.; Reich, P.B.; Trost, J.J.; Hobbie, S.E. Elevated CO2 stimulates grassland soil respiration by increasing carbon inputs rather than by enhancing soil moisture. Glob. Change Biol. 2011, 17, 3546–3563. [Google Scholar] [CrossRef]
- Bréchet, L.M.; Lopez-Sangil, L.; George, C.; Birkett, A.J.; Baxendale, C.; Castro Trujillo, B.; Sayer, E.J. Distinct responses of soil respiration to experimental litter manipulation in temperate woodland and tropical forest. Ecol. Evol. 2018, 8, 3787–3796. [Google Scholar] [CrossRef]
- Bengtson, P.; Barker, J.; Grayston, S.J. Evidence of a strong coupling between root exudation, C and N availability, and stimulated SOM decomposition caused by rhizosphere priming effects. Ecol. Evol. 2012, 2, 1843–1852. [Google Scholar] [CrossRef]
- Jansson, C.; Vogel, J.; Hazen, S.; Brutnell, T.; Mockler, T. Climate-smart crops with enhanced photosynthesis. J. Exp. Bot. 2018, 69, 3801–3809. [Google Scholar] [CrossRef]
- Hayden, H.L.; Mele, P.M.; Bougoure, D.S.; Allan, C.Y.; Norng, S.; Piceno, Y.M.; Brodie, E.L.; DeSantis, T.Z.; Andersen, G.L.; Williams, A.L. Changes in the microbial community structure of bacteria, archaea and fungi in response to elevated CO2 and warming in an A ustralian native grassland soil. Environ. Microbiol. 2012, 14, 3081–3096. [Google Scholar] [CrossRef]
- Qiao, N.; Schaefer, D.; Blagodatskaya, E.; Zou, X.; Xu, X.; Kuzyakov, Y. Labile carbon retention compensates for CO2 released by priming in forest soils. Glob. Change Biol. 2014, 20, 1943–1954. [Google Scholar] [CrossRef]
- Van Groenigen, K.J.; Qi, X.; Osenberg, C.W.; Luo, Y.; Hungate, B.A. Faster decomposition under increased atmospheric CO2 limits soil carbon storage. Science 2014, 344, 508–509. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.E.; Gallet-Budynek, A.; Hofmockel, K.S.; Bernhardt, E.S.; Billings, S.A.; Jackson, R.B.; Johnsen, K.S.; Lichter, J.; McCarthy, H.R.; McCormack, M.L. Increases in the flux of carbon belowground stimulate nitrogen uptake and sustain the long-term enhancement of forest productivity under elevated CO2. Ecol. Lett. 2011, 14, 349–357. [Google Scholar] [CrossRef]
- Lal, R. Soil carbon sequestration impacts on global climate change and food security. Science 2004, 304, 1623–1627. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y. Priming effects: Interactions between living and dead organic matter. Soil. Biol. Biochem. 2010, 42, 1363–1371. [Google Scholar] [CrossRef]
- Scharlemann, J.P.; Tanner, E.V.; Hiederer, R.; Kapos, V. Global soil carbon: Understanding and managing the largest terrestrial carbon pool. Carbon. Manag. 2014, 5, 81–91. [Google Scholar] [CrossRef]
- Clemmensen, K.; Bahr, A.; Ovaskainen, O.; Dahlberg, A.; Ekblad, A.; Wallander, H.; Stenlid, J.; Finlay, R.; Wardle, D.; Lindahl, B. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 2013, 339, 1615–1618. [Google Scholar] [CrossRef]
- Allison, S.D.; Treseder, K.K. Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Glob. Change Biol. 2008, 14, 2898–2909. [Google Scholar] [CrossRef]
- Melillo, J.M.; Frey, S.D.; DeAngelis, K.M.; Werner, W.J.; Bernard, M.J.; Bowles, F.P.; Pold, G.; Knorr, M.A.; Grandy, A.S. Long-term pattern and magnitude of soil carbon feedback to the climate system in a warming world. Science 2017, 358, 101–105. [Google Scholar] [CrossRef]
- Bradford, M.A.; Davies, C.A.; Frey, S.D.; Maddox, T.R.; Melillo, J.M.; Mohan, J.E.; Reynolds, J.F.; Treseder, K.K.; Wallenstein, M.D. Thermal adaptation of soil microbial respiration to elevated temperature. Ecol. Lett. 2008, 11, 1316–1327. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, E.M.; Mau, R.L.; Hayer, M.; Liu, X.-J.A.; Schwartz, E.; Dijkstra, P.; Koch, B.J.; Allen, K.; Blazewicz, S.J.; Hofmockel, K. Evolutionary history constrains microbial traits across environmental variation. Nat. Ecol. Evol. 2019, 3, 1064–1069. [Google Scholar] [CrossRef]
- Yu, H.; Deng, Y.; He, Z.; Van Nostrand, J.D.; Wang, S.; Jin, D.; Wang, A.; Wu, L.; Wang, D.; Tai, X. Elevated CO2 and warming altered grassland microbial communities in soil top-layers. Front. Microbiol. 2018, 9, 1790. [Google Scholar] [CrossRef] [PubMed]
- Sheik, C.S.; Beasley, W.H.; Elshahed, M.S.; Zhou, X.; Luo, Y.; Krumholz, L.R. Effect of warming and drought on grassland microbial communities. ISME J. 2011, 5, 1692–1700. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, S.; He, X.; Liu, W.; Zhao, Q.; Zhao, L.; Tian, C. Responses of soil microbial communities to experimental warming in alpine grasslands on the Qinghai-Tibet Plateau. PLoS ONE 2014, 9, e103859. [Google Scholar] [CrossRef]
- Friedlingstein, P.; Cox, P.; Betts, R.; Bopp, L.; von Bloh, W.; Brovkin, V.; Cadule, P.; Doney, S.; Eby, M.; Fung, I. Climate–carbon cycle feedback analysis: Results from the C4MIP model intercomparison. J. Clim. 2006, 19, 3337–3353. [Google Scholar] [CrossRef]
- Heimann, M.; Reichstein, M. Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 2008, 451, 289–292. [Google Scholar] [CrossRef]
- Mackelprang, R.; Saleska, S.R.; Jacobsen, C.S.; Jansson, J.K.; Taş, N. Permafrost meta-omics and climate change. Annu. Rev. Earth Planet. Sci. 2016, 44, 439–462. [Google Scholar] [CrossRef]
- Hultman, J.; Waldrop, M.P.; Mackelprang, R.; David, M.M.; McFarland, J.; Blazewicz, S.J.; Harden, J.; Turetsky, M.R.; McGuire, A.D.; Shah, M.B. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 2015, 521, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Taş, N.; Prestat, E.; Wang, S.; Wu, Y.; Ulrich, C.; Kneafsey, T.; Tringe, S.G.; Torn, M.S.; Hubbard, S.S.; Jansson, J.K. Landscape topography structures the soil microbiome in arctic polygonal tundra. Nat. Commun. 2018, 9, 777. [Google Scholar] [CrossRef] [PubMed]
- Woodcroft, B.J.; Singleton, C.M.; Boyd, J.A.; Evans, P.N.; Emerson, J.B.; Zayed, A.A.; Hoelzle, R.D.; Lamberton, T.O.; McCalley, C.K.; Hodgkins, S.B. Genome-centric view of carbon processing in thawing permafrost. Nature 2018, 560, 49–54. [Google Scholar] [CrossRef]
- Johnston, E.R.; Hatt, J.K.; He, Z.; Wu, L.; Guo, X.; Luo, Y.; Schuur, E.A.; Tiedje, J.M.; Zhou, J.; Konstantinidis, K.T. Responses of tundra soil microbial communities to half a decade of experimental warming at two critical depths. Proc. Natl. Acad. Sci. USA 2019, 116, 15096–15105. [Google Scholar] [CrossRef] [PubMed]
- Bottos, E.M.; Kennedy, D.W.; Romero, E.B.; Fansler, S.J.; Brown, J.M.; Bramer, L.M.; Chu, R.K.; Tfaily, M.M.; Jansson, J.K.; Stegen, J.C. Dispersal limitation and thermodynamic constraints govern spatial structure of permafrost microbial communities. FEMS Microbiol. Ecol. 2018, 94, fiy110. [Google Scholar] [CrossRef]
- Mackelprang, R.; Waldrop, M.P.; DeAngelis, K.M.; David, M.M.; Chavarria, K.L.; Blazewicz, S.J.; Rubin, E.M.; Jansson, J.K. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 2011, 480, 368–371. [Google Scholar] [CrossRef]
- Müller, O.; Bang-Andreasen, T.; White, R.A., III; Elberling, B.; Taş, N.; Kneafsey, T.; Jansson, J.K.; Øvreås, L. Disentangling the complexity of permafrost soil by using high resolution profiling of microbial community composition, key functions and respiration rates. Environ. Microbiol. 2018, 20, 4328–4342. [Google Scholar] [CrossRef]
- Singleton, C.M.; McCalley, C.K.; Woodcroft, B.J.; Boyd, J.A.; Evans, P.N.; Hodgkins, S.B.; Chanton, J.P.; Frolking, S.; Crill, P.M.; Saleska, S.R. Methanotrophy across a natural permafrost thaw environment. ISME J. 2018, 12, 2544–2558. [Google Scholar] [CrossRef]
- Emerson, J.B.; Roux, S.; Brum, J.R.; Bolduc, B.; Woodcroft, B.J.; Jang, H.B.; Singleton, C.M.; Solden, L.M.; Naas, A.E.; Boyd, J.A. Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 2018, 3, 870–880. [Google Scholar] [CrossRef]
- Trubl, G.; Jang, H.B.; Roux, S.; Emerson, J.B.; Solonenko, N.; Vik, D.R.; Solden, L.; Ellenbogen, J.; Runyon, A.T.; Bolduc, B. Soil viruses are underexplored players in ecosystem carbon processing. MSystems 2018, 3, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Penton, C.R.; StLouis, D.; Cole, J.R.; Luo, Y.; Wu, L.; Schuur, E.G.; Zhou, J.; Tiedje, J.M. Fungal diversity in permafrost and tallgrass prairie soils under experimental warming conditions. Appl. Environ. Microbiol. 2013, 79, 7063–7072. [Google Scholar] [CrossRef] [PubMed]
- Schütte, U.M.; Henning, J.A.; Ye, Y.; Bowling, A.; Ford, J.; Genet, H.; Waldrop, M.P.; Turetsky, M.R.; White, J.R.; Bever, J.D. Effect of permafrost thaw on plant and soil fungal community in a boreal forest: Does fungal community change mediate plant productivity response? J. Ecol. 2019, 107, 1737–1752. [Google Scholar] [CrossRef]
- McHugh, T.A.; Compson, Z.; van Gestel, N.; Hayer, M.; Ballard, L.; Haverty, M.; Hines, J.; Irvine, N.; Krassner, D.; Lyons, T. Climate controls prokaryotic community composition in desert soils of the southwestern United States. FEMS Microbiol. Ecol. 2017, 93, fix116. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Santoyo, G.; Yadav, A.N.; Babalola, O.O. Efforts toward overcoming drought stress in crops: Revisiting the mechanisms employed by plant growth-promoting bacteria. Front. Microbiol. 2022, 13, 962427. [Google Scholar] [CrossRef] [PubMed]
- Schimel, J.P. Life in dry soils: Effects of drought on soil microbial communities and processes. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 409–432. [Google Scholar] [CrossRef]
- Hyvönen, R.; Ågren, G.I.; Linder, S.; Persson, T.; Cotrufo, M.F.; Ekblad, A.; Freeman, M.; Grelle, A.; Janssens, I.A.; Jarvis, P.G. The likely impact of elevated [CO2], nitrogen deposition, increased temperature and management on carbon sequestration in temperate and boreal forest ecosystems: A literature review. New Phytologist. 2007, 173, 463–480. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef]
- Boot, C.M.; Schaeffer, S.M.; Schimel, J.P. Static osmolyte concentrations in microbial biomass during seasonal drought in a California grassland. Soil. Biol. Biochem. 2013, 57, 356–361. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Cantrell, C.L.; Williams, M.A. Microbial community response to varying magnitudes of desiccation in soil: A test of the osmolyte accumulation hypothesis. Soil. Biol. Biochem. 2013, 57, 644–653. [Google Scholar] [CrossRef]
- Waldrop, M.; Firestone, M. Response of microbial community composition and function to soil climate change. Microb. Ecol. 2006, 52, 716–724. [Google Scholar] [CrossRef]
- Bouskill, N.J.; Wood, T.E.; Baran, R.; Hao, Z.; Ye, Z.; Bowen, B.P.; Lim, H.C.; Nico, P.S.; Holman, H.-Y.; Gilbert, B. Belowground response to drought in a tropical forest soil. II. Change in microbial function impacts carbon composition. Front. Microbiol. 2016, 7, 323. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.; DeGraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [PubMed]
- Roy Chowdhury, T.; Lee, J.-Y.; Bottos, E.M.; Brislawn, C.J.; White, R.A., III; Bramer, L.M.; Brown, J.; Zucker, J.D.; Kim, Y.-M.; Jumpponen, A. Metaphenomic responses of a native prairie soil microbiome to moisture perturbations. Msystems 2019, 4, 10–1128. [Google Scholar] [CrossRef]
- Gedney, N.; Cox, P.; Huntingford, C. Climate feedback from wetland methane emissions. Geophys. Res. Lett. 2004, 31, L20503. [Google Scholar] [CrossRef]
- Moomaw, W.R.; Chmura, G.; Davies, G.T.; Finlayson, C.; Middleton, B.A.; Natali, S.M.; Perry, J.; Roulet, N.; Sutton-Grier, A.E. Wetlands in a changing climate: Science, policy and management. Wetlands 2018, 38, 183–205. [Google Scholar] [CrossRef]
- Chambers, L.G.; Osborne, T.Z.; Reddy, K.R. Effect of salinity-altering pulsing events on soil organic carbon loss along an intertidal wetland gradient: A laboratory experiment. Biogeochemistry 2013, 115, 363–383. [Google Scholar] [CrossRef]
- Steinmuller, H.E.; Chambers, L.G. Can saltwater intrusion accelerate nutrient export from freshwater wetland soils? An experimental approach. Soil. Sci. Soc. Am. J. 2018, 82, 283–292. [Google Scholar] [CrossRef]
- Sjøgaard, K.S.; Valdemarsen, T.B.; Treusch, A.H. Responses of an agricultural soil microbiome to flooding with seawater after managed coastal realignment. Microorganisms 2018, 6, 12. [Google Scholar] [CrossRef]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Viruses and mobile elements as drivers of evolutionary transitions. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150442. [Google Scholar] [CrossRef]
- Ren, B.; Wang, X.; Duan, J.; Ma, J. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 2019, 365, 919–922. [Google Scholar] [CrossRef]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef]
- Pfeifer, E.; Moura de Sousa, J.A.; Touchon, M.; Rocha, E.P. Bacteria have numerous distinctive groups of phage–plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res. 2021, 49, 2655–2673. [Google Scholar] [CrossRef] [PubMed]
- Galetti, R.; Andrade, L.N.; Varani, A.M.; Darini, A.L.C. A phage-like plasmid carrying bla KPC-2 gene in carbapenem-resistant Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 572. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brüssow, H. Prophage genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef] [PubMed]
- Quispe-Huamanquispe, D.G.; Gheysen, G.; Kreuze, J.F. Horizontal gene transfer contributes to plant evolution: The case of Agrobacterium T-DNAs. Front. Plant Sci. 2017, 8, 2015. [Google Scholar] [CrossRef]
- Zupan, J.R.; Zambryski, P. Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 1995, 107, 1041. [Google Scholar] [CrossRef]
- Tzfira, T.; Li, J.; Lacroix, B.; Citovsky, V. Agrobacterium T-DNA integration: Molecules and models. TRENDS Genet. 2004, 20, 375–383. [Google Scholar] [CrossRef]
- Richards, T.A.; Soanes, D.M.; Foster, P.G.; Leonard, G.; Thornton, C.R.; Talbot, N.J. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell 2009, 21, 1897–1911. [Google Scholar] [CrossRef]
- Li, M.; Zhao, J.; Tang, N.; Sun, H.; Huang, J. Horizontal gene transfer from bacteria and plants to the arbuscular mycorrhizal fungus Rhizophagus irregularis. Front. Plant Sci. 2018, 9, 701. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.-L.; Zhao, J.-H.; Wang, S.; Jin, Y.; Chen, Z.-Q.; Fang, Y.-Y.; Hua, C.-L.; Ding, S.-W.; Guo, H.-S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.-M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Maheshwari, M.; Abulreesh, H.H.; Khan, M.S.; Ahmad, I.; Pichtel, J. Horizontal gene transfer in soil and the rhizosphere: Impact on ecological fitness of bacteria. In Agriculturally Important Microbes for Sustainable Agriculture: Volume I: Plant-Soil-Microbe Nexus; Springer: Berlin/Heidelberg, Germany, 2017; pp. 111–130. [Google Scholar]
- Van Elsas, J.D.; Bailey, M.J. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 2002, 42, 187–197. [Google Scholar] [CrossRef]
- Haritha, A.; Sagar, K.P.; Tiwari, A.; Kiranmayi, P.; Rodrigue, A.; Mohan, P.M.; Singh, S.S. MrdH, a novel metal resistance determinant of Pseudomonas putida KT2440, is flanked by metal-inducible mobile genetic elements. J. Bacteriol. 2009, 191, 5976–5987. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, 10–1128. [Google Scholar] [CrossRef]
- Shintani, M.; Nour, E.; Elsayed, T.; Blau, K.; Wall, I.; Jechalke, S.; Spröer, C.; Bunk, B.; Overmann, J.; Smalla, K. Plant species-dependent increased abundance and diversity of IncP-1 plasmids in the rhizosphere: New insights into their role and ecology. Front. Microbiol. 2020, 11, 590776. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Mavrodi, D.V.; Loper, J.E.; Paulsen, I.T.; Thomashow, L.S. Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol. 2009, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Bruto, M.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 2014, 4, 6261. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.E.; Banfield, J.F. Community genomics in microbial ecology and evolution. Nat. Rev. Microbiol. 2005, 3, 489–498. [Google Scholar] [CrossRef]
- Werf, M.J.v.d.; Overkamp, K.M.; Muilwijk, B.; Coulier, L.; Hankemeier, T. Microbial metabolomics: Toward a platform with full metabolome coverage. Anal. Biochem. 2007, 370, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Maron, P.-A.; Ranjard, L.; Mougel, C.; Lemanceau, P. Metaproteomics: A New Approach for Studying Functional Microbial Ecology. Microb. Ecol. 2007, 53, 486–493. [Google Scholar] [CrossRef]
- Shakya, M.; Lo, C.-C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Dinesen, C.; Pioppi, A.; Kovács, Á.T.; Lozano-Andrade, C.N. Composing a microbial symphony: Synthetic communities for promoting plant growth. Trends Microbiol. 2025, 33, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, Y. The Function of Root Exudates in the Root Colonization by Beneficial Soil Rhizobacteria. Biology 2024, 13, 95. [Google Scholar] [CrossRef]
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef]
- Hartmann, M.; Six, J. Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Env. 2023, 4, 4–18. [Google Scholar] [CrossRef]
- Adedayo, A.A.; Babalola, O.O. The potential of biostimulants on soil microbial community: A review. Front. Ind. Microbiol. 2023, 1, 1308641. [Google Scholar] [CrossRef]
- Wang, Y.; Dall’Agnol, R.F.; Bertani, I.; Bez, C.; Venturi, V. Identification of synthetic consortia from a set of plant-beneficial bacteria. Microb. Biotechnol. 2024, 17, e14330. [Google Scholar] [CrossRef]
- Shah, A.M.; Khan, I.M.; Shah, T.I.; Bangroo, S.A.; Kirmani, N.A.; Nazir, S.; Malik, A.R.; Aezum, A.M.; Mir, Y.H.; Hilal, A.; et al. Soil Microbiome: A Treasure Trove for Soil Health Sustainability under Changing Climate. Land 2022, 11, 1887. [Google Scholar] [CrossRef]
- Baltrus, D.A. Adaptation, specialization, and coevolution within phytobiomes. Curr. Opin. Plant Biol. 2017, 38, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Netherway, T. Fungi as mediators linking organisms and ecosystems. FEMS Microbiol. Rev. 2022, 46, fuab058. [Google Scholar] [CrossRef]
- Barnes, C.J.; Bahram, M.; Nicolaisen, M.; Gilbert, M.T.P.; Vestergård, M. Microbiome selection and evolution within wild and domesticated plants. Trends Microbiol. 2025, 33, 447–458. [Google Scholar] [CrossRef]
- Nerva, L.; Sandrini, M.; Moffa, L.; Velasco, R.; Balestrini, R.; Chitarra, W. Breeding toward improved ecological plant-microbiome interactions. Trends Plant Sci. 2022, 27, 1134–1143. [Google Scholar] [CrossRef]
- Batista, B.D.; Singh, B.K. Realities and hopes in the application of microbial tools in agriculture. Microb. Biotechnol. 2021, 14, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, M.; Ballard, R.A.; Wright, D. Soil microbial inoculants for sustainable agriculture: Limitations and opportunities. Soil. Use Manag. 2022, 38, 1340–1369. [Google Scholar] [CrossRef]
- Zilli, J.É.; Pacheco, R.S.; Gianluppi, V.; Smiderle, O.J.; Urquiaga, S.; Hungria, M. Biological N2 fixation and yield performance of soybean inoculated with Bradyrhizobium. Nutr. Cycl. Agroecosystems 2021, 119, 323–336. [Google Scholar] [CrossRef]
- Thilakarathna, M.S.; Raizada, M.N. A meta-analysis of the effectiveness of diverse rhizobia inoculants on soybean traits under field conditions. Soil. Biol. Biochem. 2017, 105, 177–196. [Google Scholar] [CrossRef]
- Raymond, N.S.; Gómez-Muñoz, B.; van der Bom, F.J.; Nybroe, O.; Jensen, L.S.; Müller-Stöver, D.S.; Oberson, A.; Richardson, A.E. Phosphate-solubilising microorganisms for improved crop productivity: A critical assessment. New Phytol. 2021, 229, 1268–1277. [Google Scholar] [CrossRef]
- Timmis, K.; Hallsworth, J.E.; McGenity, T.J.; Armstrong, R.; Colom, M.F.; Karahan, Z.C.; Chavarría, M.; Bernal, P.; Boyd, E.S.; Ramos, J.L. A concept for international societally relevant microbiology education and microbiology knowledge promulgation in society. Microb. Biotechnol. 2024, 17, e14456. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Lakshmanan, V.; Labbé, J.L.; Craven, K.D. Microbe to microbiome: A paradigm shift in the application of microorganisms for sustainable agriculture. Front. Microbiol. 2020, 11, 622926. [Google Scholar] [CrossRef] [PubMed]
- Gourlay, S.; Aynekulu, E.; Carletto, C.; Shepherd, K.D. Spectral Soil Analysis & Household Surveys: A Guidebook for Integration; The World Bank: Washington, DC, USA, 2017. [Google Scholar]
- Bacco, M.; Barsocchi, P.; Ferro, E.; Gotta, A.; Ruggeri, M. The digitisation of agriculture: A survey of research activities on smart farming. Array 2019, 3, 100009. [Google Scholar] [CrossRef]
- Sharma, A.; Jain, A.; Gupta, P.; Chowdary, V. Machine learning applications for precision agriculture: A comprehensive review. IEEE Access 2020, 9, 4843–4873. [Google Scholar] [CrossRef]
- Bennett, G.M.; Lloyd, J. Seed Inoculation, Coating and Precision Pelleting: Science, Technology and Practical Applications; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Jones, J.W.; Antle, J.M.; Basso, B.; Boote, K.J.; Conant, R.T.; Foster, I.; Godfray, H.C.J.; Herrero, M.; Howitt, R.E.; Janssen, S. Toward a new generation of agricultural system data, models, and knowledge products: State of agricultural systems science. Agric. Syst. 2017, 155, 269–288. [Google Scholar] [CrossRef]
- Schut, M.; Klerkx, L.; Sartas, M.; Lamers, D.; Mc Campbell, M.; Ogbonna, I.; Kaushik, P.; Atta-Krah, K.; Leeuwis, C. Innovation platforms: Experiences with their institutional embedding in agricultural research for development. Exp. Agric. 2016, 52, 537–561. [Google Scholar] [CrossRef]
- Ochieng, R. Towards a Regulatory Framework for Increased and Sustainable Use of Bio-Fertilizers in Kenya. Ph.D. Thesis, University of Nairobi, Nairobi, Kenya, 2015. [Google Scholar]
- Olmo, R.; Wetzels, S.U.; Armanhi, J.S.L.; Arruda, P.; Berg, G.; Cernava, T.; Cotter, P.D.; Araujo, S.C.; de Souza, R.S.C.; Ferrocino, I. Microbiome research as an effective driver of success stories in agrifood systems–a selection of case studies. Front. Microbiol. 2022, 13, 834622. [Google Scholar] [CrossRef]
- Vapnek, J.; Purnhagen, K.; Hillel, B. Regulatory and legislative framework for novel foods. In Food Formulation: Novel Ingredients and Processing Techniques; John Wiley & Sons: Hoboken, NJ, USA, 2021; pp. 285–308. [Google Scholar]
- Elnahal, A.S.; El-Saadony, M.T.; Saad, A.M.; Desoky, E.-S.M.; El-Tahan, A.M.; Rady, M.M.; AbuQamar, S.F.; El-Tarabily, K.A. The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: A review. Eur. J. Plant Pathol. 2022, 162, 759–792. [Google Scholar] [CrossRef]
- DeGenring, L.; Poleatewich, A. The Effect of Tomato Cultivar on Pythium Root Rot and Efficacy of Biopesticides. PhytoFrontiers™ 2023, 3, 594–601. [Google Scholar] [CrossRef]
- Michaud, I. The Effect of Wood-Fiber Substrates on Natural Disease Suppression and the Efficacy of Rootshield® Wp against Rhizoctonia solani. Master’s Thesis, University of New Hampshire, Durham, NH, USA, 2021. [Google Scholar]
- Braun, R.C.; Braithwaite, E.T.; Kowalewski, A.R.; Watkins, E.; Hollman, A.B.; Patton, A.J. Nitrogen fertilizer and clover inclusion effects on the establishment of fine fescue taxa. Crop Sci. 2022, 62, 947–957. [Google Scholar] [CrossRef]
- Arthurs, S.; Dara, S.K. Microbial biopesticides for invertebrate pests and their markets in the United States. J. Invertebr. Pathol. 2019, 165, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.; Hartung, J.; Perkons, U.; Möller, K.; Zikeli, S. Plant and soil N of different winter cover crops as green manure for subsequent organic white cabbage. Nutr. Cycl. Agroecosystems 2023, 127, 285–298. [Google Scholar] [CrossRef]
- Radwan, E.M.; Taha, H.S. Effects of dipel 2x and littovir bioinsecticides on larvae of the cotton leafworm Spodoptera littoralis (Lepidoptera: Noctuidae). J. Plant Prot. Res. Inst. 2020, 3, 743–760. [Google Scholar]
- Stockwell, V.; Johnson, K.; Johnson, V.W. Colonization of flowers by Pseudomonas fluorescens A506 formulated in a biopolymer gel. In Proceedings of the X International Workshop on Fire Blight, Bologna, Italy, 5–9 July 2004; Volume 704, pp. 293–300. [Google Scholar]
- Becker, M.F.; Deichmann, M.; Klueken, A.M.; Knief, C. Effects of plant health protecting product applications on the root-associated microbiota of apple saplings and strawberries. Phytobiomes J. 2024, 9, 189–202. [Google Scholar] [CrossRef]
- Marine, S.C.; Newark, M.J.; Korir, R.C.; Everts, K.L. Evaluation of rotational biopesticide programs for disease management in organic cucurbit production. Plant Dis. 2016, 100, 2226–2233. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.A. Efficacy of Biofungicides and Their Application Timing in Managing Botrytis Bunch Rot of Wine Grape. Master’s Thesis, California Polytechnic State University, San Luis Obispo, CA, USA, 2023. [Google Scholar]
- Karise, R.; Muljar, R.; Smagghe, G.; Kaart, T.; Kuusik, A.; Dreyersdorff, G.; Williams, I.H.; Mänd, M. Sublethal effects of kaolin and the biopesticides Prestop-Mix and BotaniGard on metabolic rate, water loss and longevity in bumble bees (Bombus terrestris). J. Pest. Sci. 2016, 89, 171–178. [Google Scholar] [CrossRef]
- Jensen, J.P.; Kalwa, U.; Pandey, S.; Tylka, G.L. Avicta and clariva affect the biology of the soybean cyst nematode, Heterodera glycines. Plant Dis. 2018, 102, 2480–2486. [Google Scholar] [CrossRef]
- Pethybridge, S.J.; Gugino, B.K.; Kikkert, J.R. Efficacy of Double Nickel LC (Bacillus amyloliquefaciens D747 Strain) for management of white mold in snap and dry bean. Plant Health Prog. 2019, 20, 61–66. [Google Scholar] [CrossRef]
- Sullivan, C.F.; Parker, B.L.; Davari, A.; Lee, M.R.; Kim, J.S.; Skinner, M. Pathogenicity of Metarhizium anisopliae and Metarhizium brunneum isolates and efficacy of Met52 G against winter tick larvae, 2019. Arthropod Manag. Tests 2020, 45, tsaa100. [Google Scholar] [CrossRef]
- Nwachukwu, B.C.; Babalola, O.O. Perspectives for sustainable agriculture from the microbiome in plant rhizosphere. Plant Biotechnol. Rep. 2021, 15, 1–20. [Google Scholar] [CrossRef]
- Anklam, E.; Bahl, M.I.; Ball, R.; Beger, R.D.; Cohen, J.; Fitzpatrick, S.; Girard, P.; Halamoda-Kenzaoui, B.; Hinton, D.; Hirose, A. Emerging technologies and their impact on regulatory science. Exp. Biol. Med. 2022, 247, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.; Szymanski, E. Who Knows What a Microbe is?: The Variable Texture of Microbial Identity in Agricultural Products, Regulations, and Fields. Sci. Technol. Stud. 2023, 37, 21–39. [Google Scholar] [CrossRef]
- Havelaar, A.H.; Brul, S.; De Jong, A.; De Jonge, R.; Zwietering, M.H.; Ter Kuile, B.H. Future challenges to microbial food safety. Int. J. Food Microbiol. 2010, 139, S79–S94. [Google Scholar] [CrossRef]
- Ghorui, M.; Chowdhury, S.; Balu, P.; Burla, S. Arbuscular Mycorrhizal inoculants and its regulatory landscape. Heliyon 2024, 10, e30359. [Google Scholar] [CrossRef]
- Huttenhower, C.; Finn, R.; McHardy, A. Challenges and opportuni ties in sharing microbiome data and analyses. Nat. Microbiol. 2023, 8, 1960–1970. [Google Scholar] [CrossRef]
- Knoppers, T. Protecting Privacy in the Postgenomic Era: Ensuring Responsible Data Governance by Epigenetic; McGill University: Montreal, QC, Canada, 2020. [Google Scholar]
- Jansson, J.K.; McClure, R.; Egbert, R.G. Soil microbiome engineering for sustainability in a changing environment. Nat. Biotechnol. 2023, 41, 1716–1728. [Google Scholar] [CrossRef]
- Noman, M.; Ahmed, T.; Ijaz, U.; Shahid, M.; Azizullah; Li, D.; Manzoor, I.; Song, F. Plant–Microbiome crosstalk: Dawning from composition and assembly of microbial community to improvement of disease resilience in plants. Int. J. Mol. Sci. 2021, 22, 6852. [Google Scholar] [CrossRef]
- Priyadarshini, P.; Abhilash, P.C. Policy recommendations for enabling transition towards sustainable agriculture in India. Land Use Policy 2020, 96, 104718. [Google Scholar] [CrossRef]
- Schelkle, B.; Galland, Q. Microbiome research: Open communication today, microbiome applications in the future. Microorganisms 2020, 8, 1960. [Google Scholar] [CrossRef] [PubMed]
- Ratner, R.K.; Riis, J. Communicating science-based recommendations with memorable and actionable guidelines. Proc. Natl. Acad. Sci. USA 2014, 111, 13634–13641. [Google Scholar] [CrossRef] [PubMed]
- McGloin, A.F.; Eslami, S. Digital and social media opportunities for dietary behaviour change. Proc. Nutr. Soc. 2015, 74, 139–148. [Google Scholar] [CrossRef]
- Warner, K.D. 22 Understanding Public Risk Perceptio n for the Use of Beneficial Microorganisms. In Beneficial Microorganisms in Agriculture, Food and the Environment: Safety Assessment and Regulation; CABI: Oxfordshire, UK, 2012; p. 322. [Google Scholar]
- Trivedi, P.; Mattupalli, C.; Eversole, K.; Leach, J.E. Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 2021, 230, 2129–2147. [Google Scholar] [CrossRef]
- Adenle, A.A.; Wedig, K.; Azadi, H. Sustainable agriculture and food security in Africa: The role of innovative technologies and international organizations. Technol. Soc. 2019, 58, 101143. [Google Scholar] [CrossRef]
- Bano, S.; Wu, X.; Zhang, X. Towards sustainable agriculture: Rhizosphere microbiome engineering. Appl. Microbiol. Biotechnol. 2021, 105, 7141–7160. [Google Scholar] [CrossRef]
- Andreasen, L.; Collison, M.; Couco, E.; de Porras Acuna, M.A.; Früh, B.; Gernert, M.; Haller, L.; Melby Jespersen, L.; Keijzer, P.; Kongsted, A.G. Strategic Research & Innovation Agenda for Organics and Agroecology; FiBL: Brussels, Belgium, 2019. [Google Scholar]
- Thakur, N.; Nigam, M.; Mann, N.A.; Gupta, S.; Hussain, C.M.; Shukla, S.K.; Shah, A.A.; Casini, R.; Elansary, H.O.; Khan, S.A. Host-mediated gene engineering and microbiome-based technology optimization for sustainable agriculture and environment. Funct. Integr. Genom. 2023, 23, 57. [Google Scholar] [CrossRef] [PubMed]
- Harman, G.; Khadka, R.; Doni, F.; Uphoff, N. Benefits to plant health and productivity from enhancing plant microbial symbionts. Front. Plant Sci. 2021, 11, 610065. [Google Scholar] [CrossRef]
- Nwachukwu, B.C.; Ayangbenro, A.S.; Babalola, O.O. Elucidating the rhizosphere associated bacteria for environmental sustainability. Agriculture 2021, 11, 75. [Google Scholar] [CrossRef]
- Samantaray, A.; Chattaraj, S.; Mitra, D.; Ganguly, A.; Kumar, R.; Gaur, A.; Mohapatra, P.K.D.; de los Santos-Villalobos, S.; Rani, A.; Thatoi, H. Advances in microbial based bio-inoculum for amelioration of soil health and sustainable crop production. Curr. Res. Microb. Sci. 2024, 7, 100251. [Google Scholar] [CrossRef] [PubMed]
- Saiz, A. Negotiating ROI (Return on Investment) for ROI (Return on Impact) A Pre-Feasibility Study of Socio-Eco Resort Development in Eastern Indonesia. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 2024. [Google Scholar]
- Qiu, Z.; Egidi, E.; Liu, H.; Kaur, S.; Singh, B.K. New frontiers in agriculture productivity: Optimised microbial inoculants and in situ microbiome engineering. Biotechnol. Adv. 2019, 37, 107371. [Google Scholar] [CrossRef] [PubMed]
- Dukare, A.; Paul, S.; Kumar, R.; Sharma, V. Microbial-based inoculants in sustainable agriculture: Current perspectives and future prospects. In Biofertilizers; Elsevier: Amsterdam, The Netherlands, 2021; pp. 167–181. [Google Scholar]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.-C. Perspectives and challenges of microbial application for crop improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef]
- Durham, T.C.; Mizik, T. Comparative economics of conventional, organic, and alternative agricultural production systems. Economies 2021, 9, 64. [Google Scholar] [CrossRef]
- Tahat, M.M.; Alananbeh, K.M.; Othman, Y.A.; Leskovar, D.I. Soil health and sustainable agriculture. Sustainability 2020, 12, 4859. [Google Scholar] [CrossRef]
- Frazier, A.N. Application of Biotechnology in Agriculture: A Bioinformatic and Market Analysis of Novel Intervention Methods. Ph.D. Thesis, Colorado State University, Fort Collins, CO, USA, 2021. [Google Scholar]
- Makate, C. Effective scaling of climate smart agriculture innovations in African smallholder agriculture: A review of approaches, policy and institutional strategy needs. Environ. Sci. Policy 2019, 96, 37–51. [Google Scholar] [CrossRef]
- Mahmud, K.; Missaoui, A.; Lee, K.; Ghimire, B.; Presley, H.W.; Makaju, S. Rhizosphere microbiome manipulation for sustainable crop production. Curr. Plant Biol. 2021, 27, 100210. [Google Scholar] [CrossRef]
- Callens, K.; Fontaine, F.; Sanz, Y.; Bogdanski, A.; D ‘Hondt, K.; Lange, L.; Smidt, H.; Van Overbeek, L.; Kostic, T.; Maguin, E. Microbiome-based solutions to address new and existing threats to food security, nutrition, health and agrifood systems’ sustainability. Front. Sustain. Food Syst. 2022, 6, 1047765. [Google Scholar] [CrossRef]
- Thiele, G.; Devaux, A.; Reinoso, I.; Pico, H.; Montesdeoca, F.; Pumisacho, M.; Andrade-Piedra, J.; Velasco, C.; Flores, P.; Esprella, R. Multi-stakeholder platforms for linking small farmers to value chains: Evidence from the Andes. Int. J. Agric. Sustain. 2011, 9, 423–433. [Google Scholar] [CrossRef]
- Nguyen, T.H.N.; Yeh, Q.J.; Huang, C.Y. Understanding consumer’switching intention toward traceable agricultural products: Push-pull-mooring perspective. Int. J. Consum. Stud. 2022, 46, 870–888. [Google Scholar] [CrossRef]
- Mitter, E.K.; Tosi, M.; Obregón, D.; Dunfield, K.E.; Germida, J.J. Rethinking crop nutrition in times of modern microbiology: Innovative biofertilizer technologies. Front. Sustain. Food Syst. 2021, 5, 606815. [Google Scholar] [CrossRef]
- Singh, B.K.; Trivedi, P.; Egidi, E.; Macdonald, C.A.; Delgado-Baquerizo, M. Crop microbiome and sustainable agriculture. Nat. Rev. Microbiol. 2020, 18, 601–602. [Google Scholar] [CrossRef]
- Peng, B.; Guan, K.; Tang, J.; Ainsworth, E.A.; Asseng, S.; Bernacchi, C.J.; Cooper, M.; Delucia, E.H.; Elliott, J.W.; Ewert, F. Towards a multiscale crop modelling framework for climate change adaptation assessment. Nat. Plants 2020, 6, 338–348. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Babalola, O.O.; Santoyo, G.; Perazzolli, M. The potential role of microbial biostimulants in the amelioration of climate change-associated abiotic stresses on crops. Front. Microbiol. 2022, 12, 829099. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Xiong, C.; Egidi, E.; Singh, B.K. Formulation challenges associated with microbial biofertilizers in sustainable agriculture and paths forward. J. Sustain. Agric. Environ. 2024, 3, e70006. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Saad, A.M.; Soliman, S.M.; Salem, H.M.; Ahmed, A.I.; Mahmood, M.; El-Tahan, A.M.; Ebrahim, A.A.; Abd El-Mageed, T.A.; Negm, S.H. Plant growth-promoting microorganisms as biocontrol agents of plant diseases: Mechanisms, challenges and future perspectives. Front. Plant Sci. 2022, 13, 923880. [Google Scholar] [CrossRef]
- Das, P.P.; Singh, K.R.; Nagpure, G.; Mansoori, A.; Singh, R.P.; Ghazi, I.A.; Kumar, A.; Singh, J. Plant-soil-microbes: A tripartite interaction for nutrient acquisition and better plant growth for sustainable agricultural practices. Environ. Res. 2022, 214, 113821. [Google Scholar] [CrossRef] [PubMed]
| Product Name | Company Name | Microbial Species | Uses | References |
|---|---|---|---|---|
| RootShield | BioWorks, Victor, NY, USA | Trichoderma harzianum | Control plant diseases and growth promotion | DeGenring and Poleatewich [207,208] |
| N-Dure | Verdesian Life Sciences, Cary, NC, USA | Various Bradyrhizobium and Rhizobium strains | Used as bioinoculants for legumes to improve nitrogen fixation | Braun, et al. [209] |
| XenTari | Biofa AG, Münsingen, Germany | Bacillus thuringiensis | Different strains of Bacillus thuringiensis are employed as pesticides | Arthurs and Dara [210], Stein et al. [211] |
| Dipel | Valent BioSciences, Libertyville, IL, USA | Bacillus thuringiensis | Widely used as biopesticides | Radwan and Taha [212] |
| BlightBan A506 | Nufarm, Melbourne, Australia | Pseudomonas fluorescens | Control phytopathogens and plant growth promotion | Stockwell, et al. [213] |
| Serenade | Bayer, Leverkusen, Germany | Bacillus subtilis | Suppress plant disease and boost plant growth | Becker, et al. [214] |
| Actinovate | Novozymes, Bagsværd, Denmark | Streptomyces lydicus | Manage diseases in a variety of crops | Marine, et al. [215,216] |
| BotaniGard | BioWorks, Victor, NY, USA | Beauveria bassiana | Entomopathogenic fungus used for insect management | Karise, et al. [217] |
| Clariva | Syngenta, Basel, Switzerland | Pasteuria nishizawae | P. nishizawae spores are used to control nematode | Jensen, et al. [218] |
| Double Nickel 55 | Certis USA, Columbia, MD, USA | Bacillus amyloliquefaciens | Used to control plant diseases and improve plant growth | Pethybridge, et al. [219] |
| Met52 | Novozymes, Bagsværd, Denmark | Metarhizium anisopliae | Entomopathogenic fungus employed for insect control | Sullivan, et al. [220] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fadiji, A.E.; Adeniji, A.; Lanrewaju, A.A.; Adedayo, A.A.; Chukwuneme, C.F.; Nwachukwu, B.C.; Aderibigbe, J.; Omomowo, I.O. Key Challenges in Plant Microbiome Research in the Next Decade. Microorganisms 2025, 13, 2546. https://doi.org/10.3390/microorganisms13112546
Fadiji AE, Adeniji A, Lanrewaju AA, Adedayo AA, Chukwuneme CF, Nwachukwu BC, Aderibigbe J, Omomowo IO. Key Challenges in Plant Microbiome Research in the Next Decade. Microorganisms. 2025; 13(11):2546. https://doi.org/10.3390/microorganisms13112546
Chicago/Turabian StyleFadiji, Ayomide Emmanuel, Adegboyega Adeniji, Adedayo Ayodeji Lanrewaju, Afeez Adesina Adedayo, Chinenyenwa Fortune Chukwuneme, Blessing Chidinma Nwachukwu, Joshua Aderibigbe, and Iyabo Olunike Omomowo. 2025. "Key Challenges in Plant Microbiome Research in the Next Decade" Microorganisms 13, no. 11: 2546. https://doi.org/10.3390/microorganisms13112546
APA StyleFadiji, A. E., Adeniji, A., Lanrewaju, A. A., Adedayo, A. A., Chukwuneme, C. F., Nwachukwu, B. C., Aderibigbe, J., & Omomowo, I. O. (2025). Key Challenges in Plant Microbiome Research in the Next Decade. Microorganisms, 13(11), 2546. https://doi.org/10.3390/microorganisms13112546









