Integrated Assessment of Benthic Bacterial Community Physiology, Structure, and Function Across C, N, P, and S Gradients in Lake Villarrica Sediments, Chile
Abstract
1. Introduction
2. Materials and Methods
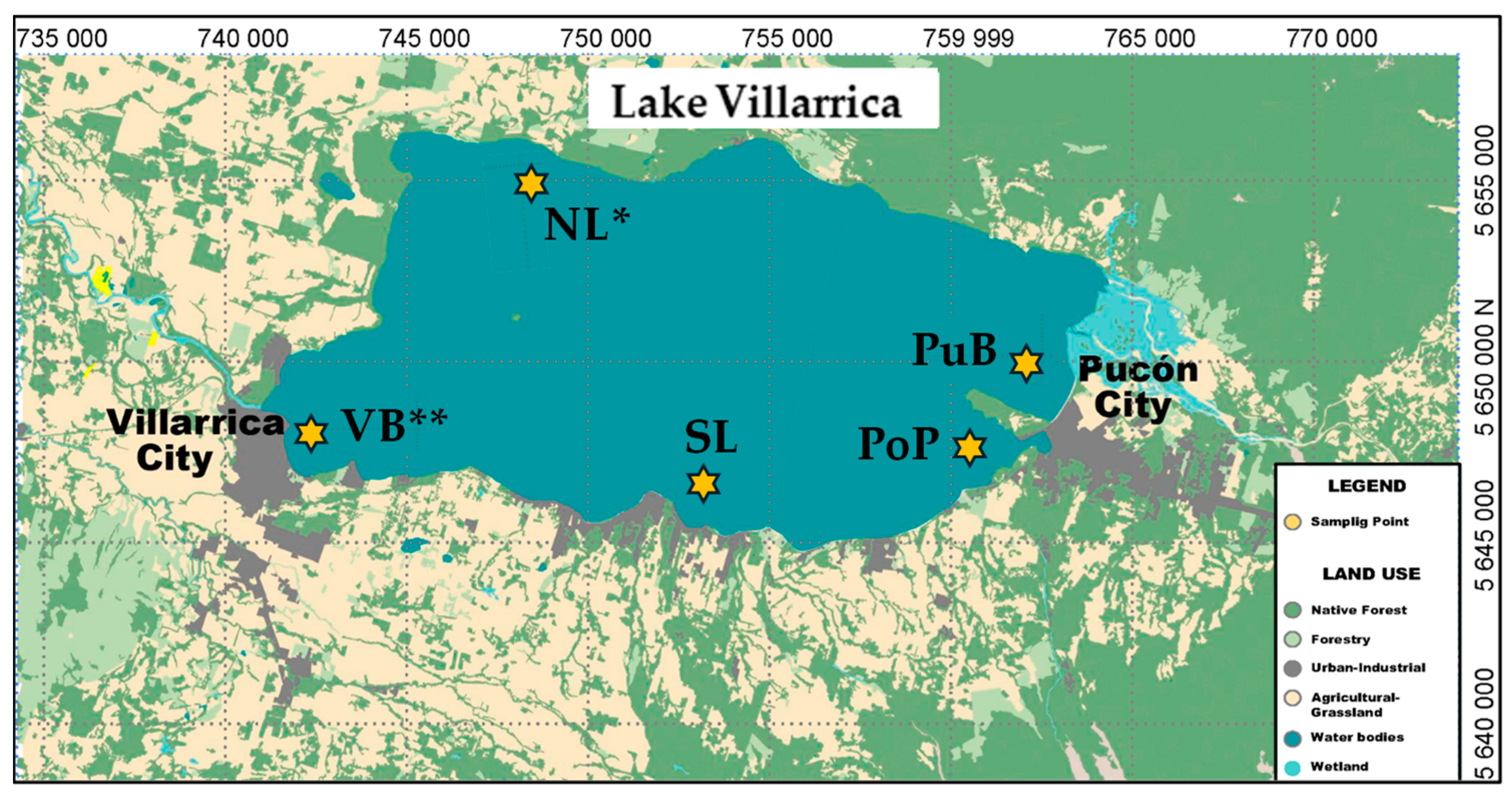
2.1. Lake Description
2.2. Sediment Sampling and Characterization of In-Situ Physicochemical Properties
2.3. Nutrient Content of Sediments
2.4. Community-Level Physiological Profile (CLPP) of Bacterial Communities
2.5. Genomic DNA Extraction
2.6. Absolute Quantification of Total Bacterial Community and Nutrient-Cycling Functional Genes
2.7. Assessment of Bacterial Community Composition, Nutrient Content, and Functional Gene Abundance
2.8. Statistical Analysis
3. Results
3.1. Physicochemical Parameters and Nutrient Contents
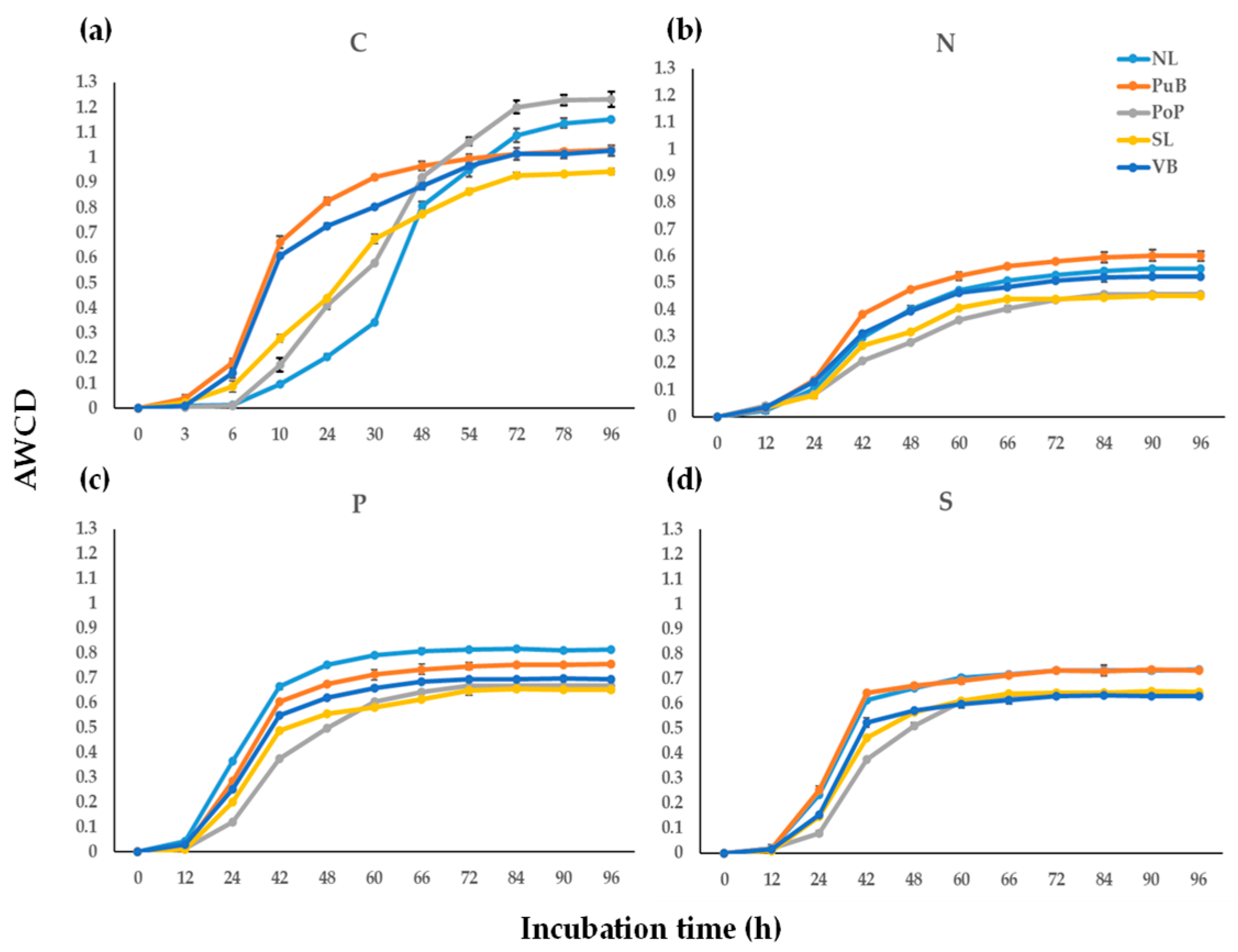
3.2. Community-Level Physiological Profile (CLPP) Responses During Sediment Incubations
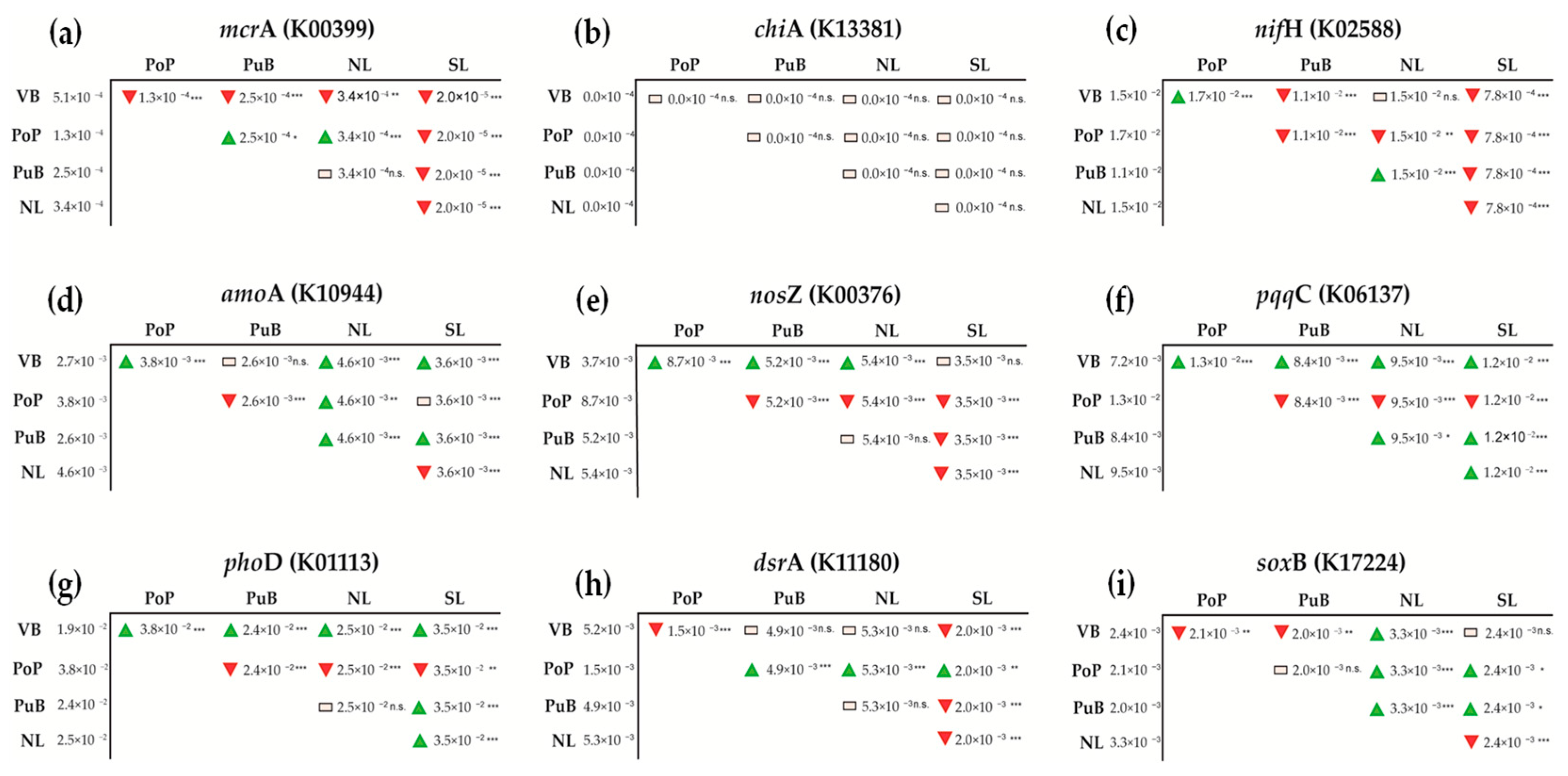
3.3. Variation of Total Bacterial Community and Functional Nutrient-Cycling Genes
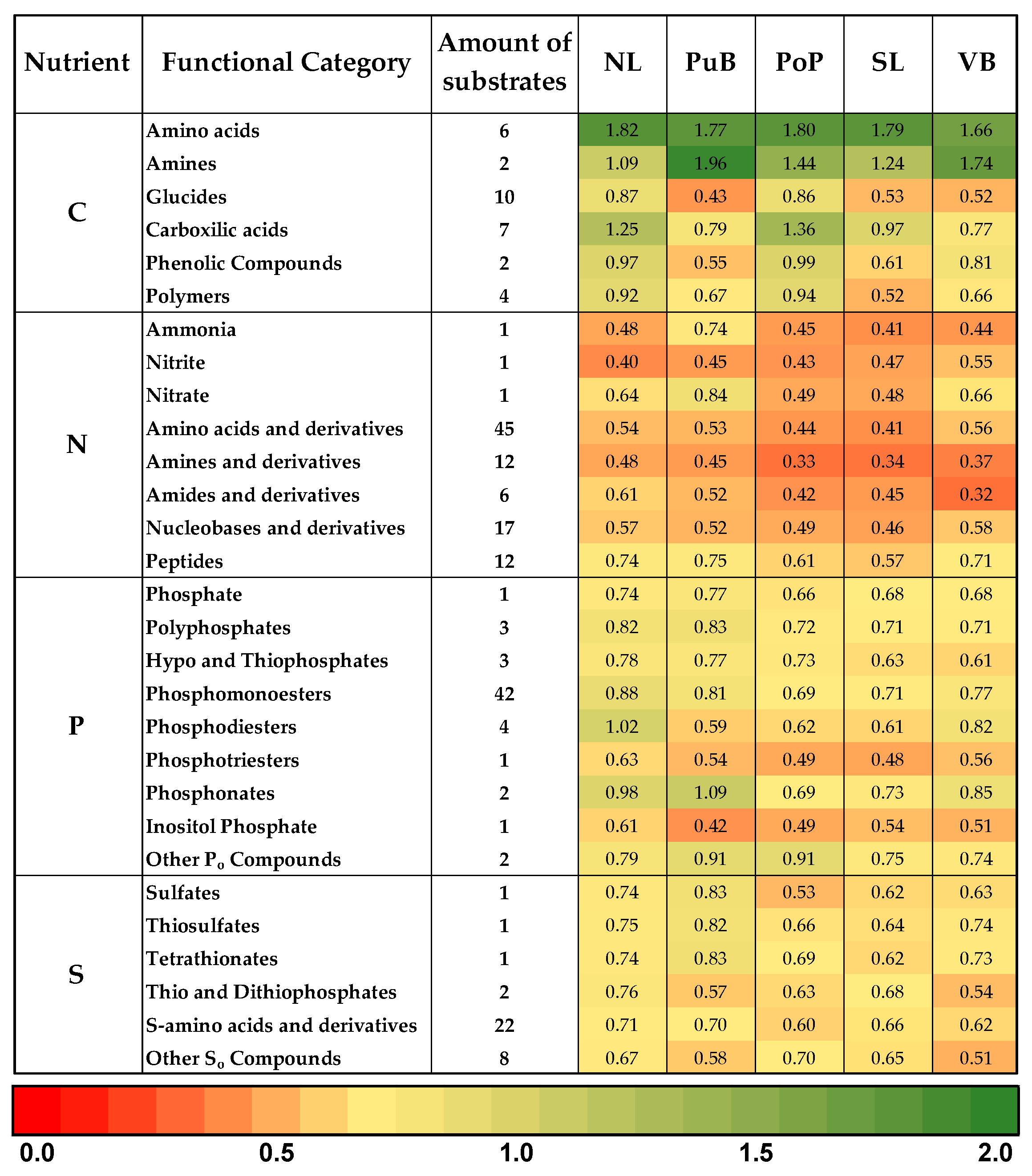
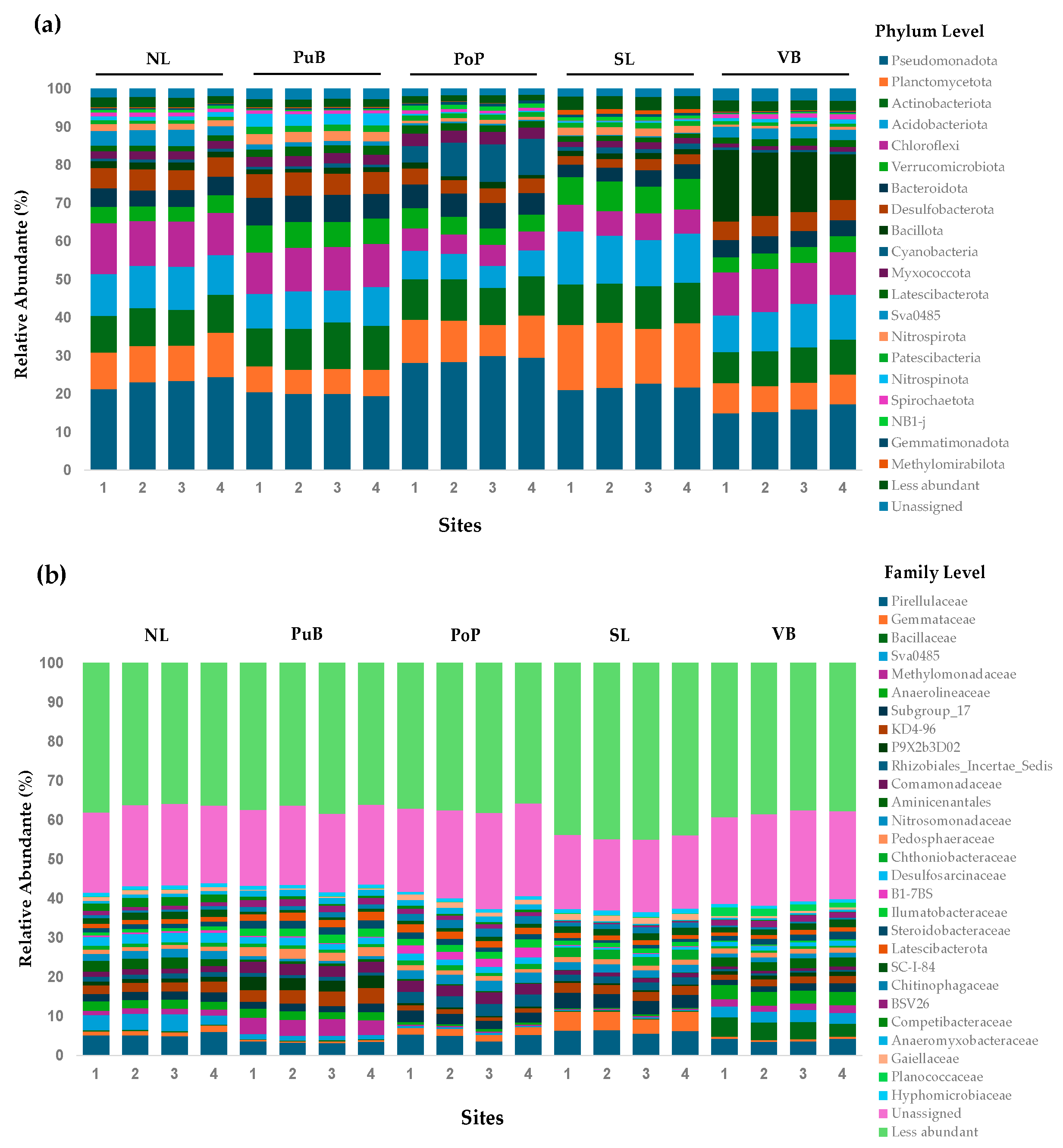
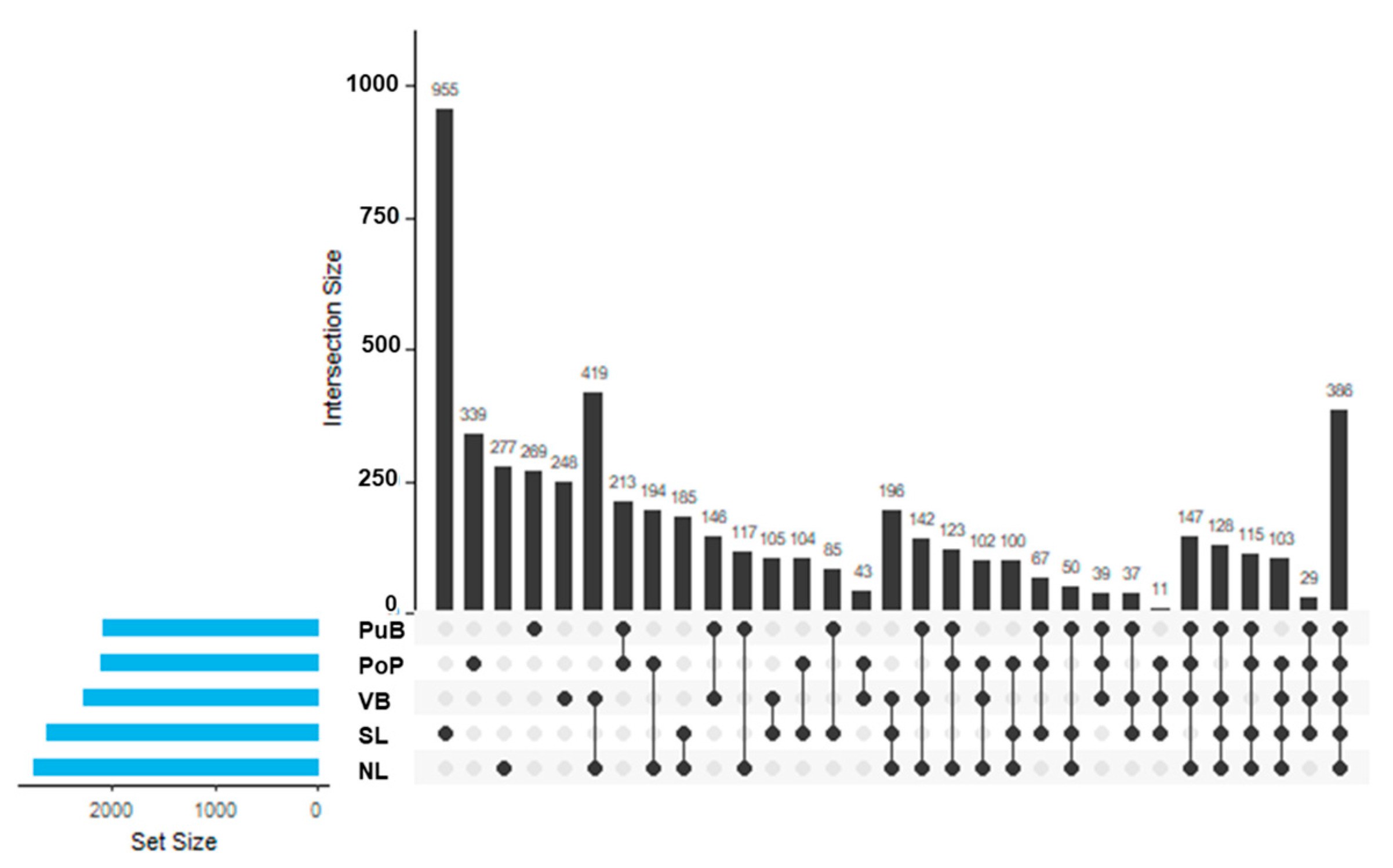
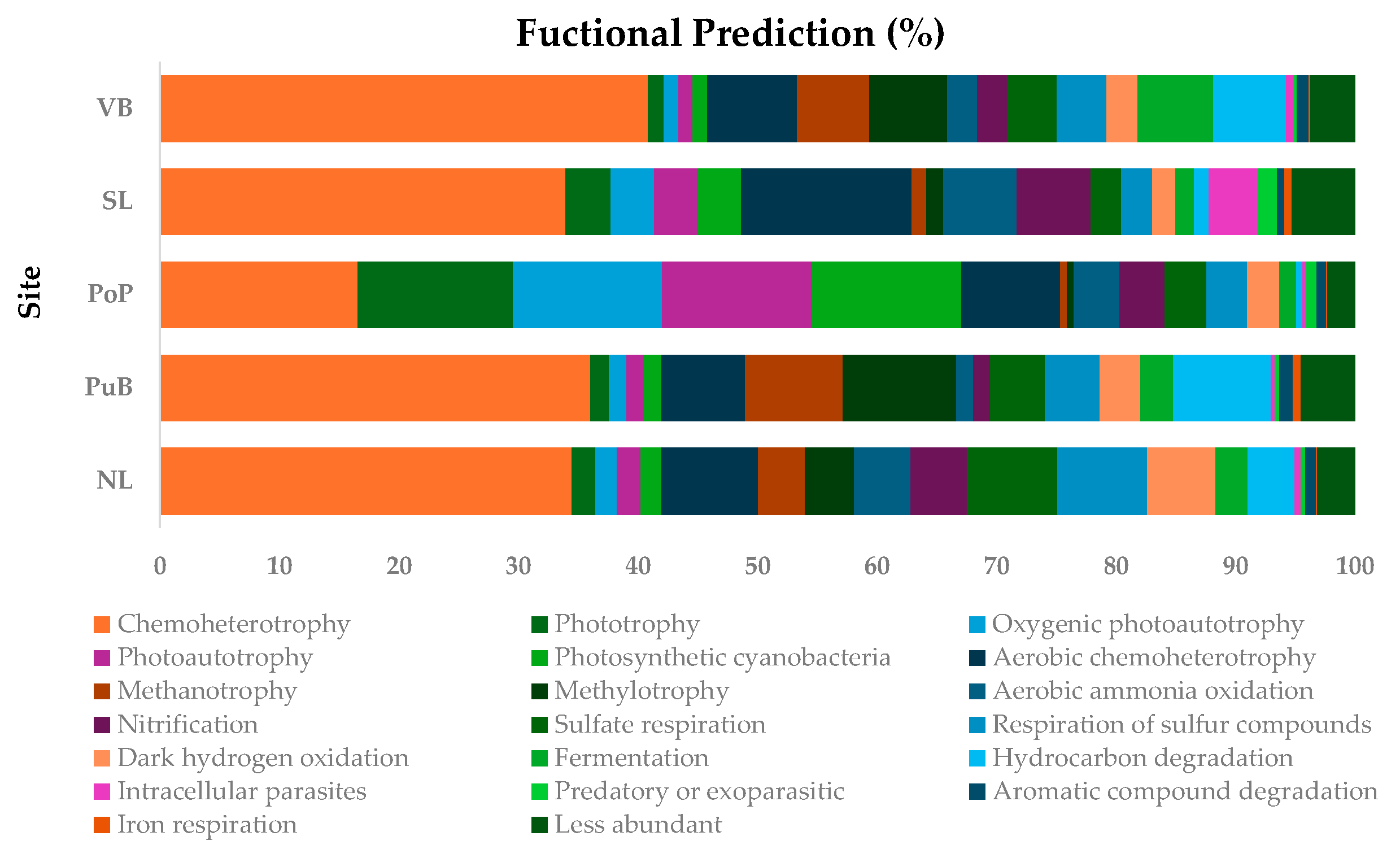
3.4. Changes in Bacterial Community Composition and Functionality, and the Relationship of Nutrient Content with Specific Functional-Cycling-Gene Abundances Among Sites
4. Discussion
4.1. Physicochemical Status and Nutrient Contents Among Sites
4.2. Relationships and Differences of Community-Level Physiological Profile (CLPP) in Sediment Sites
4.3. Total Bacterial Community and Functional Nutrient-Cycling Gene
4.4. Relationship of Bacterial Community Composition and Their Predicted Functionality, and Nutrient Content and Specific Functional-Cycling-Gene Abundances
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reinl, K.L.; Harris, T.D.; Elfferich, I.; Coker, A.; Zhan, Q.; De Senerpont Domis, L.N.; Morales-Williams, A.M.; Bhattacharya, R.; Grossart, H.P.; North, R.L.; et al. The Role of Organic Nutrients in Structuring Freshwater Phytoplankton Communities in a Rapidly Changing World. Water Res. 2022, 219, 118573. [Google Scholar] [CrossRef]
- Demin, K.A.; Prazdnova, E.V.; Minkina, T.M.; Gorovtsov, A.V.; Demin, K.A.; Prazdnova, E.V.; Minkina, T.M.; Gorovtsov, A.V. Diverse Prokaryotic Group in Terrestrial Environments. Appl. Environ. Microbiol. 2024, 90, e01390-23. [Google Scholar] [CrossRef]
- Tranvik, L.J.; Downing, J.A.; Cotner, J.B.; Loiselle, S.A.; Striegl, R.G.; Ballatore, T.J.; Dillon, P.; Finlay, K.; Fortino, K.; Knoll, L.B.; et al. Lakes Reservoirs as Regulators of Carbon Cycling and Climate. Limnol. Oceanogr. 2009, 54, 2298–2314. [Google Scholar] [CrossRef]
- Zakem, E.J.; Polz, M.F.; Follows, M.J. Redox-Informed Models of Global Biogeochemical Cycles. Nat. Commun. 2020, 11, 5680. [Google Scholar] [CrossRef]
- Campos, M.; Acuña, J.J.; Rilling, J.I.; González–González, S.; Peña–Cortés, F.; Jaisi, D.P.; Hollenback, A.; Ogram, A.; Bai, J.; Zhang, L.; et al. Spatiotemporal Distributions and Relationships of Phosphorus Content, Phosphomonoesterase Activity, and Bacterial Phosphomonoesterase Genes in Sediments from a Eutrophic Brackish Water Lake in Chile. J. Environ. Manag. 2022, 320, 115906. [Google Scholar] [CrossRef] [PubMed]
- Hessen, D.O.; Ågren, G.I.; Anderson, T.R.; Elser, J.J.; De Ruiter, P.C. Carbon Sequestration in Ecosystems: The Role of Stoichiometry. Ecology 2004, 85, 1179–1192. [Google Scholar] [CrossRef]
- Bao, K.; Zhang, Y.; Zaccone, C.; Meadows, M.E. Human Impact on C/N/P Accumulation in Lake Sediments from Northeast China during the Last 150 Years. Environ. Pollut. 2021, 271, 116345. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Paerl, H.W.; Brookes, J.D.; Liu, J.; Jeppesen, E.; Zhu, G.; Zhang, Y.; Xu, H.; Shi, K.; Deng, J. Why Lake Taihu Continues to Be Plagued with Cyanobacterial Blooms through 10 years (2007–2017) Efforts. Sci. Bull. 2019, 64, 354–356. [Google Scholar] [CrossRef]
- Wang, Z.; Ruan, X.; Li, R.; Zhang, Y. Microbial Interaction Patterns and Nitrogen Cycling Regularities in Lake Sediments under Different Trophic Conditions. Sci. Total Environ. 2024, 907, 167926. [Google Scholar] [CrossRef]
- Wrage-Mönnig, N.; Horn, M.A.; Well, R.; Müller, C.; Velthof, G.; Oenema, O. The Role of Nitrifier Denitrification in the Production of Nitrous Oxide Revisited. Soil Biol. Biochem. 2018, 123, A3–A16. [Google Scholar] [CrossRef]
- Anderson, H.S.; Johengen, T.H.; Godwin, C.M.; Purcell, H.; Alsip, P.J.; Ruberg, S.A.; Mason, L.A. Continuous In Situ Nutrient Analyzers Pinpoint the Onset and Rate of Internal P Loading under Anoxia in Lake Erie’s Central Basin. ACS EST Water 2021, 1, 774–781. [Google Scholar] [CrossRef]
- Rohwer, R.R.; Ladwig, R.; Hanson, P.C.; Walsh, J.R.; Vander Zanden, M.J.; Dugan, H.A. Increased Anoxia Following Species Invasion of a Eutrophic Lake. Limnol. Oceanogr. Lett. 2024, 9, 33–42. [Google Scholar] [CrossRef]
- Missimer, T.M.; Thomas, S.; Rosen, B.H. Legacy Phosphorus in Lake Okeechobee (Florida, USA) Sediments: A Review and New Perspective. Water 2021, 13, 39. [Google Scholar] [CrossRef]
- Wassenaar, L.I. Dissolved Oxygen Status of Lake Winnipeg: Spatio-Temporal and Isotopic (δ 18O-O2) Patterns. J. Great Lakes Res. 2012, 38, 123–134. [Google Scholar] [CrossRef]
- Steinman, A.D.; Spears, B.M. (Eds.) Internal Phosphorus Loading in Lakes: Causes, Case Studies, and Management; J. Ross Publishing: Plantation, FL, USA, 2020; ISBN 9781604271447. [Google Scholar]
- Schultz, P.; Urban, N.R. Effects of Bacterial Dynamics on Organic Matter Decomposition and Nutrient Release from Sediments: A Modeling Study. Ecol. Modell. 2008, 210, 1–14. [Google Scholar] [CrossRef]
- Burgin, A.J.; Hamilton, S.K.; Jones, S.E.; Lennon, J.T. Denitrification by Sulfur-Oxidizing Bacteria in a Eutrophic Lake. Aquat. Microb. Ecol. 2012, 66, 283–293. [Google Scholar] [CrossRef]
- Cojean, A.N.Y.; Lehmann, M.F.; Robertson, E.K.; Thamdrup, B.; Zopfi, J. Controls of H2S, Fe2+, and Mn2+ on Microbial NO3–-Reducing Processes in Sediments of an Eutrophic Lake. Front. Microbiol. 2020, 11, 1158. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, H.; Zhang, T.; Zhou, Y.; Dong, A.; Huang, R.; Zeng, Q.; Yuan, H. Seasonal Variation Regulate the Endogenous Phosphorus Release in Sediments of Shijiuhu Lake via Water-Level Fluctuation. Environ. Res. 2023, 238, 117247. [Google Scholar] [CrossRef]
- Burgin, A.J.; Yang, W.H.; Hamilton, S.K.; Silver, W.L. Beyond Carbon and Nitrogen: How the Microbial Energy Economy Couples Elemental Cycles in Diverse Ecosystems. Front. Ecol. Environ. 2011, 9, 44–52. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, Z.; Sun, Q.; Ding, Y.; Ding, Z. Response of Chemical Properties, Microbial Community Structure and Functional Genes Abundance to Seasonal Variations and Human Disturbance in Nanfei River Sediments. Ecotoxicol. Environ. Saf. 2019, 183, 109601. [Google Scholar] [CrossRef]
- Sofo, A.; Ricciuti, P. A Standardized Method for Estimating the Functional Diversity of Soil Bacterial Community by Biolog® EcoPlatesTM Assay-The Case Study of a Sustainable Olive Orchard. Appl. Sci. 2019, 9, 4035. [Google Scholar] [CrossRef]
- Gryta, A.; Frąc, M.; Oszust, K. The Application of the Biolog EcoPlate Approach in Ecotoxicological Evaluation of Dairy Sewage Sludge. Appl. Biochem. Biotechnol. 2014, 174, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Teng, Z.; Fan, W.; Wang, H.; Cao, X.; Xu, X. Monitoring Soil Microorganisms with Community-Level Physiological Profiles Using Biolog EcoPlatesTM in Chaohu Lakeside Wetland, East China. Eurasian Soil Sci. 2020, 53, 1142–1153. [Google Scholar] [CrossRef]
- Rivera, M.; Sepúlveda, L.; Silva, C. Climate Change and Its Influence on Tourism Fluctuation in the Araucania Region of Chile. Interam. J. Environ. Tour. 2022, 18, 118–136. [Google Scholar] [CrossRef]
- Rodríguez-López, L.; Duran-Llacer, I.; Bravo Alvarez, L.; Lami, A.; Urrutia, R. Recovery of Water Quality and Detection of Algal Blooms in Lake Villarrica through Landsat Satellite Images and Monitoring Data. Remote Sens. 2023, 15, 1929. [Google Scholar] [CrossRef]
- División de Fiscalización de la Superintendencia del Medio Ambiente (SMA). Informe Técnico de Cumplimiento de Normas de Calidad Del Agua, Periodo 2014-2015. Expediente SNIFA N° DFZ-2016-4695-IX-NC-EI; Superintendencia del Medio Ambiente—Gobierno de Chile: Santiago, Chile, 2017. [Google Scholar]
- Nimptsch, J.; Woelfl, S.; Osorio, S.; Valenzuela, J.; Moreira, C.; Ramos, V.; Castelo-Branco, R.; Leão, P.N.; Vasconcelos, V. First Record of Toxins Associated with Cyanobacterial Blooms in Oligotrophic North Patagonian Lakes of Chile—A Genomic Approach. Int. Rev. Hydrobiol. 2016, 101, 57–68. [Google Scholar] [CrossRef]
- Almanza, V.; Pedreros, P.; Dail Laughinghouse, H.; Félez, J.; Parra, O.; Azócar, M.; Urrutia, R. Association between Trophic State, Watershed Use, and Blooms of Cyanobacteria in South-Central Chile. Limnologica 2019, 75, 30–41. [Google Scholar] [CrossRef]
- De los Rios-Escalante, P.; Contreras, A.; Lara, G.; Latsague, M.; Esse, C. First Reports of Associations between Spectral Properties, Chlorophyll, Bacterial and Zooplankton in Two Chilean North Patagonian Lakes (Villarrica and Caburgua, 38° S, Araucania Region, Chile). J. King Saud Univ.—Sci. 2020, 32, 3167–3173. [Google Scholar] [CrossRef]
- Campos, M.A.; Zhang, Q.; Acuña, J.J.; Rilling, J.I.; Ruiz, T.; Carrazana, E.; Reyno, C.; Hollenback, A.; Gray, K.; Jaisi, D.P.; et al. Structure and Functional Properties of Bacterial Communities in Surface Sediments of the Recently Declared Nutrient-Saturated Lake Villarrica in Southern Chile. Microb. Ecol. 2023, 86, 1513–1533. [Google Scholar] [CrossRef] [PubMed]
- Ministerio del Medio Ambiente (MMA). Evaluación de Medidas de Reducción de Nutrientes (Nitrógeno y Fósforo) para Incorporar al Plan de Descontaminación del Lago Villarrica; Temuco, Chile, 2019. Available online: https://planesynormas.mma.gob.cl/archivos/2020/proyectos/Informe_Ufro_Evaluacion_de_Medidas_de_N_y_P_como_Anexo_a_Minuta_Carga_Critica.pdf (accessed on 11 April 2025).
- Bruning, M.B. Estudio de Aporte de Carga de Nutrientes por Fuentes Contaminantes y Análisis de Escenarios de Descontaminación Mediante un Modelo de Calidad de Aguas en el Lago Villarrica; Santiago, Chile, 2018. Available online: https://repositorio.uchile.cl/bitstream/handle/2250/168255/Estudio-de-aporte-de-carga-de-nutrientes-por-fuentes-contaminantes-y-an%C3%A1lisis-de-escenarios-de.pdf?sequence=1 (accessed on 11 April 2025).
- Campos, M.; Rilling, J.I.; Acuña, J.J.; Valenzuela, T.; Larama, G.; Peña-cortés, F.; Ogram, A.; Jaisi, D.P.; Jorquera, M.A. Spatiotemporal Variations and Relationships of Phosphorus, Phosphomonoesterases, and Bacterial Communities in Sediments from Two Chilean Rivers. Sci. Total Environ. 2021, 776, 145782. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis; SSSA Book Series; Soil Science Society of America: Madison, WI, USA, 1996; pp. 961–1010. ISBN 9780891188667. [Google Scholar]
- Bremner, J.M. Nitrogen-Total. In Methods of Soil Analysis; SSSA Book Series; Soil Science Society of America: Madison, WI, USA, 1996; pp. 1085–1121. ISBN 9780891188667. [Google Scholar]
- U.S. Environmental Protection Agency (U.S. EPA). Method 3051A (SW-846): Microwave Assisted Acid Digestion of Sediments, Sludges, and Oils; U.S. Environmental Protection Agency (U.S. EPA): Washington, DC, USA, 2007; Volume 7. [Google Scholar]
- Garland, J.L.; Mills, A.L. Classification and Characterization of Heterotrophic Microbial Communities on the Basis of Patterns of Community-Level Sole-Carbon-Source Utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; McManus, P.S.; Handelsman, J. Unexpected Diversity during Community Succession in the Apple Flower Microbiome. mBio 2013, 4, 1–12. [Google Scholar] [CrossRef]
- Köllner, K.E.; Carstens, D.; Keller, E.; Vazquez, F.; Schubert, C.J.; Zeyer, J.; Bürgmann, H. Bacterial Chitin Hydrolysis in Two Lakes with Contrasting Trophic Statuses. Appl. Environ. Microbiol. 2012, 78, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Cisek, A.A.; Bąk, I.; Cukrowska, B. Improved Quantitative Real-Time PCR Protocol for Detection and Quantification of Methanogenic Archaea in Stool Samples. Microorganisms 2023, 11, 660. [Google Scholar] [CrossRef] [PubMed]
- Poly, F.; Monrozier, L.J.; Bally, R. Improvement in the RFLP Procedure for Studying the Diversity of nifH Genes in Communities of Nitrogen Fixers in Soil. Res. Microbiol. 2001, 152, 95–103. [Google Scholar] [CrossRef]
- Rotthauwe, J.-H.; Witzel, K.-P.; Liesack, W. The Ammonia Monooxygenase Structural Gene Amoa as a Functional Marker: Molecular Fine-Scale Analysis of Natural Ammonia-Oxidizing Populations. Microbiology 1997, 63, 4704–4712. [Google Scholar] [CrossRef]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative Detection of the nosZ Gene, Encoding Nitrous Oxide Reductase, and Comparison of the Abundances of 16S RRNA, narG, nirK, and nosZ Genes in Soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef]
- Sakurai, M.; Wasaki, J.; Tomizawa, Y.; Shinano, T.; Osaki, M. Analysis of Bacterial Communities on Alkaline Phosphatase Genes in Soil Supplied with Organic Matter. Soil Sci. Plant Nutr. 2008, 54, 62–71. [Google Scholar] [CrossRef]
- Zheng, B.; Hao, X.; Ding, K.; Zhou, G.; Chen, Q. Long-Term Nitrogen Fertilization Decreased the Abundance of Inorganic Phosphate Solubilizing Bacteria in an Alkaline Soil. Nat. Publ. Gr. 2017, 7, 42284. [Google Scholar] [CrossRef]
- Tourna, M.; Maclean, P.; Condron, L.; O’Callaghan, M.; Wakelin, S.A. Links between Sulphur Oxidation and Sulphur-Oxidising Bacteria Abundance and Diversity in Soil Microcosms Based on soxB Functional Gene Analysis. FEMS Microbiol. Ecol. 2014, 88, 538–549. [Google Scholar] [CrossRef]
- Carolan, M.T.; Smith, J.M.; Beman, J.M. Transcriptomic Evidence for Microbial Sulfur Cycling in the Eastern Tropical North Pacific Oxygen Minimum Zone. Front. Microbiol. 2015, 6, 334. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Duan, J.L.; Sun, J.W.; Ji, M.M.; Ma, Y.; Cui, Z.T.; Tian, R.K.; Xu, P.C.; Sun, W.L.; Yuan, X.Z. Indicatory Bacteria and Chemical Composition Related to Sulfur Distribution in the River-Lake Systems. Microbiol. Res. 2020, 236, 126453. [Google Scholar] [CrossRef]
- Zhang, L.; Shen, T.; Cheng, Y.; Zhao, T.; Li, L.; Qi, P. Temporal and Spatial Variations in the Bacterial Community Composition in Lake Bosten, a Large, Brackish Lake in China. Sci. Rep. 2020, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, T.; Wang, Q.; Li, L.; Shen, T.; Gao, G. Bacterial Community Composition in Aquatic and Sediment Samples with Spatiotemporal Dynamics in Large, Shallow, Eutrophic Lake Chaohu, China. J. Freshw. Ecol. 2019, 34, 575–589. [Google Scholar] [CrossRef]
- Huang, W.; Chen, X.; Jiang, X.; Zheng, B. Characterization of Sediment Bacterial Communities in Plain Lakes with Different Trophic Statuses. Microbiol. Open 2017, 6, e503. [Google Scholar] [CrossRef]
- Avramidis, P.; Samiotis, A.; Kalimani, E.; Papoulis, D.; Lampropoulou, P.; Bekiari, V. Sediment Characteristics and Water Physicochemical Parameters of the Lysimachia Lake, Western Greece. Environ. Earth Sci. 2013, 70, 383–392. [Google Scholar] [CrossRef]
- El Zokm, G.M.; Okbah, M.A.; El-Said, G.F.; Sinoussy, K.S. Ecological Implications of Boron and Sulfur in Sediments from a Lake in the Nile Delta: Remote Sensing, Fractionation and Ecotoxicity. J. Soils Sediments 2024, 24, 1361–1376. [Google Scholar] [CrossRef]
- Torres, I.C.; Turner, B.L.; Ramesh Reddy, K. The Chemical Nature of Phosphorus in Subtropical Lake Sediments. Aquat. Geochem. 2014, 20, 437–457. [Google Scholar] [CrossRef]
- Hezhong, Y.; Liang, C.; Enfeng, L.; Qi, L.; Cheng, W.; Enlou, Z. Fractions and Transformation of Organic Phosphorus in Sediments from a Eutrophic Lake in China. Environ. Sci. Pollut. Res. 2017, 24, 27314–27325. [Google Scholar] [CrossRef]
- Zhang, W.; Jin, X.; Liu, D.; Tang, W.; Shan, B. Assessment of the Sediment Quality of Freshwater Ecosystems in Eastern China Based on Spatial and Temporal Variation of Nutrients. Environ. Sci. Pollut. Res. 2017, 24, 19412–19421. [Google Scholar] [CrossRef] [PubMed]
- Imre, N.; Moln, S.; Vaszita, E. The Biolog EcoPlate TM Technique for Assessing the Effect of Metal Oxide Nanoparticles on Freshwater Microbial Communities. Nanomaterials 2021, 11, 2–23. [Google Scholar] [CrossRef]
- Costa, P.S.; Reis, M.P.; Ávila, M.P.; Leite, L.R.; De Araújo, F.M.G.; Salim, A.C.M.; Oliveira, G.; Barbosa, F.; Chartone-Souza, E.; Nascimento, A.M.A. Metagenome of a Microbial Community Inhabiting a Metal-Rich Tropical Stream Sediment. PLoS ONE 2015, 10, e0119465. [Google Scholar] [CrossRef] [PubMed]
- Button, M.; Weber, K.; Nivala, J.; Aubron, T.; Müller, R.A. Community-Level Physiological Profiling of Microbial Communities in Constructed Wetlands: Effects of Sample Preparation. Appl. Biochem. Biotechnol. 2016, 178, 960–973. [Google Scholar] [CrossRef]
- Miao, L.; Wang, C.; Adyel, T.M.; Wu, J.; Liu, Z.; You, G.; Meng, M.; Qu, H.; Huang, L.; Yu, Y.; et al. Microbial Carbon Metabolic Functions of Biofilms on Plastic Debris Influenced by the Substrate Types and Environmental Factors. Environ. Int. 2020, 143, 106007. [Google Scholar] [CrossRef]
- Cook, K.L.; Garland, J.L.; Layton, A.C.; Dionisi, H.M.; Levine, L.H.; Sayler, G.S. Effect of Microbial Species Richness on Community Stability and Community Function in a Model Plant-Based Wastewater Processing System. Microb. Ecol. 2006, 52, 725–737. [Google Scholar] [CrossRef]
- Kirchman, D.L. The Uptake of Inorganic Nutrients by Heterotrophic Bacteria. Microb. Ecol. 1994, 28, 255–271. [Google Scholar] [CrossRef]
- Durand, S.; Guillier, M. Transcriptional and Post-Transcriptional Control of the Nitrate Respiration in Bacteria. Front. Mol. Biosci. 2021, 8, 667758. [Google Scholar] [CrossRef]
- Mena-Rivera, L.; Lloyd, C.E.M.; Reay, M.K.; Goodall, T.; Read, D.S.; Johnes, P.J.; Evershed, R.P. Tracing Carbon and Nitrogen Microbial Assimilation in Suspended Particles in Freshwaters. Biogeochemistry 2023, 164, 277–293. [Google Scholar] [CrossRef]
- Sieradzki, E.T.; Nuccio, E.E.; Pett-Ridge, J.; Firestone, M.K. Expression of Macromolecular Organic Nitrogen Degrading Enzymes Identifies Potential Mediators of Soil Organic N Availability to an Annual Grass. ISME J. 2023, 17, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Andersen, K.H.; Merico, A.; Riemann, L. Particle-Associated N2 Fixation by Heterotrophic Bacteria in the Global Ocean. Sci. Adv. 2025, 11, eadq4693. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Ren, M.; Xie, E.; Ding, A.; Liu, Y.; Deng, S.; Zhang, D. Roles of Phosphorus Sources in Microbial Community Assembly for the Removal of Organic Matters and Ammonia in Activated Sludge. Front. Microbiol. 2019, 10, 1023. [Google Scholar] [CrossRef] [PubMed]
- Bai, F.; Liu, R.; Yang, Y.; Ran, X.; Shi, J.; Wu, Z. Dissolved Organic Phosphorus Use by the Invasive Freshwater Diazotroph Cyanobacterium, Cylindrospermopsis raciborskii. Harmful Algae 2014, 39, 112–120. [Google Scholar] [CrossRef]
- Rippka, R.; Coursin, T.; Hess, W.; Lichtle, C.; Scanlan, D.J.; Palinska, K.A.; Iteman, I.; Partensky, F.; Houmard, J.; Herdman, M. Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. Strain PCC 9511, the First Axenic Chlorophyll a2/b2-Containing Cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evol. Microbiol. 2000, 50, 1833–1847. [Google Scholar] [CrossRef]
- Gorski, L.; Noriega, A.A. Comparison of Phenotype Nutritional Profiles and Phosphate Metabolism Genes in Four Serovars of Salmonella enterica from Water Sources. Microorganisms 2023, 11, 2109. [Google Scholar] [CrossRef]
- Tiwari, R.; Kumar, K.; Singh, S.; Nain, L.; Shukla, P. Molecular Detection and Environment-Specific Diversity of Glycosyl Hydrolase Family 1 β-Glucosidase in Different Habitats. Front. Microbiol. 2016, 7, 1597. [Google Scholar] [CrossRef]
- Mazur, A.; Stasiak, G.; Wielbo, J.; Koper, P.; Kubik-Komar, A.; Skorupska, A. Phenotype Profiling of Rhizobium leguminosarum Bv. Trifolii Clover Nodule Isolates Reveal Their Both Versatile and Specialized Metabolic Capabilities. Arch. Microbiol. 2013, 195, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Cha, J.; Song, B.; Huang, Y.; Kim, S.; Kim, S.; Jo, E.; Fortin, S.; An, S. Total Microbial Activity and Sulfur Cycling Microbe Changes in Response to the Development of Hypoxia in a Shallow Estuary. Ocean Sci. J. 2020, 55, 165–181. [Google Scholar] [CrossRef]
- Raimundo, I.; Silva, R.; Meunier, L.; Valente, S.M.; Lago-Lestón, A.; Keller-Costa, T.; Costa, R. Functional Metagenomics Reveals Differential Chitin Degradation and Utilization Features across Free-Living and Host-Associated Marine Microbiomes. Microbiome 2021, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.; Holmes, M.E.; Chanton, J.P.; Reddy, K.R. Distribution, activities, and interactions of methanogens and sulfate-reducing prokaryotes in the Florida Everglades. Appl. Environ. Microbiol. 2015, 81, 7431–7442. [Google Scholar] [CrossRef]
- Fan, Y.Y.; Li, B.B.; Yang, Z.C.; Cheng, Y.Y.; Liu, D.F.; Yu, H.Q. Mediation of Functional Gene and Bacterial Community Profiles in the Sediments of Eutrophic Chaohu Lake by Total Nitrogen and Season. Environ. Pollut. 2019, 250, 233–240. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, J.; Tong, T.; Xie, S.; Liu, Y. Influences of Eutrophication on Methanogenesis Pathways and Methanogenic Microbial Community Structures in Freshwater Lakes. Environ. Pollut. 2020, 260, 114106. [Google Scholar] [CrossRef]
- Kim, H.; Bae, H.S.; Reddy, K.R.; Ogram, A. Distributions, Abundances and Activities of Microbes Associated with the Nitrogen Cycle in Riparian and Stream Sediments of a River Tributary. Water Res. 2016, 106, 51–61. [Google Scholar] [CrossRef]
- Shi, W. Diverse Responses of pqqC- and phoD-Harbouring Bacterial Communities to Variation in Soil Properties of Moso Bamboo Forests. Microb. Biotechnol. 2022, 15, 2097–2111. [Google Scholar] [CrossRef]
- Zheng, B.X.; Zhang, D.P.; Wang, Y.; Hao, X.L.; Wadaan, M.A.M.; Hozzein, W.N.; Peñuelas, J.; Zhu, Y.G.; Yang, X.R. Responses to Soil pH Gradients of Inorganic Phosphate Solubilizing Bacteria Community. Sci. Rep. 2019, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Anzuay, M.S.; Frola, O.; Angelini, J.G.; Ludueña, L.M.; Ibañez, F.; Fabra, A.; Taurian, T. Effect of Pesticides Application on Peanut (Arachis hypogaea L.) Associated Phosphate Solubilizing Soil Bacteria. Appl. Soil Ecol. 2015, 95, 31–37. [Google Scholar] [CrossRef]
- Luo, J.; Tan, X.; Liu, K.; Lin, W. Survey of Sulfur-Oxidizing Bacterial Community in the Pearl River Water Using soxB, sqr, and dsrA as Molecular Biomarkers. 3 Biotech 2018, 8, 73. [Google Scholar] [CrossRef]
- Liang, J.; Yan, M.; Zhu, Z.; Lu, L.; Ding, J.; Zhou, Q.; Gao, X.; Tang, N.; Li, S.; Li, X.; et al. The Role of Microorganisms in Phosphorus Cycling at River-Lake Confluences: Insights from a Study on Microbial Community Dynamics. Water Res. 2025, 268, 122556. [Google Scholar] [CrossRef]
- Rissanen, A.J.; Peura, S.; Mpamah, P.A.; Taipale, S.; Tiirola, M.; Biasi, C.; Mäki, A.; Nykänen, H. Vertical Stratification of Bacteria and Archaea in Sediments of a Small Boreal Humic Lake. FEMS Microbiol. Lett. 2019, 366, fnz044. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Jones, R.B.; Fodor, A.A. Inference-Based Accuracy of Metagenome Prediction Tools Varies across Sample Types and Functional Categories. Microbiome 2020, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; van der Zaan, B.; Geerlings, G.; Zandt, M.I.; Lufiandi; Roosmini, D.; Ariesyady, H.D.; Lüke, C.; Jetten, M. Decrease in Microbial Diversity Along a Pollution Gradient in Citarum River Sediment. bioRxiv 2018, 357111. [Google Scholar] [CrossRef]
- Emerson, J.B.; Varner, R.K.; Wik, M.; Parks, D.H.; Neumann, R.B.; Johnson, J.E.; Singleton, C.M.; Woodcroft, B.J.; Tollerson, R.; Owusu-Dommey, A.; et al. Diverse Sediment Microbiota Shape Methane Emission Temperature Sensitivity in Arctic Lakes. Nat. Commun. 2021, 12, 5815. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]

| Site | pH | Temp (°C) | DO (%) | EC (µS cm−1) | ORP (mV) | TC (%) | TOC (%) | OM (%) | TN (%) | TP (mg kg−1) | TS (mg kg−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NL | 6.7 ± 0.2 A | 18.4 ± 0.3 B | 2.0 ± 0.0 C | 57.4 ± 0.5 B | −108.6 ± 7.7 AB | 3.0 ± 0.2 B | 2.6 ± 0.2 B | 4.5 ± 0.2 B | 0.3 ± 0.02 B | 677.4 ± 11.4 B | 568.3 ± 14.4 B |
| PuB | 7.3 ± 0.4 A | 20.7 ± 0.3 A | 2.5 ± 0.1 B | 61.3 ± 0.9 A | −141.3 ± 16 B | 0.2 ± 0.0 D | 0.2 ± 0.0 D | 0.8 ± 0.1 D | 0.02 ± 0.0 C | 159.8 ± 4.4 D | 81.5 ± 2.0 C |
| PoP | 7.4 ± 0.2 A | 20.2 ± 0.7 A | 3.0 ± 0.1 A | 59 ± 0.5 B | −86.9 ± 11 A | 0.3 ± 0.0 CD | 0.3 ± 0.0 CD | 3.7 ± 0.2 BC | 0.03 ± 0.0 C | 276.9 ± 8.0 C | 75.7 ± 1.4 C |
| SL | 6.7 ± 0.5 A | 18.8 ± 0.2 B | 2.2 ± 0.1 C | 58.2 ± 1.4 B | −111.1 ± 29 AB | 0.6 ± 0.1 C | 0.5 ± 0.0 C | 1.1 ± 0.1 CD | 0.06 ± 0.01 C | 267.7 ± 9.6 C | 85.8 ± 1.5 C |
| VB | 6.7 ± 0.3 A | 17.5 ± 0.2 B | 1.5 ± 0.2 D | 58.1 ± 0.3 B | −100.3 ± 22 AB | 5.4 ± 0.2 A | 5.3 ± 0.2 A | 14 ± 2.3 A | 0.54 ± 0.01 A | 1302.8 ± 12.5 A | 854.1 ± 15.1 A |
| Absolute Quantification (Gene Copy g−1 dw of Sediment) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | 16S rRNA | C | N | P | S | ||||||
| chiA | mcrA | nifH | amoA | nosZ | phoD | pqqC | soxB | dsrA | |||
| NL | Mean | 7.2 × 1010 BC | 1.2 × 103 A | 3.8 × 103 A | 2.3 × 104 A | 8.5 × 101 A | 2.4 × 104 A | 2.6 × 105 A | 1.1 × 105 A | 6.0 × 103 A | 3.0 × 104 C |
| SD | ± 2 × 1010 | ± 2 × 102 | ± 1 × 103 | ± 7 × 103 | ± 2 × 101 | ± 4 × 103 | ± 6 × 104 | ± 8 × 104 | ± 2 × 103 | ± 3 × 103 | |
| PuB | Mean | 7.4 × 109 C | 4.1 × 102 C | 8.0 ×102 B | 2.6 × 103 B | 4.3 × 101 B | 6.4 × 102 C | 1.9 × 104 B | 4.2 × 104 A | 9.1 × 102 B | 8.8 × 103 C |
| SD | ± 6 × 109 | ± 6 × 101 | ± 1 × 102 | ± 1 × 103 | ± 1 × 101 | ± 9 × 100 | ± 8 × 103 | ± 1 × 104 | ± 8 × 101 | ± 1 × 103 | |
| PoP | Mean | 1.5 × 1011 B | 7.8 × 102 B | 3.2 × 103 B | 1.5 × 104 AB | 4.0 × 101 B | 6.3 × 103 B | 3.0 × 105 A | 1.2 × 105 A | 8.1 × 103 A | 2.3 × 104 A |
| SD | ± 8 × 1010 | ± 5 × 100 | ± 1 × 102 | ± 3 × 103 | ± 9 × 100 | ± 8 × 101 | ± 1 × 105 | ± 1 × 104 | ± 1 × 103 | ± 3 × 103 | |
| SL | Mean | 8.1 × 1010 BC | 6.0 × 102 BC | 5.8 × 102 C | 1.5 × 104 AB | 6.7 × 101 B | 5.1 × 103 BC | 2.5 × 105 A | 3.9 × 104 A | 2.5 × 103 B | 1.5 × 104 B |
| SD | ± 3 × 109 | ± 8 × 101 | ± 5 × 101 | ± 7 × 103 | ± 2 × 101 | ± 1 × 102 | ± 4 × 104 | ± 1 × 104 | ± 4 × 102 | ± 3 × 103 | |
| VB | Mean | 2.9 × 1011 A | 9.1 × 101 D | 2.1 × 103 AB | 2.7 × 103 B | 1.4 × 101 B | 1.3 × 103 BC | 3.6 × 103 B | 2.4 × 104 A | 1.5 × 102 B | 4.7 × 103 C |
| SD | ± 6 × 1010 | ± 2 × 101 | ± 8 × 102 | ± 2 × 102 | ± 2 × 100 | ± 7 × 101 | ± 2 × 103 | ± 1 × 104 | ± 6 × 101 | ± 3 × 102 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Gil, T.; Elgueta, S.; Larama, G.; Rilling, J.-I.; Hollenback, A.; Jaisi, D.P.; Valdebenito, D.; Spears, B.M.; Campos, M.A. Integrated Assessment of Benthic Bacterial Community Physiology, Structure, and Function Across C, N, P, and S Gradients in Lake Villarrica Sediments, Chile. Microorganisms 2025, 13, 2544. https://doi.org/10.3390/microorganisms13112544
Ruiz-Gil T, Elgueta S, Larama G, Rilling J-I, Hollenback A, Jaisi DP, Valdebenito D, Spears BM, Campos MA. Integrated Assessment of Benthic Bacterial Community Physiology, Structure, and Function Across C, N, P, and S Gradients in Lake Villarrica Sediments, Chile. Microorganisms. 2025; 13(11):2544. https://doi.org/10.3390/microorganisms13112544
Chicago/Turabian StyleRuiz-Gil, Tay, Sebastián Elgueta, Giovanni Larama, Joaquín-Ignacio Rilling, Anthony Hollenback, Deb P. Jaisi, Diego Valdebenito, Bryan M. Spears, and Marco A. Campos. 2025. "Integrated Assessment of Benthic Bacterial Community Physiology, Structure, and Function Across C, N, P, and S Gradients in Lake Villarrica Sediments, Chile" Microorganisms 13, no. 11: 2544. https://doi.org/10.3390/microorganisms13112544
APA StyleRuiz-Gil, T., Elgueta, S., Larama, G., Rilling, J.-I., Hollenback, A., Jaisi, D. P., Valdebenito, D., Spears, B. M., & Campos, M. A. (2025). Integrated Assessment of Benthic Bacterial Community Physiology, Structure, and Function Across C, N, P, and S Gradients in Lake Villarrica Sediments, Chile. Microorganisms, 13(11), 2544. https://doi.org/10.3390/microorganisms13112544






