Unveiling Species Diversity Within Early-Diverging Fungi from China XI: Eight New Species of Cunninghamella (Mucoromycota)
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Morphology
2.2. DNA Extraction and Amplification
2.3. Phylogenetic Analyses
3. Result
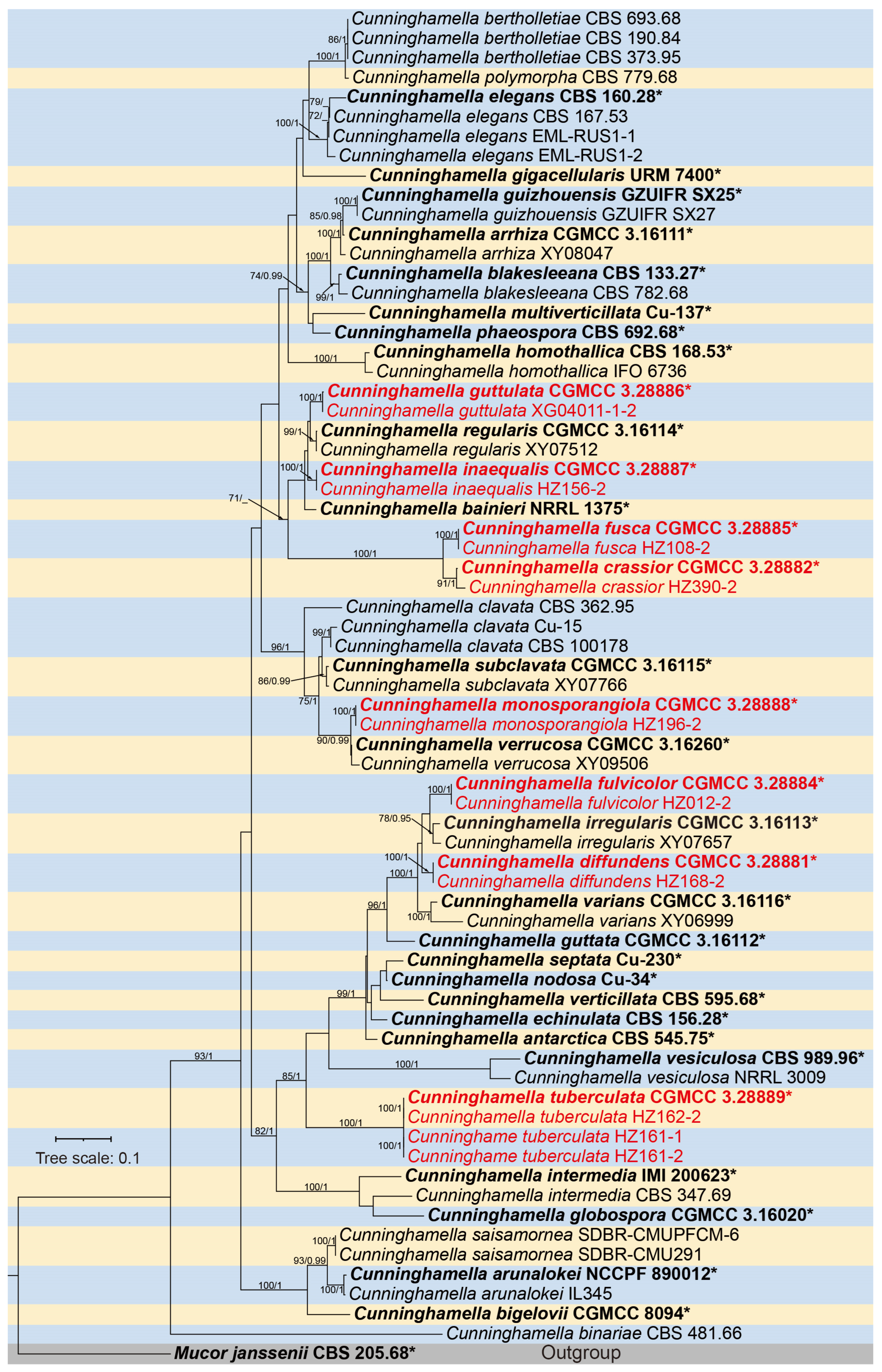
3.1. Molecular Phylogeny
3.2. Taxonomy
3.2.1. Cunninghamella Crassior Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 2

3.2.2. Cunninghamella diffundens Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 3
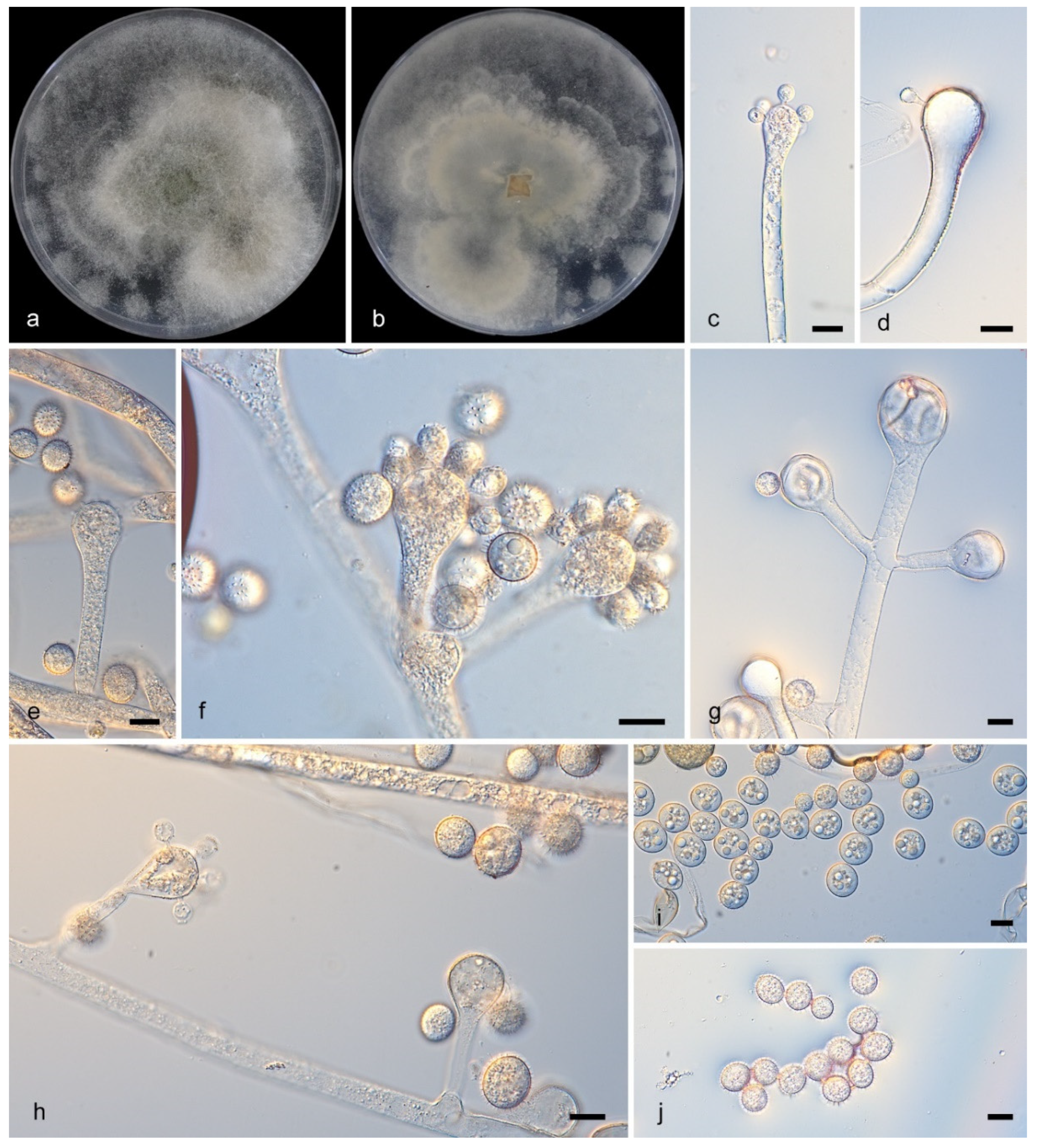
3.2.3. Cunninghamella fulvicolor Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 4

3.2.4. Cunninghamella fusca Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 5
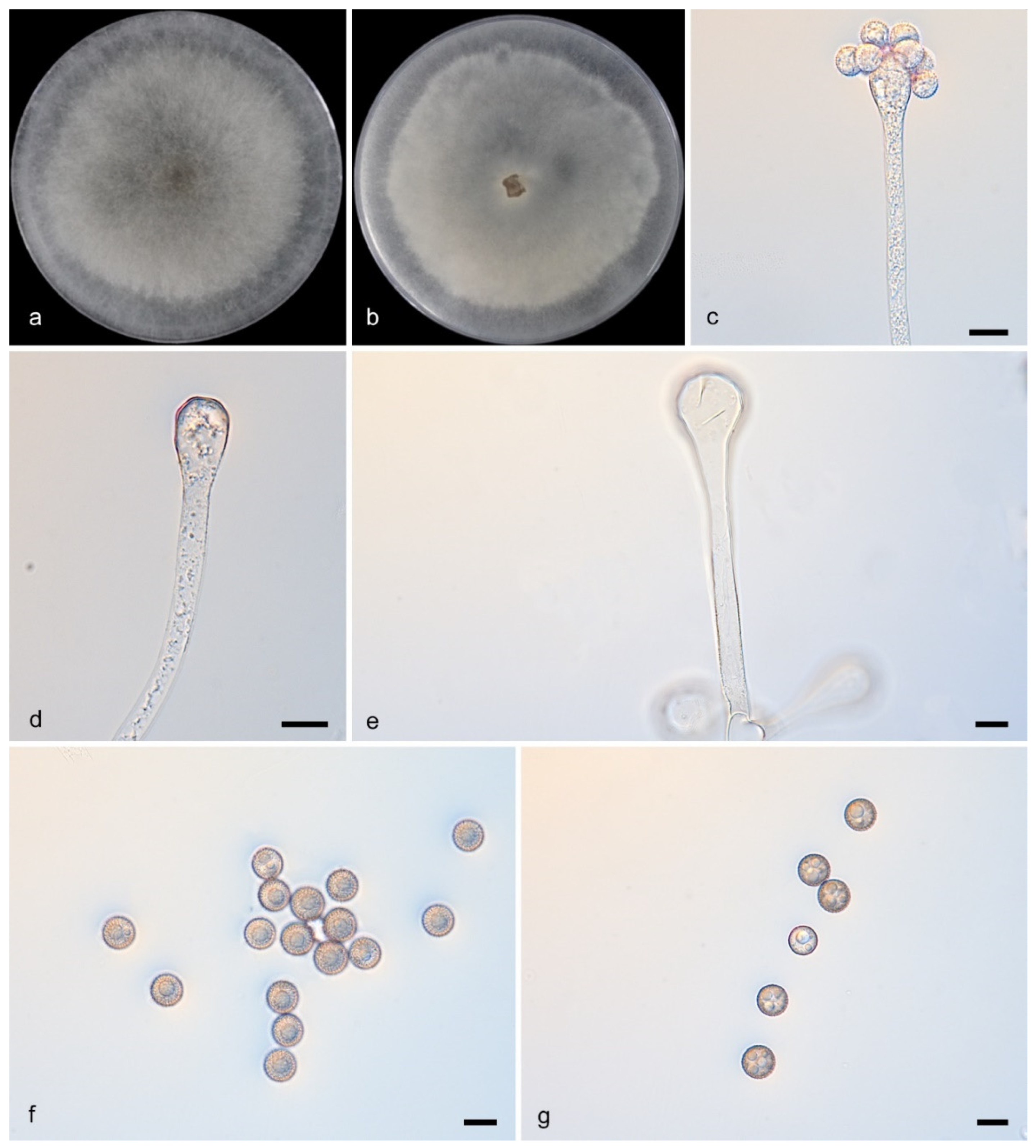
3.2.5. Cunninghamella guttulata Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 6
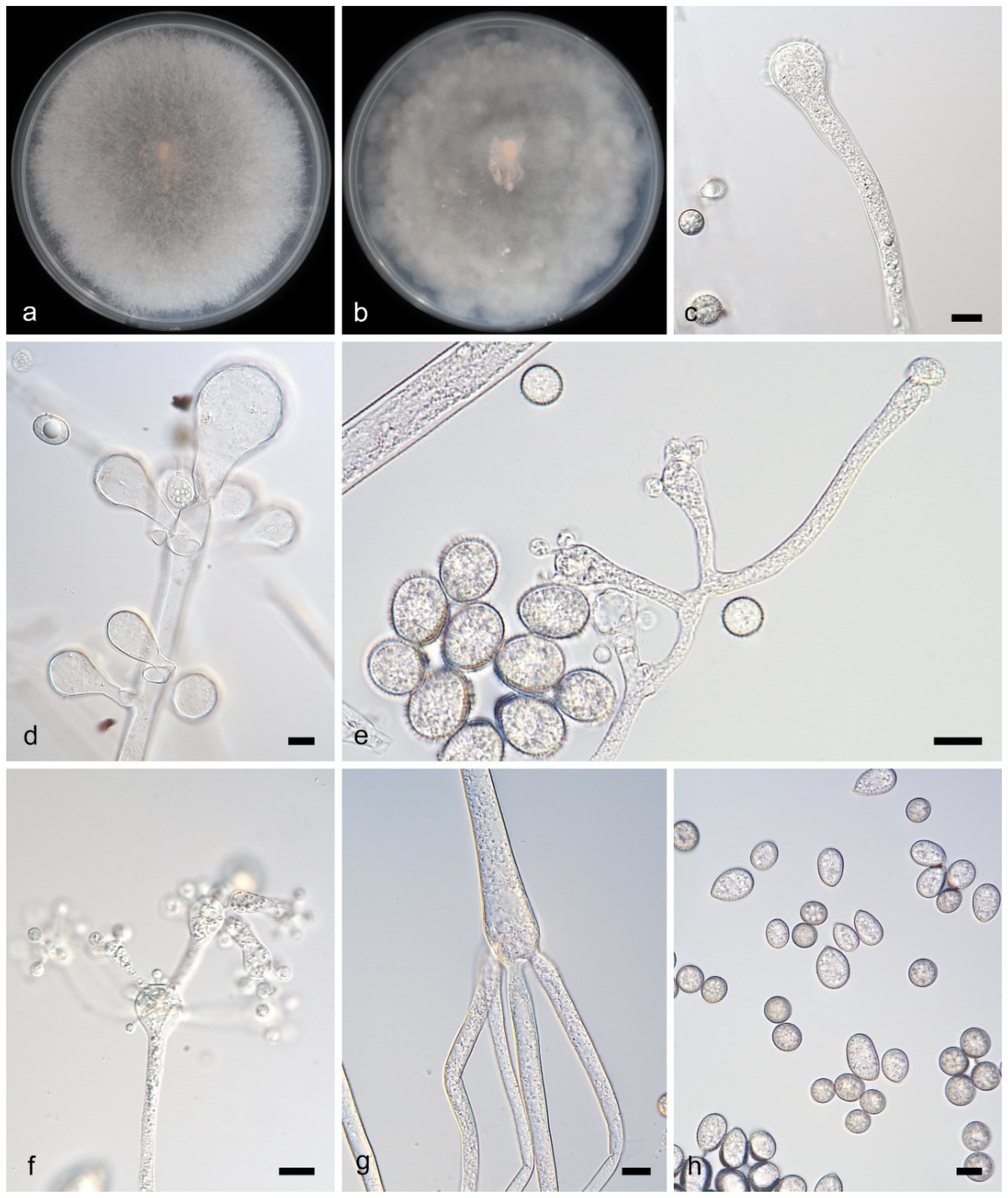
3.2.6. Cunninghamella inaequalis Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 7

3.2.7. Cunninghamella monosporangiola Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 8

3.2.8. Cunninghamella tuberculata Y. Jiang, H. Zhao, Z. Meng & X.Y. Liu. sp. nov. Figure 9

4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Zheng, R.Y.; Chen, G.Q. A monograph of Cunninghamella. Mycotaxon 2001, 80, 1–57. [Google Scholar]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Zhao, H.; Zhu, J.; Zong, T.K.; Liu, X.L.; Ren, L.Y.; Lin, Q.; Qiao, M.; Nie, Y.; Zhang, Z.D.; Liu, X.Y. Two new species in the family Cunninghamellaceae from China. Mycobiology 2021, 49, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Zhao, Y.X.; Shen, X.; Chen, W.H.; Han, Y.F.; Huang, J.Z.; Liang, Z.Q. Molecular phylogeny and morphology of Cunninghamella guizhouensis sp. nov. (Cunninghamellaceae, Mucorales), from soil in Guizhou, China. Phytotaxa 2020, 455, 31–39. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Choi, Y.J.; Lee, H.B. Zygomycete fungi in Korea: Cunninghamella bertholletiae, Cunninghamella echinulata, and Cunninghamella elegans. Mycobiology 2017, 45, 318–326. [Google Scholar] [CrossRef]
- Liu, C.W.; Liou, G.Y.; Chien, C.Y. New records of the genus Cunninghamella (Mucorales) in Taiwan. Fungal Sci. 2005, 20, 1–9. [Google Scholar]
- Alves, A.L.; de Souza, C.A.; de Oliveira, R.J.; Cordeiro, T.R.; de Santiago, A.L. Cunninghamella clavata from Brazil: A new record for the western hemisphere. Mycotaxon 2017, 132, 381–389. [Google Scholar] [CrossRef]
- Bragulat, M.R.; Castella, G.; Isidoro-Ayza, M.; Domingo, M.; Cabanes, F.J. Characterization and phylogenetic analysis of a Cunninghamella bertholletiae isolate from a bottlenose dolphin (Tursiops truncatus). Rev. Iberoam. Micol. 2017, 34, 215–219. [Google Scholar] [CrossRef]
- Suwannarach, N.; Kumla, J.; Supo, C.; Honda, Y.; Nakazawa, T.; Khanongnuch, C.; Wongputtisin, P. Cunninghamella saisamornae (Cunninghamellaceae, Mucorales), a new soil fungus from northern Thailand. Phytotaxa 2021, 509, 291–300. [Google Scholar] [CrossRef]
- Yu, J.; Walther, G.; Van Diepeningen, A.D.; Gerrits Van Den Ende, A.H.G.; Li, R.Y.; Moussa, T.A.A.; Almaghrabi, O.A.; De Hoog, G.S. DNA barcoding of clinically relevant Cunninghamella species. Med. Mycol. 2015, 53, 99–106. [Google Scholar] [CrossRef]
- Wang, Y.J.; Zhao, T.; Wu, W.Y.; Wang, M.; Liu, X.Y. Cunninghamella verrucosa sp. nov. (Mucorales, Mucoromycota) from Guangdong province in China. Phytotaxa 2022, 560, 274–284. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Zong, T.; Wang, K.; Lv, M.; Cui, Y.; Tohtirjap, A.; Chen, J.; Zhao, C.; Wu, F.; et al. Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Divers. 2023, 123, 49–157. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J. Taxonomy of Fungi Causing Mucormycosis and Entomophthoramycosis (Zygomycosis) and Nomenclature of the Disease: Molecular Mycologic Perspectives. Clin. Infect. Dis. 2012, 54, S8–S15. [Google Scholar] [CrossRef]
- Matruchot, L. Une Mucorinée purement conidienne, Cunninghamella africana. Ann. Mycologici. 1903, 1, 45–60. [Google Scholar]
- Waren, L.; Freitas, S.D.; Souza-Motta, C.M.; Barbosa, R.D.N.; Melo, R.F.R.; Santiago, A.L.C.M.D.A. A new homothallic species of Cunninghamella (Mucorales, Mucoromycota) with a synoptic key for the genus in the Neotropics. Sydowia 2024, 77, 119–127. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2017, 108, 1028–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Wang, H.; Liu, T.X.; Xin, Z.H. The individual lipid compositions produced by Cunninghamella sp. Salicorn 5, an endophytic oleaginous fungus from Salicornia bigelovii Torr. Eur. Food Res. Technol. 2014, 238, 621–633. [Google Scholar] [CrossRef]
- Fakas, S.; Papanikolaou, S.; Galiotou-Panayotou, M.; Komaitis, M.; Aggelis, G. Organic nitrogen of tomato waste hydrolysate enhances glucose uptake and lipid accumulation in Cunninghamella echinulata. J. Appl. Microbiol. 2008, 105, 1062–1070. [Google Scholar] [CrossRef]
- Alakhras, R.; Bellou, S.; Fotaki, G.; Stephanou, G.; Demopoulos, N.A.; Papanikolaou, S.; Aggelis, G. Fatty acid lithium salts from Cunninghamella echinulata have cytotoxic and genotoxic effects on HL-60 human leukemia cells. Eng. Life. Sci. 2015, 15, 243–253. [Google Scholar] [CrossRef]
- Guo, J.; Wang, H.; Liu, D.; Zhang, J.N.; Zhao, Y.H.; Liu, T.X.; Xin, Z.H. Isolation of Cunninghamella bigelovii sp. nov. CGMCC 8094 as a new endophytic oleaginous fungus from Salicornia bigelovii. Mycol. Prog. 2015, 14, 11. [Google Scholar] [CrossRef]
- Liu, X.Y.; Huang, H.; Zheng, R.Y. Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon 2001, 80, 77–95. [Google Scholar]
- Zhao, H.; Nie, Y.; Huang, B.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China I: Three new species of Backusella (Backusellaceae, Mucoromycota). MycoKeys 2024, 109, 285–304. [Google Scholar] [CrossRef]
- Tao, M.F.; Ding, Z.Y.; Wang, Y.X.; Zhang, Z.X.; Zhao, H.; Meng, Z.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China II: Three new species of Absidia (Cunninghamellaceae, Mucoromycota) from Hainan Province. MycoKeys 2024, 110, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Zhao, H.; Jiang, Y.; Liu, X.Y.; Tao, M.F.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China III: Six new species and a new record of Gongronella (Cunninghamellaceae, Mucoromycota). MycoKeys 2024, 110, 287–317. [Google Scholar] [CrossRef]
- Ding, Z.Y.; Ji, X.Y.; Tao, M.F.; Jiang, Y.; Liu, W.X.; Wang, Y.X.; Meng, Z.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China IV: Four new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys 2025, 117, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.Y.; Ding, Z.Y.; Nie, Y.; Zhao, H.; Wang, S.; Huang, B.; Liu, X.Y. Unveiling species diversity: Within early-diverging fungi. From China V: Five new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys 2025, 117, 267–288. [Google Scholar] [CrossRef]
- Wang, Y.X.; Ding, Z.Y.; Ji, X.Y.; Meng, Z.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China VI: Four Absidia sp. nov. (Mucorales) in Guizhou and Hainan. Microorganisms 2025, 13, 1315. [Google Scholar] [CrossRef]
- Ding, Z.Y.; Tao, M.F.; Ji, X.Y.; Jiang, Y.; Liu, W.X.; Wang, S.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China VII: Seven new species of Cunninghamella (Cunninghamellaceae, Mucoromycota). J. Fungi 2025, 11, 417. [Google Scholar] [CrossRef]
- Ji, X.Y.; Jiang, Y.; Li, F.; Ding, Z.Y.; Meng, Z.; Liu, X.Y. Unveiling Species Diversity Within Early-Diverging Fungi from China VIII: Four New Species in Mortierellaceae (Mortierellomycota). Microorganisms 2025, 13, 1330. [Google Scholar] [CrossRef]
- Ding, Z.Y.; Ji, X.Y.; Li, F.; Liu, W.X.; Wang, S.; Zhao, H.; Liu, X.Y. Unveiling species diversity within early-diverging fungi from China IX: Four new species of Mucor (Mucoromycota). J. Fungi 2025, 11, 682. [Google Scholar] [CrossRef]
- Corry, J.E.L. Rose bengal chloramphenicol (RBC) agar. In Progress in Industrial Microbiology; Elsevier: Amsterdam, The Netherlands, 1995; pp. 431–433. [Google Scholar]
- Zhao, H.; Nie, Y.; Zong, T.K.; Dai, Y.C.; Liu, X.Y. Three New Species of Absidia (Mucoromycota) from China Based on Phylogeny, Morphology and Physiology. Diversity 2022, 14, 132. [Google Scholar] [CrossRef]
- Zheng, R.Y.; Chen, G.Q.; Huang, H.; Liu, X.Y. A monograph of Rhizopus. Sydowia 2007, 59, 273–372. [Google Scholar]
- Zheng, R.Y.; Liu, X.Y.; Li, R.Y. More Rhizomucor causing human mucormycosis from China: R. chlamydosporus sp. nov. Sydowia 2009, 61, 135–147. [Google Scholar]
- Zheng, R.Y.; Liu, X.Y. Taxa of Pilaira (Mucorales, Zygomycota) from China. Nova Hedwig. 2009, 88, 255–267. [Google Scholar] [CrossRef]
- Zong, T.K.; Zha, H.; Liu, X.L.; Ren, L.Y.; Zhao, C.L.; Liu, X.Y. Taxonomy and phylogeny of four new species in Absidia (Cunninghamellaceae, Mucorales) from China. Front. Microbiol. 2021, 12, 677836. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar] [CrossRef]
- Guo, L.D.; Hyde, K.D.; Liew, E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Lutzoni, F.M.; Ward, T.J.; Benny, G.L. Evolutionary relationships among mucoralean fungi Zygomycota: Evidence for family polyphyly on a large scale. Mycologia 2001, 93, 286–297. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest V2: Program distributed by the author. Bioinformatics 2004, 24, 581–583. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Hu, D.M.; Wang, M.; Cai, L. Phylogenetic assessment and taxonomic revision of Mariannaea. Mycol. Prog. 2016, 16, 271–283. [Google Scholar] [CrossRef]
- Zou, Y.; Hou, J.; Guo, S.; Li, C.; Li, Z.; Stephenson, S.L.; Pavlov, I.N.; Liu, P.; Li, Y. Diversity of Dictyostelid Cellular Slime Molds, Including Two Species New to Science, in Forest Soils of Changbai Mountain, China. Microbiol. Spectr. 2022, 10, e02402-22. [Google Scholar] [CrossRef]
- Hurdeal, V.G.; Gentekaki, E.; Lee, H.B.; Jeewon, R.; Hyde, K.D.; Tibpromma, S.; Mortimer, P.E.; Xu, J.C. Mucoralean fungi in Thailand: Novel species of Absidia from tropical forest soil cryptogamie. Mycologie 2021, 42, 39–61. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Zong, T.K.; Wang, Y.J.; Wang, M.; Dai, Y.C.; Liu, X.Y. Species Diversity and Ecological Habitat of Absidia (Cunninghamellaceae, Mucorales) with Emphasis on Five New Species from Forest and Grassland Soil in China. J. Fungi 2022, 8, 471. [Google Scholar] [CrossRef]
- Zhao, H.; Lv, M.L.; Liu, Z.; Zhang, M.Z.; Wang, Y.N.; Ju, X.; Song, Z.; Ren, L.Y.; Jia, B.S.; Qiao, M.; et al. High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. BioEnergy Res. 2021, 14, 1196–1206. [Google Scholar] [CrossRef]
- Weitzman, I.; Crist, M.Y. Studies with clinical isolates of Cunninghamella II. 654 Physiological and morphological studies. Mycologia 1980, 72, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Alors, D.; Lumbsch, H.T.; Divakar, P.K.; Leavitt, S.D.; Crespo, A. An integrative approach for understanding diversity in the Punctelia rudecta Species Complex (Parmeliaceae, Ascomycota). PLoS ONE 2016, 11, e0146537. [Google Scholar] [CrossRef] [PubMed]
| Loci | PCR Primers | Primer Sequences (5′–3′) | PCR Cycles | References |
|---|---|---|---|---|
| ITS | ITS5 | GGA AGT AAA AGT CGT AAC AAG G | 94 °C 5 min; (94 °C 30 s, 55 °C 30 s, 72 °C 1 min) × 35 cycles; 72 °C 10 min | [40] |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | |||
| LSU | LR0R | GTA CCC GCT GAA CTT AAG C | 94 °C 5 min; (94 °C 30 s, 51 °C 30 s, 72 °C 1 min) × 35 cycles; 72 °C 10 min | [41] |
| LR7 | TAC TAC CAC CAA GAT CT | |||
| TEF1α | EF1-728F | ATG TGC AAG GCC GGT TTC GC | 94 °C 5 min; (94 °C 30 s, 48 °C 30 s, 72 °C 1 min) × 35 cycles; 72 °C 10 min | [42,43] |
| EF-2 | GGA RGT ACC AGT SAT CAT GTT |
| Species | Strains | GenBank Accession Numbers | ||
|---|---|---|---|---|
| ITS | LSU | TEF1α | ||
| C. antarctica | CBS 545.75 T | JN205893 | JN206597 | KJ156492 |
| C. arrhiza | CGMCC 3.16111 T | OL678142 | PQ399916 | NA |
| XY08047 | OL678143 | NA | NA | |
| C. arunalokei | IL3459 | MN431159 | MN431158 | NA |
| NCCPF 890012 T | NR_177485 | NG153887 | NA | |
| C. bainieri | CBS 481.66 | MH858865 | MH870507 | KJ156495 |
| CGMCC 8094 T | KJ013403 | KJ013405 | KJ395944 | |
| NRRL 1375 T | AF254935 | NA | NA | |
| C. bertholletiae | CBS 190.84 | JN205878 | HM849701 | NA |
| CBS 373.95 | JN205873 | NA | KJ156497 | |
| CBS 693.68 | AF254931 | MH870924 | KJ156490 | |
| C. bigelovii | CGMCC 8094 T | KJ013403 | KJ013405 | KJ395944 |
| C. binariae | CBS 481.66 | MH858865 | MH870507 | KJ156495 |
| C. blakesleeana | CBS 133.27 T | NR119974 | MH866397 | KJ156479 |
| CBS 782.68 | JN205869 | MH870950 | KJ156478 | |
| C. clavata | CBS 100178 | JN205890 | JN206604 | KJ156477 |
| Cu-15 | AF254942 | NA | NA | |
| CBS 362.95 | JN205891 | NA | NA | |
| C. crassior | CGMCC3.28882 T | PV235924 | PV239680 | PV254891 |
| HZ390-2 | PV235925 | PV239681 | PV254892 | |
| C. diffundens | CGMCC3.28881 T | PV235920 | PV239676 | PV254889 |
| HZ168-2 | PV235921 | PV239677 | PV254890 | |
| C. echinulata | CBS 156.28 T | JN205895 | JN939199.1 | KJ156500 |
| C. elegans | CBS 160.28 T | AF254928 | NR_154747 | KJ156470 |
| CBS 167.53 | JN205882 | HM849700 | KJ156494 | |
| EML-RUS1-1 | MF806023 | MF806027 | NA | |
| EML-RUS1-2 | MF806021 | MF806028 | NA | |
| C. fulvicolor | CGMCC3.28884 T | PV235910 | PV239666 | PV254879 |
| HZ012-2 | PV235911 | PV239667 | PV254880 | |
| C. fusca | CGMCC3.28885 T | PV235908 | PV239664 | PV254877 |
| HZ108-2 | PV235909 | PV239665 | PV254878 | |
| C. globospora | CGMCC 3.16020 T | MW264073 | MW264132 | NA |
| C. gigacellularis | URM 7400 T | NR_168760 | NG_068773 | NA |
| C. guizhouensis | GZUIFR-SX25 T | MN908596 | MN908599 | MN912633 |
| GZUIFR-SX27 | MN908598 | MN908601 | MN912635 | |
| C. guttata | CGMCC 3.16112 T | OL678144 | PQ399917 | NA |
| C. guttulata | CGMCC3.28886 T | PV235930 | PV239686 | PV254893 |
| XG04011-1-2 | PV235931 | PV239687 | PV254894 | |
| C. homothallica | CBS 168.53 T | JN205863 | JN206605 | KJ156498 |
| IFO 6736 | AF254941 | NA | NA | |
| C. inaequalis | CGMCC3.28887 T | PV235914 | PV239670 | PV254883 |
| HZ156-2 | PV235915 | PV239671 | PV254884 | |
| C. intermedia | CBS 347.69 | JN205892 | JN206606 | NA |
| IMI 200623 T | AF254939 | NA | NA | |
| C. irregularis | CGMCC 3.16113 T | OL678145 | PQ399918 | NA |
| XY07657 | OL678146 | NA | NA | |
| C. monosporangiola | CGMCC3.28888 T | PV235922 | PV239678 | PV785980 |
| HZ196-2 | PV235923 | PV239679 | PV785981 | |
| C. multiverticillata | CBS 989.96 T | JN205897 | HM849693 | KJ156474 |
| Cu-137 T | AF254933 | NA | NA | |
| NRRL 3009 | AF254943 | NA | NA | |
| C. nodosa | Cu-34 T | AF346407 | NA | NA |
| C. phaeospora | CBS 692.68 T | JN205864 | HM849697 | NA |
| C. polymorpha | CBS 779.68 | JN205874 | JN206599 | NA |
| C. regularis | CGMCC 3.16114 T | OL678148 | PQ399919 | NA |
| XY07512 | OL678150 | NA | NA | |
| C. saisamornae | SDBR-CMUPFCM-6 | MW709394 | MW699571 | MW715866 |
| SDBR-CMU291 | MG571234 | MW699591 | MW715865 | |
| C. septata | Cu-230 T | AF346408 | NA | NA |
| C. subclavata | CGMCC 3.16115 T | OL678152 | NA | NA |
| XY07766 | OL678153 | NA | NA | |
| C. tuberculata | CGMCC3.28889 T | PV235918 | PV239674 | PV254887 |
| HZ162-2 | PV235919 | PV239675 | PV254888 | |
| C. varians | CGMCC 3.16116 T | OL678154 | PQ399920 | NA |
| XY06999 | OL678155 | NA | NA | |
| C. vesiculosa | CBS 989.96 T | JN205897 | HM849693 | KJ156474 |
| NRRL 3009 | AF254943 | NA | NA | |
| C. verrucosa | CGMCC 3.16260 | ON262555 | ON261192 | NA |
| XY09506 | ON262556 | ON261193 | NA | |
| C. verticillata | CBS595.68 T | AF254937 | NA | NA |
| M. janssenii | CBS 205.68 T | MH859119 | MH870832 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Zhao, H.; Ji, X.-Y.; Ding, Z.-Y.; Liu, W.-X.; Li, F.; Wang, S.; Liu, X.-Y.; Meng, Z. Unveiling Species Diversity Within Early-Diverging Fungi from China XI: Eight New Species of Cunninghamella (Mucoromycota). Microorganisms 2025, 13, 2508. https://doi.org/10.3390/microorganisms13112508
Jiang Y, Zhao H, Ji X-Y, Ding Z-Y, Liu W-X, Li F, Wang S, Liu X-Y, Meng Z. Unveiling Species Diversity Within Early-Diverging Fungi from China XI: Eight New Species of Cunninghamella (Mucoromycota). Microorganisms. 2025; 13(11):2508. https://doi.org/10.3390/microorganisms13112508
Chicago/Turabian StyleJiang, Yang, Heng Zhao, Xin-Yu Ji, Zi-Ying Ding, Wen-Xiu Liu, Fei Li, Shi Wang, Xiao-Yong Liu, and Zhe Meng. 2025. "Unveiling Species Diversity Within Early-Diverging Fungi from China XI: Eight New Species of Cunninghamella (Mucoromycota)" Microorganisms 13, no. 11: 2508. https://doi.org/10.3390/microorganisms13112508
APA StyleJiang, Y., Zhao, H., Ji, X.-Y., Ding, Z.-Y., Liu, W.-X., Li, F., Wang, S., Liu, X.-Y., & Meng, Z. (2025). Unveiling Species Diversity Within Early-Diverging Fungi from China XI: Eight New Species of Cunninghamella (Mucoromycota). Microorganisms, 13(11), 2508. https://doi.org/10.3390/microorganisms13112508






