Genomic Drivers of Biofilm Formation in Salmonella Enteritidis and S. Kentucky from Poultry Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Salmonella Biofilm Formation
2.1.1. Salmonella Bacterial Isolates Selection and Culture Conditions
2.1.2. Culturing Procedures
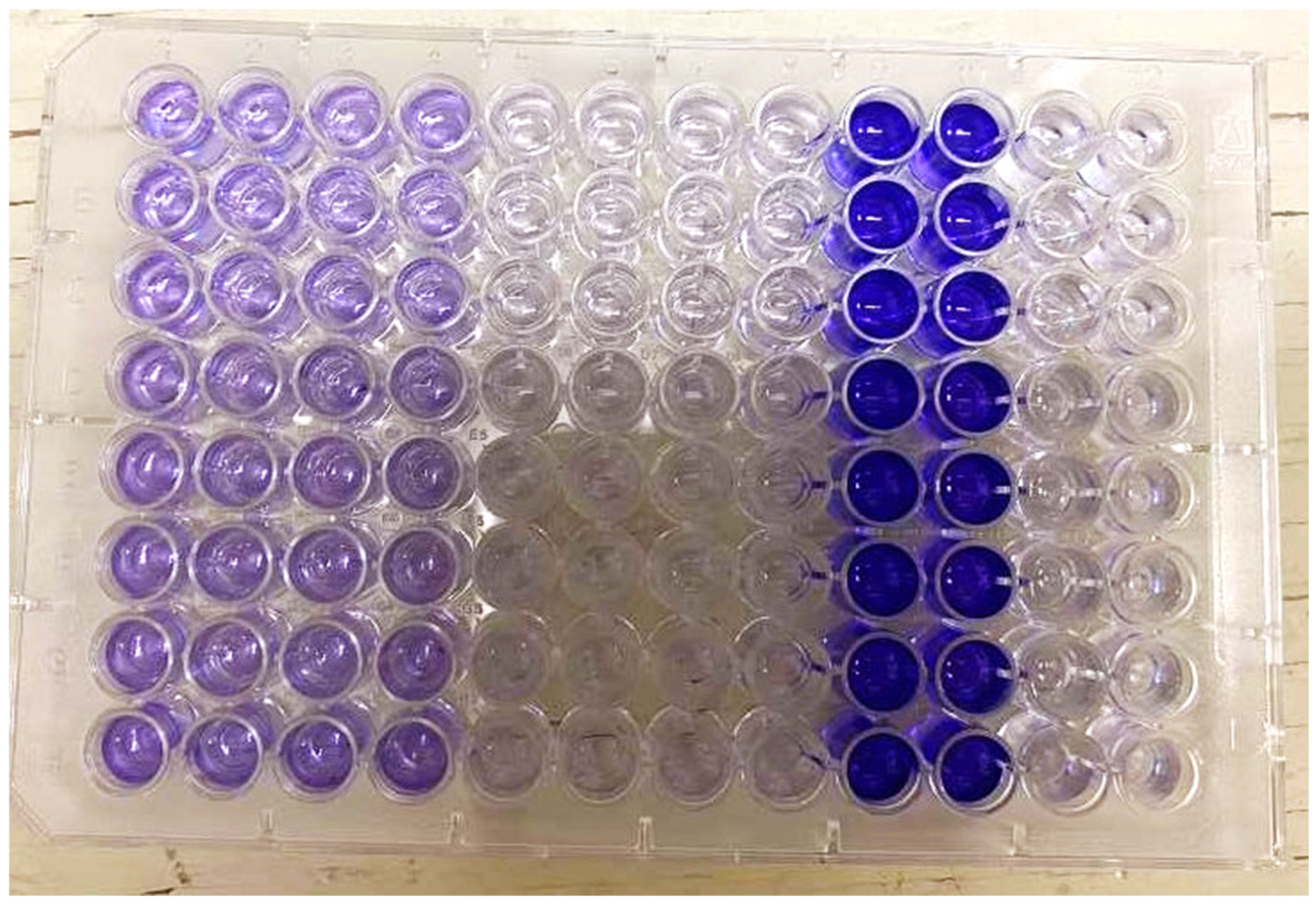
2.1.3. Biofilm Assessment Using the Crystal Violet Method
2.1.4. Curli and Cellulose Detection
2.2. Genomic Comparison of S. Enteritidis and S. Kentucky with Regard to Their Biofilm Formation Gene Information
2.2.1. Whole Genome Sequence Generation
2.2.2. Phylogenetic Tree and Heatmapping
2.2.3. SNP Variation Analysis
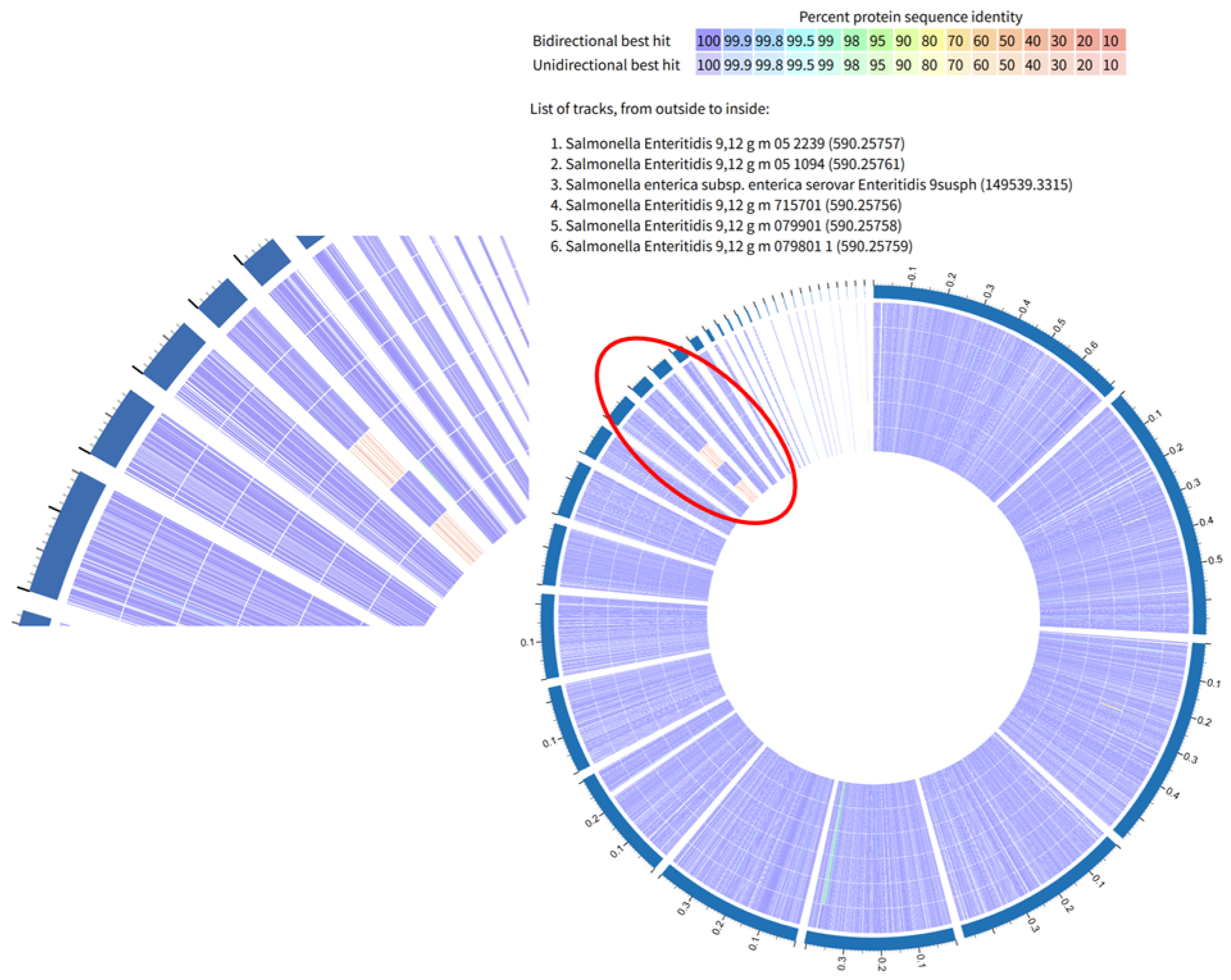
2.2.4. BLASTp (Basic Local Alignment Search Tool for Protein) Comparison
2.2.5. BLASTp (Basic Local Alignment Search Tool for Protein) Comparison
3. Results
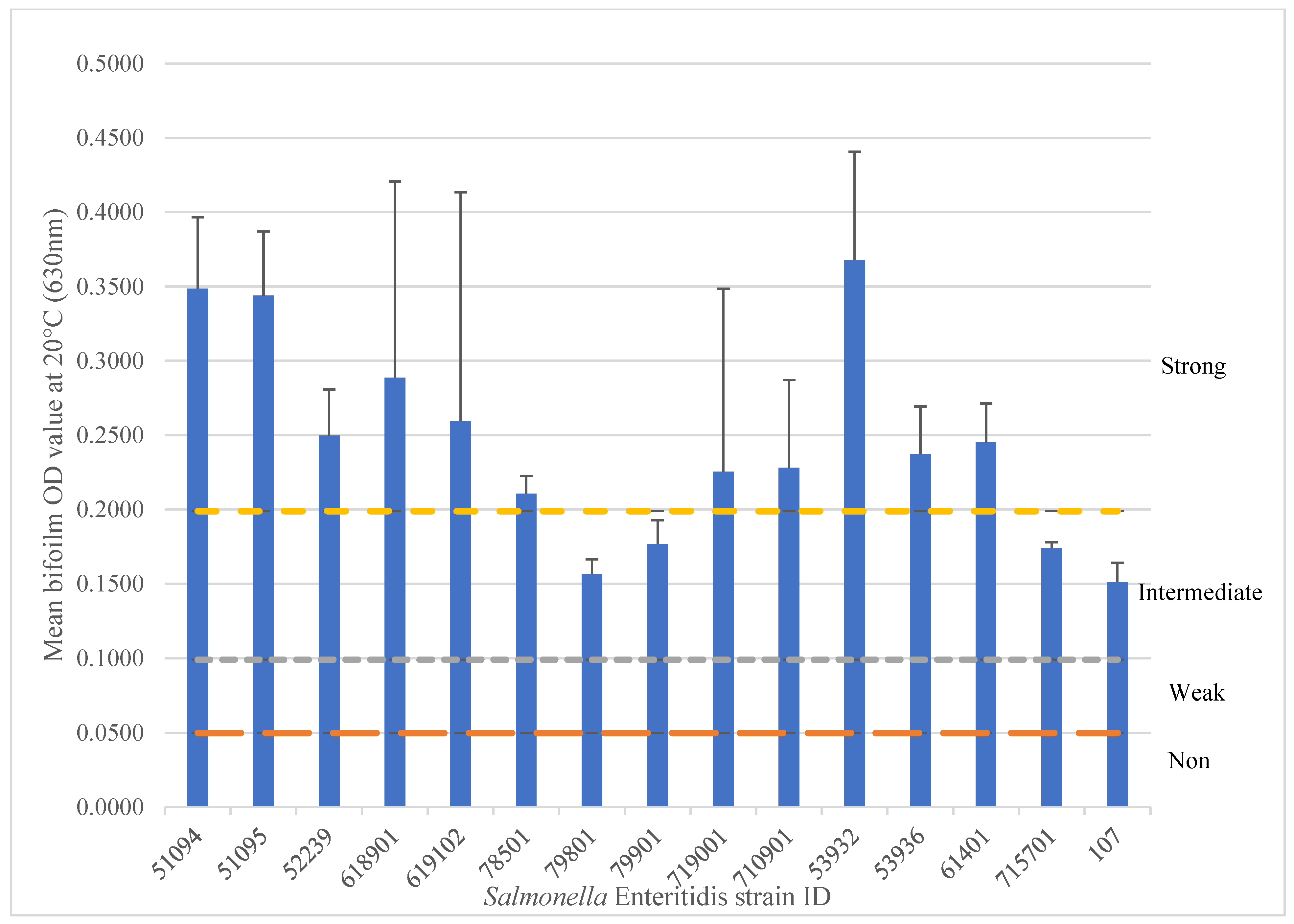
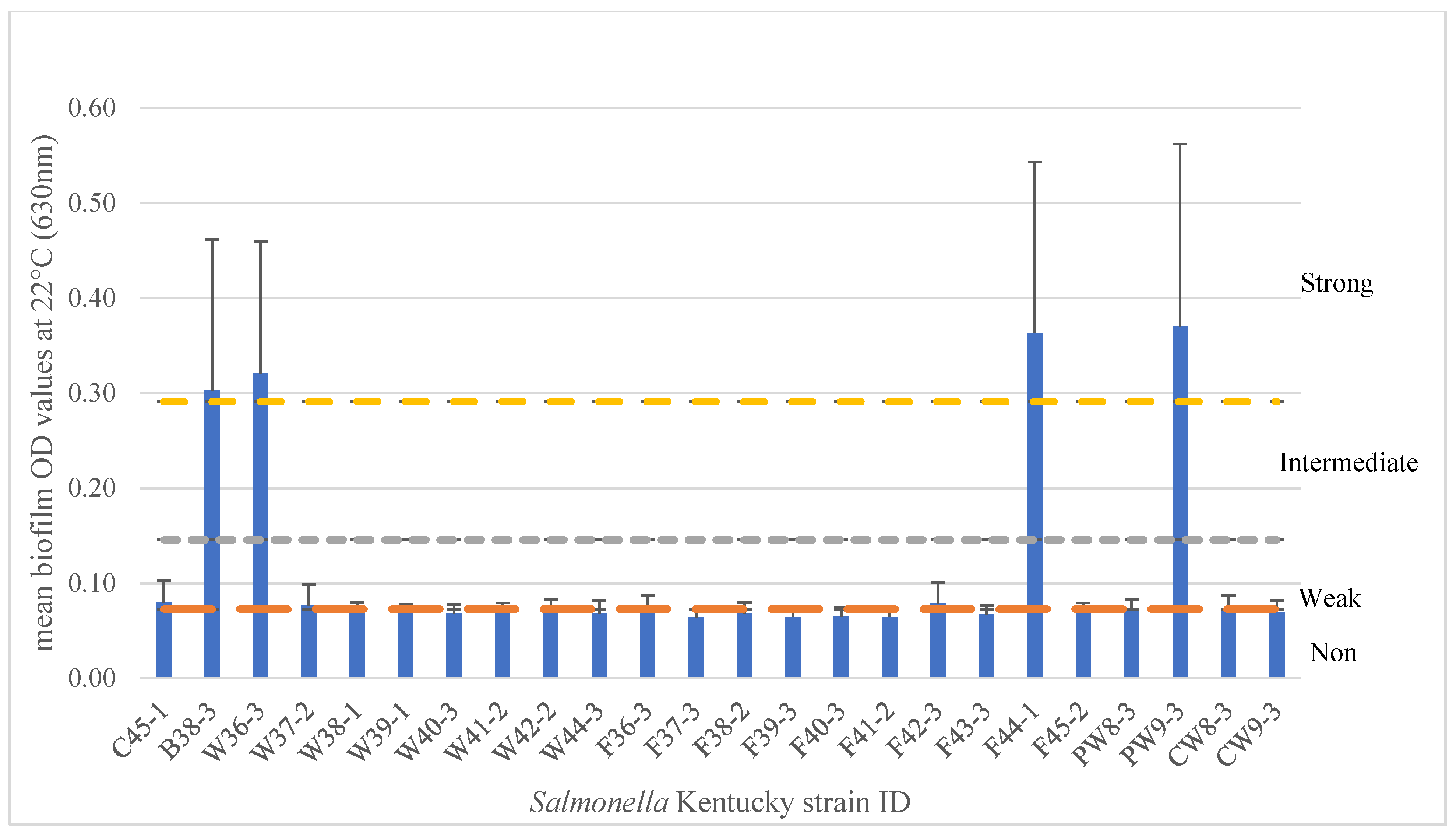
3.1. Biofilm Formation
3.2. Curli and Cellulose Phenotypic Results for SE and SK Isolates
3.3. Phylogenetic Tree of the SE and SK Isolates
3.4. Genomic Comparison Between Different Biofilm Formation Categories and Between Serotypes
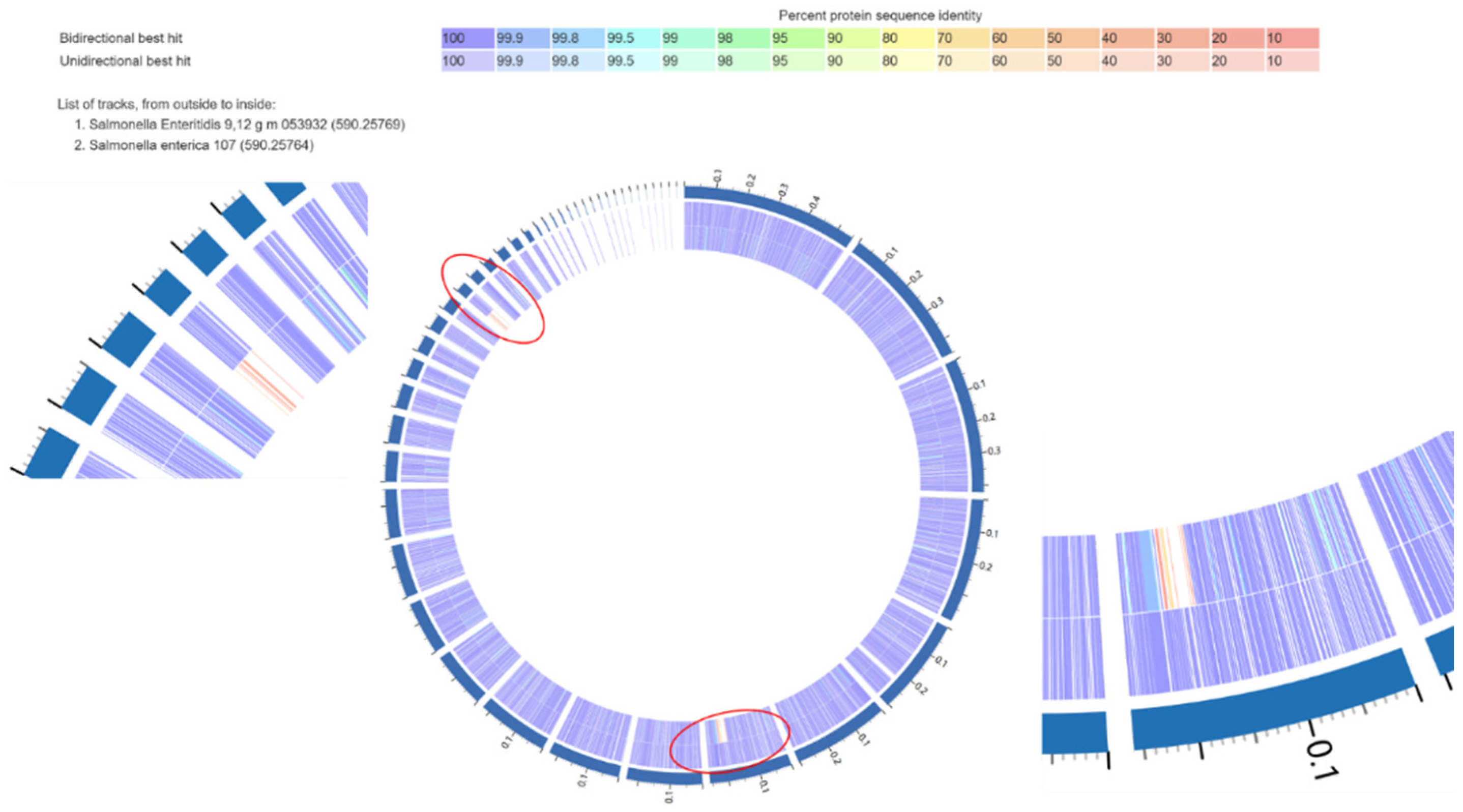
3.4.1. Comparative Genomic Analysis Results with SE and SK Isolates
3.4.2. SNP Variation Analysis
3.4.3. SNP Variation Analysis of 15 SE Isolates
3.4.4. SNP Variation Analysis of 24 SK Isolates
3.5. SE Isolates Functional Genomic Comparison (Strong vs. Intermediate Biofilm-Forming Isolates)
4. Discussion
4.1. Biofilm Formation
4.2. Genomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SNP | Single Nucleotide Polymorphism |
| BLASTp | Basic Local Alignment Search Tool for proteins |
| EPS | Extra polymeric substance |
| LB-NS | Luria Broth no-salt |
| OD | Optical density |
| CV | Crystal violet |
Appendix A
Appendix A.1
| Strains | Serotype | Country | Isolation Source | BioProject | BioSample |
|---|---|---|---|---|---|
| Salmonella Enteritidis 0618901 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784150 |
| Salmonella Enteritidis 0619102 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784151 |
| Salmonella Enteritidis 061401 | Enteritidis | Canada | Feed | PRJNA1285942 | SAMN49784152 |
| Salmonella Enteritidis 053932 | Enteritidis | Canada | Chicken breast | PRJNA1285942 | SAMN49784153 |
| Salmonella Enteritidis 053936 | Enteritidis | Canada | Chicken breast | PRJNA1285942 | SAMN49784154 |
| Salmonella Enteritidis 0710901 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784155 |
| Salmonella Enteritidis 052239 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784156 |
| Salmonella Enteritidis 051094 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784157 |
| Salmonella Enteritidis 051095 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784158 |
| Salmonella Enteritidis 078501 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784159 |
| Salmonella Enteritidis 079901 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784160 |
| Salmonella Enteritidis 079801 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784161 |
| Salmonella Enteritidis 0715701 | Enteritidis | Canada | Feed | PRJNA1285942 | SAMN49784162 |
| Salmonella Enteritidis 719001 | Enteritidis | Canada | Chicken nuggets | PRJNA1285942 | SAMN49784163 |
| Salmonella Enteritidis 107 | Enteritidis | Canada | Bovine | PRJNA1285942 | SAMN49784164 |
| Salmonella Kentucky C45-1 | Kentucky | Canada | Crate | PRJNA1290672 | SAMN49932175 |
| Salmonella Kentucky B38-3 | Kentucky | Canada | Plucking belt | PRJNA1290672 | SAMN49932176 |
| Salmonella Kentucky W36-3 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932177 |
| Salmonella Kentucky W37-2 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932178 |
| Salmonella Kentucky W38-1 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932179 |
| Salmonella Kentucky W39-1 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932180 |
| Salmonella Kentucky W40-3 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932181 |
| Salmonella Kentucky W41-2 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932182 |
| Salmonella Kentucky W42-2 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932183 |
| Salmonella Kentucky W44-3 | Kentucky | Canada | Carcass wash/Pre-chill | PRJNA1290672 | SAMN49932184 |
| Salmonella Kentucky F36-3 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932185 |
| Salmonella Kentucky F37-3 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932186 |
| Salmonella Kentucky F38-2 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932187 |
| Salmonella Kentucky F39-3 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932188 |
| Salmonella Kentucky F40-3 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932189 |
| Salmonella Kentucky F41-2 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932190 |
| Salmonella Kentucky F42-3 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932191 |
| Salmonella Kentucky F43-3 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932192 |
| Salmonella Kentucky F44-1 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932193 |
| Salmonella Kentucky F45-2 | Kentucky | Canada | Final wash/post chill | PRJNA1290672 | SAMN49932194 |
| Salmonella Kentucky PW8-3 | Kentucky | Canada | Plucking water | PRJNA1290672 | SAMN49932195 |
| Salmonella Kentucky PW9-3 | Kentucky | Canada | Plucking water | PRJNA1290672 | SAMN49932196 |
| Salmonella Kentucky CW8-3 | Kentucky | Canada | Chill water | PRJNA1290672 | SAMN49932197 |
| Salmonella Kentucky CW9-3 | Kentucky | Canada | Chill water | PRJNA1290672 | SAMN49932198 |
Appendix A.2
Appendix A.3

References
- FSIS. Roadmap to Reducing Salmonella; United States Department of Agriculture: Washington, DC, USA, 2020. [Google Scholar]
- Sher, A.A.; Mustafa, B.E.; Grady, S.C.; Gardiner, J.C.; Saeed, A.M. Outbreaks of foodborne Salmonella enteritidis in the United States between 1990 and 2015: An analysis of epidemiological and spatial-temporal trends. Int. J. Infect. Dis. 2021, 105, 54–61. [Google Scholar] [CrossRef]
- CDC. 2021 Salmonella Outbreak Linked to Raw Frozen Breaded Stuffed Chicken Products. 2012. Available online: https://archive.cdc.gov/#/details?url=https://www.cdc.gov/salmonella/enteritidis-06-21/index.html (accessed on 26 February 2025).
- CDC. 2018 Salmonella Infections Linked to Gravel Ridge Farms Shell Eggs—Final Update. 2018. Available online: https://archive.cdc.gov/www_cdc_gov/salmonella/enteritidis-09-18/index.html (accessed on 26 February 2025).
- CDC. 2018 Salmonella Braenderup Infections Linked to Rose Acre Farms Shell Eggs (Final Update). 2018. Available online: https://archive.cdc.gov/#/details?url=https://www.cdc.gov/salmonella/braenderup-04-18/index.html (accessed on 26 February 2025).
- CDC. 2018 Salmonella Infections Linked to Raw Chicken Products. 2019. Available online: https://archive.cdc.gov/#/details?url=https://www.cdc.gov/salmonella/infantis-10-18/index.html (accessed on 26 February 2025).
- CDC. Salmonella Outbreak Linked to Eggs. 2024. Available online: https://www.cdc.gov/salmonella/outbreaks/eggs-09-24/investigation.html (accessed on 26 February 2025).
- Public Health Agency of Canada. National Enteric Surveillance Program (NESP) Annual Summary 2023; Public Health Agency of Canada: Ottawa, ON, Canada, 2025. [Google Scholar]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Soltys, R.C.; Sakomoto, C.K.; Oltean, H.N.; Guard, J.; Haley, B.J.; Shah, D.H. High-resolution comparative genomics of Salmonella Kentucky aids source tracing and detection of ST198 and ST152 lineage-specific mutations. Front. Sustain. Food Syst. 2021, 5, 695368. [Google Scholar] [CrossRef]
- Shah, D.H.; Paul, N.C.; Sischo, W.C.; Crespo, R.; Guard, J. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult. Sci. 2017, 96, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; De Cesare, A.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Salmonella control in poultry flocks and its public health impact. EFSA J. 2019, 17, e05596. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Barnett-Neefs, C.; Chavez, R.A.; Kealey, E.; Wiedmann, M.; Stasiewicz, M.J. Risk Assessment Predicts Most of the Salmonellosis Risk in Raw Chicken Parts is Concentrated in Those Few Products with High Levels of High-Virulence Serotypes of Salmonella. J. Food Prot. 2024, 87, 100304. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Vestby, L.K.; Møretrø, T.; Langsrud, S.; Heir, E.; Nesse, L.L. Biofilm forming abilities of Salmonella are correlated with persistence in fish meal- and feed factories. BMC Vet. Res. 2009, 5, 20. [Google Scholar] [CrossRef]
- Shree, P.; Singh, C.K.; Sodhi, K.K.; Surya, J.N.; Singh, D.K. Biofilms: Understanding the structure and contribution towards bacterial resistance in antibiotics. Med. Microecol. 2023, 16, 100084. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Morita, Y.; Komoda, E.; Ono, K.; Kumagai, S. Survival of Biofilm-Forming Salmonella on Stainless Steel Bolt Threads under Dry Conditions. Food Hyg. Saf. Sci. (Shokuhin Eiseigaku Zasshi) 2011, 52, 299–303. [Google Scholar] [CrossRef]
- Rodrigues, L.B.; Webber, B.; Levandowski, R.; Gehlen, S.S.; Santos, L.R.d.; Pilotto, F.; Tondo, E.C.; Nascimento, V.P.d. Biofilm formation by Salmonella enteritidis at different incubation temperatures. Acta Sci. Vet. 2019, 47, 1654. [Google Scholar] [CrossRef]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by bacterial human pathogens: Clinical relevance-development, composition and regulation-therapeutical strategies. Microb. Cell 2021, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Marin, C.; Hernandiz, A.; Lainez, M. Biofilm development capacity of Salmonella strains isolated in poultry risk factors and their resistance against disinfectants. Poult. Sci. 2009, 88, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Carrascosa, C.; Raheem, D.; Ramos, F.; Saraiva, A.; Raposo, A. Microbial Biofilms in the Food Industry—A Comprehensive Review. Int. J. Environ. Res. Public. Health 2021, 18, 2014. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Ćirković, I.; Ranin, L.; Svabić-Vlahović, M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef]
- Musa, L.; Toppi, V.; Stefanetti, V.; Spata, N.; Rapi, M.C.; Grilli, G.; Addis, M.F.; Di Giacinto, G.; Franciosini, M.P.; Casagrande Proietti, P. High Biofilm-Forming Multidrug-Resistant Salmonella Infantis Strains from the Poultry Production Chain. Antibiotics 2024, 13, 595. [Google Scholar] [CrossRef]
- Kroupitski, Y.; Golberg, D.; Belausov, E.; Pinto, R.; Swartzberg, D.; Granot, D.; Sela, S. Internalization of Salmonella enterica in Leaves Is Induced by Light and Involves Chemotaxis and Penetration through Open Stomata. Appl. Environ. Microbiol. 2009, 75, 6076–6086. [Google Scholar] [CrossRef]
- Adator, E.H.; Cheng, M.; Holley, R.; McAllister, T.; Narvaez-Bravo, C. Ability of Shiga toxigenic Escherichia coli to survive within dry-surface biofilms and transfer to fresh lettuce. Int. J. Food Microbiol. 2018, 269, 52–59. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.J.; Gerdes, S.; Olsen, G.J.; Olson, R.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Wattam, A.R.; Yoo, H. PATtyFams: Protein families for the microbial genomes in the PATRIC database. Front. Microbiol. 2016, 7, 118. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- BV-BRC. Protein Families. 2023. Available online: https://www.bv-brc.org/docs/quick_references/organisms_taxon/protein_families.html (accessed on 26 February 2025).
- Brombacher, E.; Baratto, A.; Dorel, C.; Landini, P. Gene Expression Regulation by the Curli Activator CsgD Protein: Modulation of Cellulose Biosynthesis and Control of Negative Determinants for Microbial Adhesion. J. Bacteriol. 2006, 188, 2027–2037. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- NIH. Single Nucleotide Polymorphisms (SNPs). 2024. Available online: https://www.genome.gov/genetics-glossary/Single-Nucleotide-Polymorphisms-SNPs (accessed on 26 February 2025).
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Van Staaveren, N.; Decina, C.; Baes, C.F.; Widowski, T.M.; Berke, O.; Harlander-Matauschek, A. A description of laying hen husbandry and management practices in Canada. Animals 2018, 8, 114. [Google Scholar] [CrossRef]
- Jonas, K.; Tomenius, H.; Kader, A.; Normark, S.; Römling, U.; Belova, L.M.; Melefors, Ö. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 2007, 7, 70. [Google Scholar] [CrossRef]
- Anriany, Y.; Sahu, S.N.; Wessels, K.R.; McCann, L.M.; Joseph, S.W. Alteration of the rugose phenotype in waaG and ddhC mutants of Salmonella enterica serovar Typhimurium DT104 is associated with inverse production of curli and cellulose. Appl. Environ. Microbiol. 2006, 72, 5002–5012. [Google Scholar] [CrossRef]
- Turki, Y.; Ouzari, H.; Mehri, I.; Ben Aissa, R.; Hassen, A. Biofilm formation, virulence gene and multi-drug resistance in Salmonella Kentucky isolated in Tunisia. Food Res. Int. 2012, 45, 940–946. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.C.V.; Fernandes Junior, A.; Kaneno, R.; Silva, M.G.; Araujo Junior, J.P.; Silva, N.C.C.; Rall, V.L.M. Ability of Salmonella spp. to produce biofilm is dependent on temperature and surface material. Foodborne Pathog. Dis. 2014, 11, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Obe, T.; Nannapaneni, R.; Schilling, W.; Zhang, L.; Kiess, A. Antimicrobial tolerance, biofilm formation, and molecular characterization of Salmonella isolates from poultry processing equipment. J. Appl. Poult. Res. 2021, 30, 100195. [Google Scholar] [CrossRef]
- Stepanović, S.; Ćirković, I.; Mijač, V.; Švabić-Vlahović, M. Influence of the incubation temperature, atmosphere and dynamic conditions on biofilm formation by Salmonella spp. Food Microbiol. 2003, 20, 339–343. [Google Scholar] [CrossRef]
- Piras, F.; Fois, F.; Consolati, S.G.; Mazza, R.; Mazzette, R. Influence of Temperature, Source, and Serotype on Biofilm Formation of Salmonella enterica Isolates from Pig Slaughterhouses. J. Food Prot. 2015, 78, 1875–1878. [Google Scholar] [CrossRef]
- Ziech, R.E.; Perin, A.P.; Lampugnani, C.; Sereno, M.J.; Viana, C.; Soares, V.M.; Pereira, J.G.; Pinto, J.P.d.A.N.; Bersot, L.d.S. Biofilm-producing ability and tolerance to industrial sanitizers in Salmonella spp. isolated from Brazilian poultry processing plants. LWT—Food Sci. Technol. 2016, 68, 85–90. [Google Scholar] [CrossRef]
- Podolak, R.; Enache, E.; Stone, W.; Black, D.G.; Elliott, P.H. Sources and Risk Factors for Contamination, Survival, Persistence, and Heat Resistance of Salmonella in Low-Moisture Foods. J. Food Prot. 2010, 73, 1919–1936. [Google Scholar] [CrossRef]
- Ojha, K.K.; Mishra, S.; Singh, V.K. Chapter 5—Computational molecular phylogeny: Concepts and applications. In Bioinformatics; Singh, D.B., Pathak, R.K., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 67–89. [Google Scholar]
- Holmes, S. Bootstrapping phylogenetic trees: Theory and methods. Stat. Sci. 2003, 18, 241–255. [Google Scholar] [CrossRef]
- Haley, B.J.; Kim, S.W.; Pettengill, J.; Luo, Y.; Karns, J.S.; Van Kessel, J.A.S. Genomic and evolutionary analysis of two salmonella enterica serovar Kentucky sequence types isolated from bovine and poultry sources in North America. PLoS ONE 2016, 11, e0161225. [Google Scholar] [CrossRef]
- Chin, K.C.J.; Taylor, T.D.; Hebrard, M.; Anbalagan, K.; Dashti, M.G.; Phua, K.K. Transcriptomic study of Salmonella enterica subspecies enterica serovar Typhi biofilm. BMC Genom. 2017, 18, 836. [Google Scholar] [CrossRef]
- Domka, J.; Lee, J.; Wood Thomas, K. YliH (BssR) and YceP (BssS) Regulate Escherichia coli K-12 Biofilm Formation by Influencing Cell Signaling. Appl. Environ. Microbiol. 2006, 72, 2449–2459. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Yang, Z.; Pu, M.; Peti, W.; Wood, T.K. Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ. Microbiol. 2011, 13, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Jenal, U.; Malone, J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 2006, 40, 385–407. [Google Scholar] [CrossRef] [PubMed]
- Gerstel, U.; Römling, U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 2003, 154, 659–667. [Google Scholar] [CrossRef]
- Hammar, M.; Arnqvist, A.; Bian, Z.; Olsén, A.; Normark, S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995, 18, 661–670. [Google Scholar] [CrossRef]
- Bhoite, S.; van Gerven, N.; Chapman, M.R.; Remaut, H. Curli Biogenesis: Bacterial Amyloid Assembly by the Type VIII Secretion Pathway. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef]
- Römling, U.; Rohde, M.; Olsén, A.; Normark, S.; Reinköster, J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000, 36, 10–23. [Google Scholar] [CrossRef]
- Römling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [CrossRef]
- Römling, U.; Sierralta, W.D.; Eriksson, K.; Normark, S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 1998, 28, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Robbe-Saule, V.; Jaumouillé, V.; Prévost, M.-C.; Guadagnini, S.; Talhouarne, C.; Mathout, H.; Kolb, A.; Norel, F. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006, 188, 3983–3994. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.K.; Dozois, C.M.; Nickerson, C.A.; Zuppardo, A.; Terlonge, J.; Curtiss, R., III. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2001, 41, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Spurbeck, R.R.; Stapleton, A.E.; Johnson, J.R.; Walk, S.T.; Hooton, T.M.; Mobley, H.L. Fimbrial profiles predict virulence of uropathogenic Escherichia coli strains: Contribution of ygi and yad fimbriae. Infect. Immun. 2011, 79, 4753–4763. [Google Scholar] [CrossRef]
- Garnett, J.A.; Martínez-Santos, V.I.; Saldaña, Z.; Pape, T.; Hawthorne, W.; Chan, J.; Simpson, P.J.; Cota, E.; Puente, J.L.; Girón, J.A. Structural insights into the biogenesis and biofilm formation by the Escherichia coli common pilus. Proc. Natl. Acad. Sci. USA 2012, 109, 3950–3955. [Google Scholar] [CrossRef]
- Christie, P.J.; Atmakuri, K.; Krishnamoorthy, V.; Jakubowski, S.; Cascales, E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005, 59, 451–485. [Google Scholar] [CrossRef]
- Chen, C.-L.; Wang, C.-Y.; Chu, C.; Su, L.-H.; Chiu, C.-H. Functional and molecular characterization of pSE34 encoding a type IV secretion system in Salmonella enterica serotype Enteritidis phage type 34. FEMS Immunol. Med. Microbiol. 2009, 57, 274–283. [Google Scholar] [CrossRef]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef]
- Wurpel, D.J.; Beatson, S.A.; Totsika, M.; Petty, N.K.; Schembri, M.A. Chaperone-Usher Fimbriae of Escherichia coli. PLoS ONE 2013, 8, e52835. [Google Scholar] [CrossRef]
- Cascales, E.; Gavioli, M.; Sturgis, J.N.; Lloubès, R. Proton motive force drives the interaction of the inner membrane tolA and outer membrane Pal proteins in Escherichia coli. Mol. Microbiol. 2000, 38, 904–915. [Google Scholar] [CrossRef]
- Walburger, A.; Lazdunski, C.; Corda, Y. The Tol/Pal system function requires an interaction between the C-terminal domain of tolA and the N-terminal domain of tolB. Mol. Microbiol. 2002, 44, 695–708. [Google Scholar] [CrossRef]
- Su, S.; Li, Z.; Sun, Y.; Gao, S.; Gao, Q. The multifaceted role of tolA protein in promoting survival, biofilm formation and virulence of avian pathogenic Escherichia coli. Poult. Sci. 2024, 103, 104142. [Google Scholar] [CrossRef]
- Nikhil, K.C.; Priyadarsini, S.; Pashupathi, M.; Ratta, B.; Saxena, M.; Ramakrishnan, S.; Behera, P.; Kumar, A. Regulatory Role of fnr Gene in Growth and tolA Gene Expression in Salmonella Typhimurium. Indian J. Anim. Res. 2021, 55, 774–779. [Google Scholar] [CrossRef]
- Solano, C.; García, B.; Valle, J.; Berasain, C.; Ghigo, J.M.; Gamazo, C.; Lasa, I. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol. Microbiol. 2002, 43, 793–808. [Google Scholar] [CrossRef]


| Serovar | Isolate ID | Source | Serovar | Isolate ID | Source |
|---|---|---|---|---|---|
| Enteritidis | 51094 | Chicken nuggets | Kentucky | C45-1 | Crate |
| Enteritidis | 51095 | Chicken nuggets | Kentucky | B38-3 | Plucking belt |
| Enteritidis | 52239 | Chicken nuggets | Kentucky | W36-3 | Carcass wash/Pre-chill |
| Enteritidis | 618901 | Chicken nuggets | Kentucky | W37-2 | Carcass wash/Pre-chill |
| Enteritidis | 619102 | Chicken nuggets | Kentucky | W38-1 | Carcass wash/Pre-chill |
| Enteritidis | 78501 | Chicken nuggets | Kentucky | W39-1 | Carcass wash/Pre-chill |
| Enteritidis | 79801 | Chicken nuggets | Kentucky | W40-3 | Carcass wash/Pre-chill |
| Enteritidis | 79901 | Chicken nuggets | Kentucky | W41-2 | Carcass wash/Pre-chill |
| Enteritidis | 719001 | Chicken nuggets | Kentucky | W42-2 | Carcass wash/Pre-chill |
| Enteritidis | 710901 | Chicken nuggets | Kentucky | W44-3 | Carcass wash/Pre-chill |
| Enteritidis | 53932 | Chicken breast | Kentucky | F36-3 | Final wash/post chill |
| Enteritidis | 53936 | Chicken breast | Kentucky | F37-3 | Final wash/post chill |
| Enteritidis | 61401 | Feed | Kentucky | F38-2 | Final wash/post chill |
| Enteritidis | 715701 | Feed | Kentucky | F39-3 | Final wash/post chill |
| Enteritidis | 107 | Bovine | Kentucky | F40-3 | Final wash/post chill |
| Kentucky | F41-2 | Final wash/post chill | |||
| Kentucky | F42-3 | Final wash/post chill | |||
| Kentucky | F43-3 | Final wash/post chill | |||
| Kentucky | F44-1 | Final wash/post chill | |||
| Kentucky | F45-2 | Final wash/post chill | |||
| Kentucky | PW8-3 | Plucking water | |||
| Kentucky | PW9-3 | Plucking water | |||
| Kentucky | CW8-3 | Chill water | |||
| Kentucky | CW9-3 | Chill water |
| Gene Category | Protein Name | Function |
|---|---|---|
| Biofilm regulation protein-coding genes | BdcA (Yjgl) | Cyclic-di-GMP-binding biofilm dispersal mediator protein |
| BssS/BssR | Biofilm regulator | |
| YjgK | Linked to biofilm formation | |
| Biofilm structural protein | BapA | A large cell-surface protein required for biofilm formation by S. Enteritidis |
| Curli production | CsgA | Major curlin subunit precursor |
| CsgB | Minor curlin subunit, nucleation component of curlin monomers | |
| CsgC | Putative curli production protein | |
| CsgD | Transcriptional regulator for the 2nd curli operon | |
| CsgE/CsgF, CsgG | Curli production assembly/transport component | |
| OmpR | Two-component system response regulator | |
| RpoS | RNA polymerase sigma factor | |
| Regulator of RpoS | ||
| Crl | An RpoS-binding factor, binding to RpoS, facilitates RNA polymerase holoenzyme formation (EσS) | |
| Curlin gene transcriptional activator | ||
| MlrA | HTH-type transcriptional regulator; Positive regulator of CsgD expression | |
| DdhC | Involved in the synthesis of abequose | |
| WaaG | UDP-glucose:(heptosyl) LPS alpha1,3-glucosyltransferase | |
| Cellulose production | BcsA | Cellulose synthase catalytic subunit |
| BcsB | Cyclic di-GMP-binding protein | |
| BcsC | Cellulose synthase operon protein C | |
| BcsE/BcsF/BcsG/BcsQ | Cellulose biosynthesis protein | |
| AdrA (DgcC) | Diguanylate cyclase positively regulates cellulose synthesis via production of the secondary messenger signaling molecule (3′-5′)-cyclic diguanosine monophosphate (c-di-GMP) | |
| Fimbriae production | StbC */StdB */SthB */StiC * | Fimbriae usher protein |
| SthA */StiB * | Putative fimbrial chaperone | |
| StiA * | Putative fimbrial subunit | |
| SafB * | Salmonella atypical fimbria periplasmic chaperone | |
| FimI * | Fimbriae-like adhesin | |
| FimW * | Fimbriae W protein | |
| FimY */FimZ * | Transcriptional regulator of fimbriae expression | |
| BcfA */BcfD */BcfE */BcfF * | Fimbrial subunit | |
| BcfB */BcfG * | Fimbrial chaperone | |
| StfA | Major fimbrial subunit | |
| StfC | Fimbriae usher protein | |
| StfD | Periplasmic fimbrial chaperone | |
| StfE/StfF/StfG | Minor fimbrial subunit | |
| SfmA/SfmH/YehD/SfmF | Uncharacterized fimbrial-like protein | |
| SfmC/YehC | Probable fimbrial chaperone | |
| YehA/YadK | Uncharacterized fimbrial-like protein | |
| YadU | Uncharacterized protein in the stf fimbrial cluster | |
| YhcA | Uncharacterized fimbrial chaperone | |
| EcpD | Fimbria adhesin | |
| Pili | PilA | Type IV pilin |
| PilB | Type IV pilus assembly, ATPase | |
| PilC | Type IV pilus assembly protein | |
| PilM/PilN/PilO/PilP/PilQ | Type IV pilus biogenesis protein | |
| Flagella | FliC | Flagellin |
| FliD | Flagellar cap protein | |
| FliE | Flagellar hook-basal body complex protein | |
| FliF | Flagellar M-ring protein | |
| FliG | Flagellar motor switch protein | |
| FliH | Flagellar assembly protein | |
| FliI | Flagellum-specific ATP synthase | |
| FliJ | Flagellar protein | |
| FliK | Flagellar hook-length control protein | |
| FliL | Flagellar basal body-associated protein | |
| FliM/FliN | Flagellar motor switch protein | |
| FliO/FliP/FliQ/FliR/FliS/FliT | Flagellar biosynthesis protein | |
| FlgA | Flagellar basal-body P-ring formation protein | |
| FlgB/FlgC/FlgF/FlgG | Flagellar basal-body rod protein | |
| FlgD | Flagellar basal-body rod modification protein | |
| FlgE | Flagellar hook protein | |
| FlgH | Flagellar L-ring protein | |
| FlgI | Flagellar P-ring protein | |
| FlgJ/FlhE | Flagellar protein | |
| FlgK/FlgL | Flagellar hook-associated protein | |
| FlgM | Negative regulator of flagellin synthesis | |
| FlgN/FlhA/FlhB | Flagellar biosynthesis protein | |
| FlhC/FlhD | Flagellar transcriptional activator | |
| MotA/MotB | Flagellar motor rotation protein | |
| YcgR | Flagellar brake protein | |
| flk | Flagellar regulator | |
| RtsA | Type III secretion and flagellar regulator | |
| RtsB | Flagellar regulon repressor | |
| Chemotaxis regulator | Transmits chemoreceptor signals to flagellar motor components CheY | |
| RNA polymerase sigma factor for the flagellar operon | ||
| Type IV secretion system | NAD(P)H dehydrogenase (quinone), VirB4, VirB11 | ATPase is required for both the assembly of the type IV secretion complex and the secretion of the T-DNA complex |
| VirB1 | Peptidoglycan hydrolase, involved in T-DNA transfer | |
| VirB3 | Inner membrane protein forms a channel for the type IV secretion of the T-DNA complex | |
| VirB6 | Inner membrane protein of the type IV secretion of the T-DNA complex | |
| VirB10 | Inner membrane protein of type IV secretion of T-DNA complex, TonB-like | |
| VirD4 | Coupling protein, ATPase required for T-DNA transfer | |
| Serovar | Isolates | Type | Function | Upstream Feature | Downstream Feature | snpEff Type | snpEff Impact |
|---|---|---|---|---|---|---|---|
| SE | 715701 (I), 79801 (I), 79901 (I), 52239 (S), 51094 (S), 51095 (S) | Nonsyn | Cellulose synthase catalytic subunit [UDP-forming] (EC 2.4.1.12) | Cellulose biosynthesis protein BcsQ | Cyclic di-GMP-binding protein BcsB | stop gained | High |
| SE | 715701, 79801, 79901 (I) | Deletion | Cellulose synthase operon protein C | beta-1,4-glucanase (cellulase) (EC 3.2.1.4) | C-di-GMP phosphodiesterase (EC 3.1.4.52) | frameshift | High |
| SE | 715701 (I), 79801 (I), 79901 (I), 52239 (S), 51094 (S), 51095 (S) | Insertion | Uncharacterized fimbrial-like protein YehA | Outer membrane usher protein YehB | Nickel/cobalt homeostasis protein RcnB | frameshift | High |
| SK | F43-3 (N) | Nonsyn | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | missense | Moderate |
| SE | 53936 (S) | Nonsyn | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | missense | Moderate |
| SE | 52239, 51094, 53936 (S) | Nonsyn | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | missense | Moderate |
| SE | 719001 (S), 715701 (I), 52239 (S), 51094 (S), 710901 (S), 53936 (S), 618901 (S), 619102 (S) | Nonsyn | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | missense | Moderate |
| SE | 107 (I) | Nonsyn | UPF0126 inner membrane protein YadS | Vitamin B12 ABC transporter, substrate-binding protein BtuF | Iron-sulfur cluster insertion protein ErpA | missense | Moderate |
| SE | 107 (I) | Nonsyn | Glutamate-1-semialdehyde 2,1-aminomutase (EC 5.4.3.8) | FIG01048481: hypothetical protein | Uncharacterized protein YadU in the stf fimbrial cluster | missense | Moderate |
| SE | 715701, 79801, 79901 (I) | Nonsyn | Glutamate-1-semialdehyde 2,1-aminomutase (EC 5.4.3.8) | FIG01048481: hypothetical protein | Uncharacterized protein YadU in the stf fimbrial cluster | missense | Moderate |
| SE | 715701 (I), 79801 (I), 79901 (I) 52239 (S), 51094 (S), 51095 (S) | Nonsyn | Flagellar basal-body P-ring formation protein FlgA | Negative regulator of flagellin synthesis, FlgM (anti-sigma28) | Flagellar basal-body rod protein FlgB | missense | Moderate |
| SE | 107 (I) | Nonsyn | Cytoplasmic alpha-amylase (EC 3.2.1.1) | Flagellar biosynthesis protein FliT | Uncharacterized lipoprotein YedD | missense | Moderate |
| SE | 52239 (S) | Nonsyn | Curli production assembly/transport component CsgF | Curli production assembly/transport component CsgG | Curli production assembly/transport component CsgE | missense | Moderate |
| SK | CW8-3 (W), F38-2 (N), F39-3 (N), F41-2 (N), F42-3 (N), F43-3 (N), F44-1 (S), F45-2 (N), W36-3 (S), W37-2 (N), W38-1 (N), W39-1 (N), W41-2 (N), W42-2 (N), W44-3 (N), F36-3 (N) | Synon | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | synonymous | Low |
| SK | C45-1 (N), CW8-3 (W), F38-2 (N), F39-3 (N), F41-2 (N), F42-3 (N), F43-3 (N), F44-1 (S), F45-2, W36-3 (S), W37-2 (S), W38-1 (N), W39-1 (N), W41-2 (N), W44-3 (N), F36-3 (N) | Synon | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | synonymous | Low |
| SK | CW8-3 (W), W38-1 (N), W39-1 (N), W41-2 (N), F36-3 (N) | Synon | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | synonymous | Low |
| SK | W38-1, W39-1 (N) | Synon | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | synonymous | Low |
| SE | 715701 (I), 79801 (I), 52239 (S), 51094 (S), 710901 (S), 53936 (S) | Synon | TolA protein | Tol biopolymer transport system, TolR protein | Tol-Pal system beta propeller repeat protein TolB | synonymous | Low |
| SE | 107 (I) | Synon | Cellulose biosynthesis protein BcsG | Small inner membrane protein, YmgF family | Cellulose biosynthesis protein BcsF | synonymous | Low |
| SE | 107 (I) | Synon | Cellulose biosynthesis protein BcsQ | Putative cytoplasmic protein YhjR | Cellulose synthase catalytic subunit [UDP-forming] (EC 2.4.1.12) | synonymous | Low |
| SE | 107 (I) | Synon | Cellulose synthase operon protein C | beta-1,4-glucanase (cellulase) (EC 3.2.1.4) | c-di-GMP phosphodiesterase (EC 3.1.4.52) | synonymous | Low |
| SE | 107 (I) | Synon | Uncharacterized protein YadE | Putative PTS system IIA component YadI | Aspartate 1-decarboxylase (EC 4.1.1.11) | synonymous | Low |
| SE | 107 (I) | Synon | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase (EC 2.1.1.61)/FAD-dependent cmnm(5)s(2)U34 oxidoreductase | Uncharacterized protein YfcL | 3-oxoacyl-[acyl-carrier-protein] synthase, KASI (EC 2.3.1.41) | synonymous | Low |
| SE | 107 (I) | Synon | Uncharacterized protein YfcC | Transketolase, C-terminal section (EC 2.2.1.1) | BioD-like N-terminal domain/Phosphate acetyltransferase (EC 2.3.1.8) | synonymous | LOW |
| SE | 107 (I) | Synon | Flagellar motor rotation protein MotA | Flagellar motor rotation protein MotB | Flagellar transcriptional activator FlhC | synonymous | LOW |
| SE | 107 (I) | Synon | hypothetical protein | Flagellar regulon repressor RtsB | hypothetical protein | synonymous | LOW |
| Proteins Present in Strong SE 53932, but Absent in Intermediate SE 107 Excluding Hypothetical Proteins and Proteins with Unknown Functions |
|---|
| Exoenzymes regulatory protein AepA precursor |
| putative e6 protein |
| ail and ompX Homolog |
| Phage repressor protein cI |
| Phage Cox (control of excision) protein |
| Phage activator protein cII |
| FIL protein |
| Phage replication protein GpB * |
| Phage Orf80 protein * |
| Phage replication protein GpA, endonuclease * |
| Phage protein * |
| DNA-damage-inducible protein I * |
| Phage protein * |
| Phage portal vertex protein GpQ * |
| Phage terminase, ATPase subunit GpP * |
| Phage capsid scaffolding protein GpO * |
| Phage major capsid protein GpN * |
| Phage terminase, endonuclease subunit GpM * |
| Phage head completion-stabilization protein GpL * |
| Phage tail protein GpX * |
| Phage holin * |
| Phage lysis regulatory protein, LysB * |
| Phage tail completion protein GpR *, GpS * |
| Phage baseplate assembly protein GpV *, GpW *, GpJ * |
| Phage tail formation protein GpI * |
| putative inner membrane protein * |
| Phage tail sheath monomer GpFI * |
| Phage major tail tube protein GpFII * |
| Phage tail protein GpE * |
| Phage P2 GpE family protein * |
| Phage tail protein GpU * |
| Phage tail formation protein GpD * |
| Oxaloacetate decarboxylase Na(+) pump, alpha chain (EC 4.1.1.3) |
| Anaerobic sulfite reductase subunit A |
| SdiA-regulated putative outer membrane protein SrgB |
| Putative cytoplasmic protein |
| Oxaloacetate decarboxylase Na(+) pump, alpha chain (EC 4.1.1.3) |
| RelB/StbD replicon stabilization protein (antitoxin to RelE/StbE) ** |
| PI protein ** |
| DNA distortion protein 3 ** |
| Cell division protein FtsH (EC 3.4.24.-) ** |
| IncN plasmid KikA protein ** |
| Coupling protein VirD4, ATPase required for T-DNA transfer ** |
| ATPase required for both assembly of type IV secretion complex and secretion of T-DNA complex, VirB11 ** |
| Inner membrane protein of type IV secretion of T-DNA complex, TonB-like, VirB10 ** |
| Forms the bulk of type IV secretion complex that spans outer membrane and periplasm (VirB9) ** |
| putative conjugal transfer protein |
| Inner membrane protein of type IV secretion of T-DNA complex, VirB6 ** |
| IncQ plasmid conjugative transfer protein TraG ** |
| Minor pilin of type IV secretion complex (VirB5) ** |
| Inner membrane protein forms channel for type IV secretion of T-DNA complex, VirB3/ATPase required for both assembly of type IV secretion complex and secretion of T-DNA complex, VirB4 ** |
| Pilx2 protein ** |
| Peptidoglycan hydrolase VirB1, involved in T-DNA transfer ** |
| IncQ plasmid conjugative transfer DNA nicking endonuclease TraR (pTi VirD2 homolog) ** |
| DNA distortion protein 1 ** |
| putative membrane protein |
| Phage integrase |
| E3 ubiquitin-protein ligase SspH2 |
| Proteins Present in Strong SE 53932, but Absent in Intermediate SE 107 Excluding Hypothetical Proteins and Proteins with Unknown Functions |
|---|
| Phage head, head-DNA stabilization protein D * |
| Pahe protein b |
| Ail and ompX Homolog b |
| Protein KdpF a |
| Oxaloacetate decarboxylase Na(+) pump, alpha chain (EC 4.1.1.3) * |
| Kappa-fimbriae probable subunit * |
| Kappa-fimbriae major subunit * |
| Kappa-fimbriae regulatory protein * |
| RepFIB replication protein A * |
| Putative periplasmic protein * |
| Putative inner membrane protein * |
| putative invasin * |
| Transposase * |
| 27.5 kDa virulence protein * |
| Actin-ADP-ribosyltransferase, toxin SpvB * |
| Outer membrane protein * |
| Virulence genes transcriptional activator * |
| putative phosphoribulokinase/uridine kinase protein * |
| putative integrase protein * |
| RlgA * |
| Putative cytoplasmic protein * |
| Alpha-helical coiled coil protein * |
| Chromosome (plasmid) partitioning protein ParA *, ParB |
| putative ParB-like nuclease * |
| Plasmid SOS inhibition protein PsiA *, PsiB * |
| UPF0380 proteins YafZ and homologs * |
| IncF plasmid conjugative transfer mating signal transduction protein TraM * |
| IncF plasmid conjugative transfer regulator TraJ *, TraY * |
| IncF plasmid conjugative transfer pilin protein TraA * |
| IncF plasmid conjugative transfer pilus assembly protein TraB *, TraE *, TraK *, TraL * |
| Oxaloacetate decarboxylase Na(+) pump, alpha chain (EC 4.1.1.3) |
| secreted effector protein * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Ebosa, O.; Diarra, M.; Nadon, C.; McAllister, T.; Sparling, R.; Narvaez-Bravo, C. Genomic Drivers of Biofilm Formation in Salmonella Enteritidis and S. Kentucky from Poultry Production. Microorganisms 2025, 13, 2473. https://doi.org/10.3390/microorganisms13112473
Zhang J, Ebosa O, Diarra M, Nadon C, McAllister T, Sparling R, Narvaez-Bravo C. Genomic Drivers of Biofilm Formation in Salmonella Enteritidis and S. Kentucky from Poultry Production. Microorganisms. 2025; 13(11):2473. https://doi.org/10.3390/microorganisms13112473
Chicago/Turabian StyleZhang, Jiayi, Oritsetimeyin Ebosa, Moussa Diarra, Celine Nadon, Tim McAllister, Richard Sparling, and Claudia Narvaez-Bravo. 2025. "Genomic Drivers of Biofilm Formation in Salmonella Enteritidis and S. Kentucky from Poultry Production" Microorganisms 13, no. 11: 2473. https://doi.org/10.3390/microorganisms13112473
APA StyleZhang, J., Ebosa, O., Diarra, M., Nadon, C., McAllister, T., Sparling, R., & Narvaez-Bravo, C. (2025). Genomic Drivers of Biofilm Formation in Salmonella Enteritidis and S. Kentucky from Poultry Production. Microorganisms, 13(11), 2473. https://doi.org/10.3390/microorganisms13112473







