Neobacillus terrisolis sp. nov. and Neobacillus solisequens sp. nov. Isolated from Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culturing
2.2. Morphological and Physiological Analysis
2.3. 16S rRNA Gene Sequencing and Phylogenetic Analysis
2.4. Genome Sequencing and Analysis
3. Results and Discussion
3.1. Morphological and Physiological Investigation
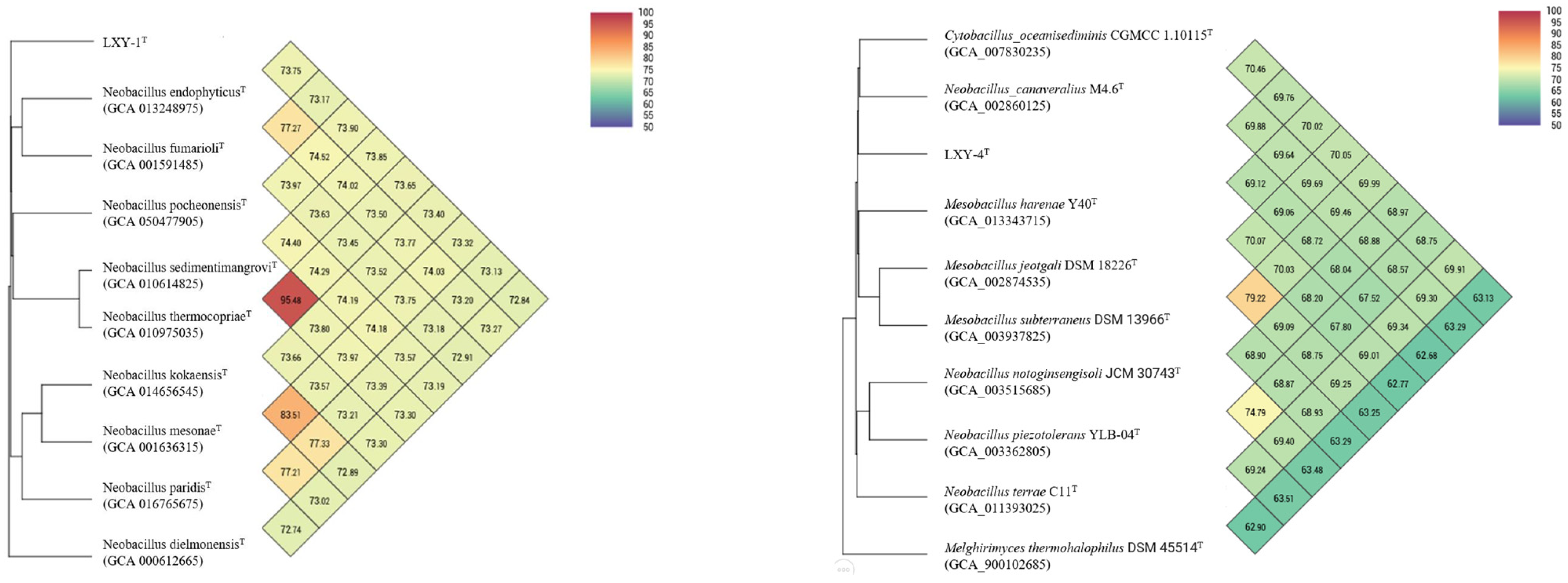
3.2. Phylogenetic Analysis
3.3. Genome Characteristics
3.4. Chemotaxonomic Characterization
4. Conclusions
4.1. Description of Neobacillus terrisolis sp. nov.
4.2. Description of Neobacillus solisequens sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ANI | Average nucleotide identity |
| dDDH | Digital DNA–DNA hybridization |
| ML | Maximum-likelihood |
| MP | Maximum parsimony |
| NJ | Neighbor-joining |
| R2A | Reasoner’s 2A |
Appendix A
References
- Patel, S.; Gupta, R.S. A Phylogenomic and Comparative Genomic Framework for Resolving the Polyphyly of the Genus Bacillus: Proposal for Six New Genera of Bacillus Species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. And Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [Google Scholar] [CrossRef]
- Jiang, L.M.; Lee, M.H.; Jeong, J.C.; Kim, D.H.; Kim, C.Y.; Kim, S.W.; Lee, J. Neobacillus endophyticus sp. nov., an Endophytic Bacterium Isolated from Selaginella Involvens Roots. Int. J. Syst. Evol. Microbiol. 2021, 71, 8. [Google Scholar] [CrossRef]
- Zhan, P.C.; Li, C.J.; Zhang, Z.; Mao, R.F.; Liu, J.R.; Jiang, X.W.; Zhi, X.Y.; Yang, L.L. Neobacillus paridis sp. nov., an Endophyte of Paris Polyphylla Smith Var. Yunnanensis. Arch. Microbiol. 2022, 204, 7. [Google Scholar] [CrossRef]
- Han, D.M.; Choi, D.G.; Baek, J.H.; Hao, L.J.; Jeon, C.O. Neobacillus terrae sp. nov., Isolated from Mountain Soil. Int. J. Syst. Evol. Microbiol. 2023, 73, 9. [Google Scholar] [CrossRef]
- Yuki, K.; Matsubara, H.; Yamaguchi, S. Neobacillus kokaensis sp. nov., Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2022, 72, 7. [Google Scholar] [CrossRef]
- Ten, L.N.; Baek, S.H.; Im, W.T.; Liu, Q.M.; Aslam, Z.; Lee, S.T. Bacillus panaciterrae sp. nov., Isolated from Soil of a Ginseng Field. Int. J. Syst. Evol. Microbiol. 2006, 56, 2861–2866. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, D.; Gugliandolo, C.; Stackebrandt, E.; Maugeri, T.L. Bacillus vulcani sp. nov., a Novel Thermophilic Species Isolated from a Shallow Marine Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2000, 50, 2009–2012. [Google Scholar] [CrossRef]
- Liu, B.; Liu, G.H.; Hu, G.H.; Chen, M.C. Bacillus mesonae sp. nov., Isolated from the Root of Mesona Chinensis. Int. J. Syst. Evol. Microbiol. 2014, 64, 3346–3352. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Busse, H.J.; Glaeser, S.P.; Kloepper, J.W.; Hu, C.H.; McInroy, J.A. Bacillus cucumis sp. nov Isolated from the Rhizosphere of Cucumber (Cucumis sativus). Int. J. Syst. Evol. Microbiol. 2016, 66, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Kaempfer, P.; Glaeser, S.P.; McInroy, J.A.; Clermont, D.; Lipski, A.; Criscuolo, A. Neobacillus rhizosphaerae sp. nov., Isolated from the Rhizosphere, and Reclassification of Bacillus dielmonensis as Neobacillus dielmonensis comb. nov. Int. J. Syst. Evol. Microbiol. 2022, 72, 9. [Google Scholar] [CrossRef]
- Han, L.C.; Yang, G.Q.; Zhou, X.M.; Yang, D.H.; Hu, P.; Lu, Q.; Zhou, S.G. Bacillus thermocopriae sp. nov., Isolated from a Compost. Int. J. Syst. Evol. Microbiol. 2013, 63, 3024–3029. [Google Scholar] [CrossRef]
- Yu, L.B.; Tang, X.X.; Wei, S.P.; Qiu, Y.K.; Xu, X.S.T.; Xu, G.X.; Wang, Q.L.; Yang, Q. Two Novel Species of the Family Bacillaceae: Oceanobacillus piezotolerans sp. nov. And Bacillus piezotolerans sp. nov., from Deep-Sea Sediment Samples of Yap Trench. Int. J. Syst. Evol. Microbiol. 2019, 69, 3022–3030. [Google Scholar] [CrossRef]
- Bittar, F.; Bibi, F.; Ramasamy, D.; Lagier, J.C.; Azhar, E.I.; Jiman-Fatani, A.A.; Al-Ghamdi, A.K.; Nguyen, T.T.; Yasir, M.; Fournier, P.E.; et al. Non Contiguous-Finished Genome Sequence and Description of Bacillus jeddahensis sp. nov. Stand. Genom. Sci. 2015, 10, 12. [Google Scholar] [CrossRef]
- El-Sapagh, S.H.; El-Zawawy, N.A.; Elshobary, M.E.; Alquraishi, M.; Zabed, H.M.; Nouh, H.S. Harnessing the Power of Neobacillus niacini Aumc-B524 for Silver Oxide Nanoparticle Synthesis: Optimization, Characterization, and Bioactivity Exploration. Microb. Cell Factories 2024, 23, 220. [Google Scholar] [CrossRef]
- Tan, Z.; Guo, X.; Tan, X.; Liang, Q.; Peng, G. New Neobacillus mesonae Fjat-13985ndj36 Strain Is Preserved in Guangdong Microbiological Culture Collection Center, Used for Preventing and Treating Rice Sheath Blight and Plant Blight by Inhibiting Rhizoctonia solani, and Fusarium oxysporum and Producing Protease; South China Agricultural University: Guangzhou, China, 2024. [Google Scholar]
- Ryu, H.W.; Yoo, S.K.; Choi, J.M.; Cho, K.S.; Cha, D.K. Thermophilic Biofiltration of H2S and Isolation of a Thermophilic and Heterotrophic H2S-Degrading Bacterium, Bacillus Sp Tso3. J. Hazard. Mater. 2009, 168, 501–506. [Google Scholar] [CrossRef]
- Tyulenev, A.; Smirnova, G.; Ushakov, V.; Kalashnikova, T.; Sutormina, L.; Oktyabrsky, O. Stress-Induced Sulfide Production by Bacillus subtilis and Bacillus megaterium. Microorganisms 2024, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Li, J.J.; Shen, Y.M.; Liu, M.H.; Liu, H.L.; Liu, H.W.; Xun, L.Y.; Xia, Y.Z. A Sulfide-Sensor and a Sulfane Sulfur-Sensor Collectively Regulate Sulfur-Oxidation for Feather Degradation by Bacillus licheniformis. Commun. Biol. 2023, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Goyal, P.; Belapurkar, P.; Kar, A. A Review on in Vitro and in Vivo Bioremediation Potential of Environmental and Probiotic Species of Bacillus and Other Probiotic Microorganisms for Two Heavy Metals, Cadmium and Nickel. Biosci. Biotechnol. Res. Asia 2019, 16, 1–13. [Google Scholar] [CrossRef]
- Belapurkar, P.; Goyal, P.; Kar, A. In Vitro Evaluation of Bioremediation Capacity of a Commercial Probiotic, Bacillus coagulans, for Chromium (Vi) and Lead (Ii) Toxicity. J. Pharm. Bioallied Sci. 2016, 8, 272–276. [Google Scholar] [CrossRef]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid-Determination of 16s Ribosomal-Rna Sequences for Phylogenetic Analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef]
- Reasoner, D.J.; Geldreich, E.E. A New Medium for the Enumeration and Subculture of Bacteria from Potable Water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16s Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. A Place for DNA-DNA Reassociation and 16s Ribosomal-Rna Sequence-Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Liu, D.M.; Zhang, Y.F.; Fan, G.M.; Sun, D.Z.; Zhang, X.J.; Yu, Z.F.; Wang, J.F.; Wu, L.H.; Shi, W.Y.; Ma, J.C. Ipga: A Handy Integrated Prokaryotes Genome and Pan-Genome Analysis Web Service. iMeta 2022, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Hucker, G.J. A New Modification and Application of the Gram Stain. J. Bacteriol. 1921, 6, 395–397. [Google Scholar] [CrossRef]
- Preston-Mafham, J.; Boddy, L.; Randerson, P.F. Analysis of Microbial Community Functional Diversity Using Sole-Carbon-Source Utilisation Profiles—A Critique. FEMS Microbiol. Ecol. 2002, 42, 1–14. [Google Scholar] [PubMed]
- Wilkins, T.D.; Abramson, I.J.; Moore, W.E.C.; Holdeman, L.V. Standardized Single-Disk Method for Antibiotic Susceptibility Testing of Anaerobic Bacteria. Antimicrob. Agents Chemother. 1972, 1, 451–459. [Google Scholar] [CrossRef]
- Wang, Y.J.; Xu, X.J.; Zhou, N.; Sun, Y.T.; Liu, C.; Liu, S.J.; You, X. Parabacteroides acidifaciens sp. nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2019, 69, 761–766. [Google Scholar] [CrossRef]
- Abdugheni, R.; Li, D.H.; Wang, Y.J.; Du, M.X.; Zhou, N.; Liu, C.; Liu, S.J. Acidaminococcus homini s sp. nov., Amedibacillus hominis sp. nov., Lientehia hominis gen. nov. sp. nov., Merdimmobilis hominis gen. nov. sp. nov., and Paraeggerthella hominis sp. nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 2023, 73, 21. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. Ncbi Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 21, baaa062. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method—A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary Trees from DNA-Sequences—A Maximum-Likelihood Approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Yan, Z.; Cao, Z.; Liu, Y.S.; Ogilvie, H.A.; Nakhleh, L. Maximum Parsimony Inference of Phylogenetic Networks in the Presence of Polyploid Complexes. Syst. Biol. 2022, 71, 706–720. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Abdugheni, R.; Liu, C.; Liu, F.L.; Zhou, N.; Jiang, C.Y.; Liu, Y.H.; Li, L.; Li, W.J.; Liu, S.J. Comparative Genomics Reveals Extensive Intra-Species Genetic Divergence of the Prevalent Gut Commensal Ruminococcus gnavu. Microb. Genom. 2023, 9, 12. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. Checkm: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. Tygs Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 10. [Google Scholar] [CrossRef]
- Heyrman, J.; Vanparys, B.; Logan, N.A.; Balcaen, A.; Rodíguez-Díaz, M.; Felske, A.; De Vos, P. Bacillus novalis sp. nov., Bacillus vireti sp. nov., Bacillus soli sp. nov., Bacillus bataviensis sp. nov. and Bacillus drentensis sp. nov., from the Drentse a Grasslands. Int. J. Syst. Evol. Microbiol. 2004, 54, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Kanso, S.; Greene, A.C.; Patel, B.K.C. Bacillus subterraneus sp. nov., an Iron- and Manganese-Reducing Bacterium from a Deep Subsurface Australian Thermal Aquifer. Int. J. Syst. Evol. Microbiol. 2002, 52, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Gittel, A.; Seidel, M.; Kuever, J.; Galushko, A.S.; Cypionka, H.; Könneke, M. Desulfopila inferna sp nov., a Sulfate-Reducing Bacterium Isolated from the Subsurface of a Tidal Sand-Flat. Int. J. Syst. Evol. Microbiol. 2010, 60, 1626–1630. [Google Scholar] [CrossRef] [PubMed]
- Hao, O.J.; Chen, J.M.; Huang, L.; Buglass, R.L. Sulfate-Reducing Bacteria. Crit. Rev. Environ. Sci. Technol. 1996, 26, 155–187. [Google Scholar] [CrossRef]

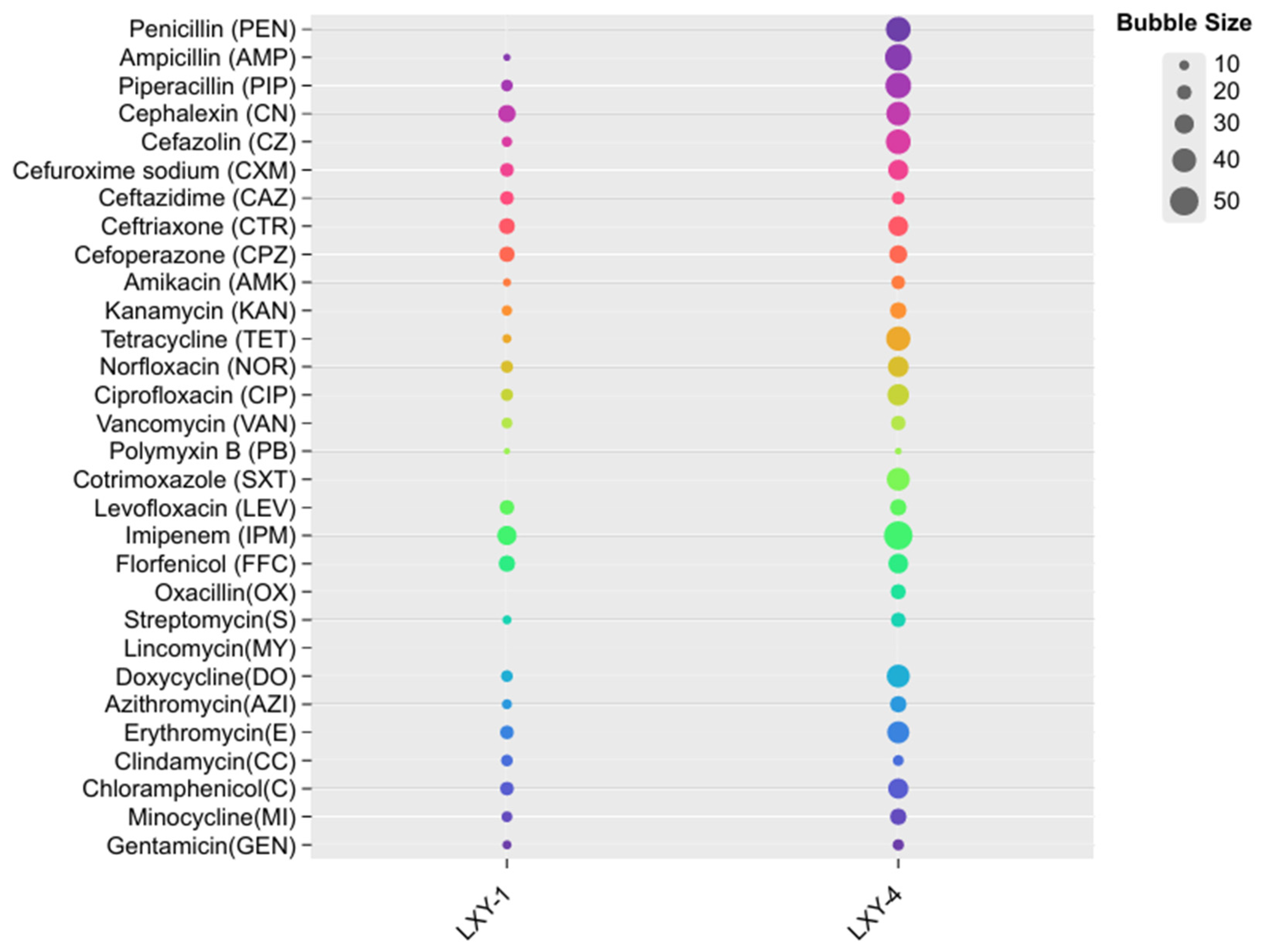
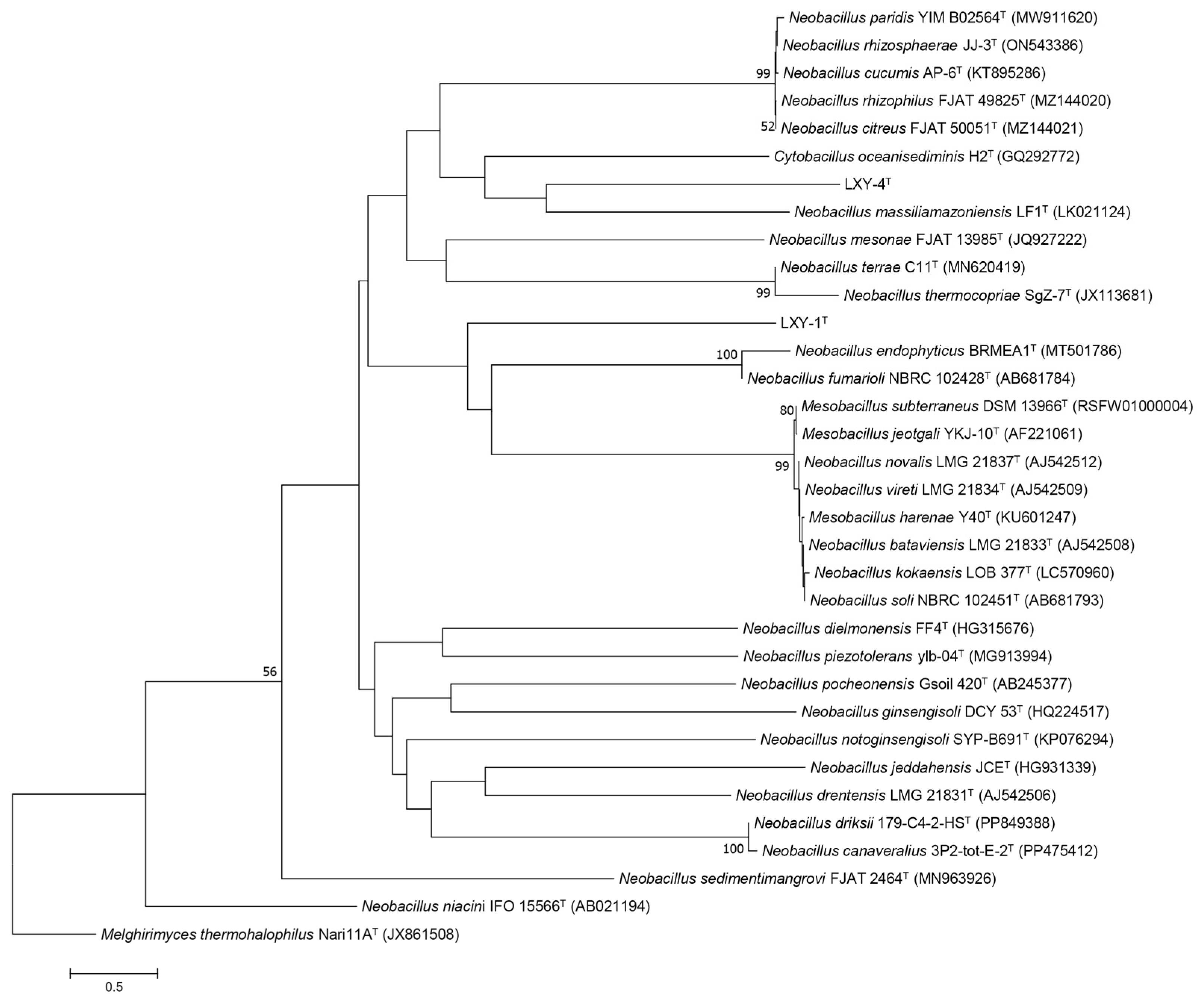
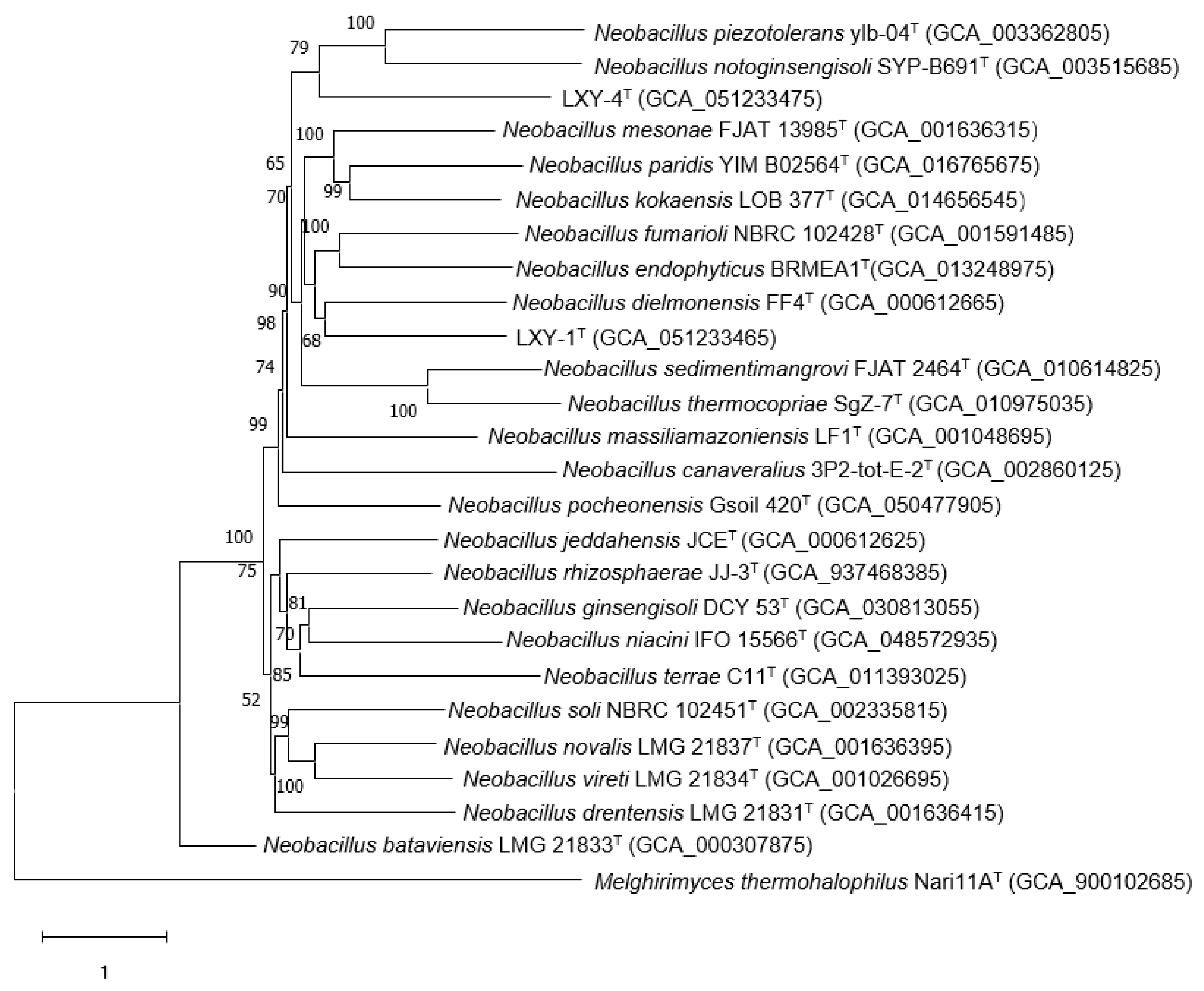
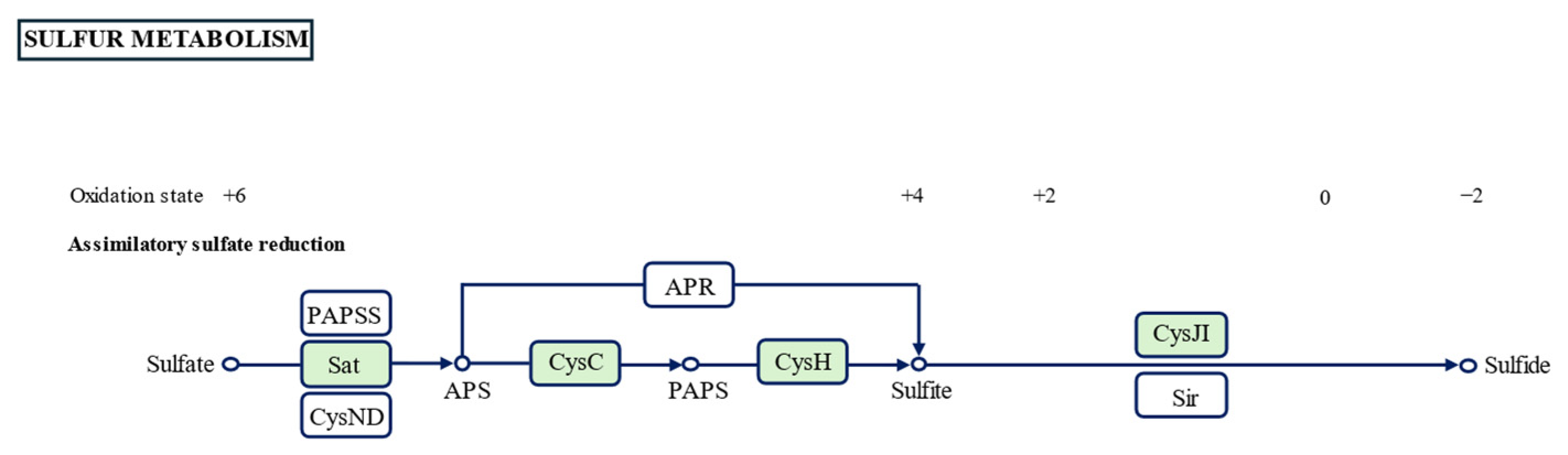
| Characteristics | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Source of Isolation | Soil | Soil | Soil | Soil |
| Cell shape | Short-oval | Rod-shaped | Tapered rod-shaped | Slightly tapered rod-shaped |
| Anaerobic growth | + | + | + | + |
| Temperature (°C) Optimal (°C) | 22–50 (22) | 22–50 (22) | Up to 50–55 (30) | Up to 50–55 (30) |
| pH | 6.0–12.0 | 7.0–12.0 | 5.5–10.0 | 4.0–10.0 |
| Gram staining | + | + | + | + |
| Major cellular fatty acids | Antéiso C15:0 | Antéiso C15:0 | Antéiso C15:0, iso-C15:0 and C16:1 ω11c | Antéiso C15:0, iso-C15:0 and C16:1 ω11c |
| β-Galactosidase (ONPG) | − | − | v | + |
| Nitrate reduction to N2 | − | + | v | + |
| Genome features | ||||
| 16S rRNA gene identities * | 97.3 | 96.7 | 95.7 | 100 |
| Mol% G+C | 38.61 | 38.75 | 39 | 39.5 |
| ANI * | 78.74 | 71.09 | 77.04 | 79.41 |
| Acid production from | ||||
| L-Fucose | − | − | − | − |
| Lactose | − | − | − | − |
| D-Mannitol | − | − | − | − |
| Melibiose | − | − | + | − |
| Raffinose | − | − | v | − |
| Salicin | − | + | + | − |
| Sucrose | − | + | v | − |
| Turanose | − | − | v | − |
| Fatty Acid | Percentage (%) | |||
|---|---|---|---|---|
| LXY-1T | LXY-4T | N. drentensis LMG 21831T | N. bataviensis LMG 21833T | |
| iso-C14:0 | - | - | 8.7 ± 2.5 | 6.9 ± 1.1 |
| C14:0 | 1.94 | - | 1.4 ± 1.3 | 1.5 ± 0.6 |
| C16:1 ω7c alcohol | - | - | 3.1 ± 0.8 | 2.3 ± 0.8 |
| C14:1 ω5c | - | 7.97 | - | - |
| C16:0 | 11.94 | 5.49 | 3.4 ± 1.6 | 7.7 ± 3.2 |
| iso-C15:0 | 14.61 | 13.93 | 32.2 ± 4.2 | 36.9 ± 4.3 |
| antéiso-C15:0 | 44.14 | 33.52 | 21.8 ± 5.6 | 20.5 ± 3.0 |
| iso-C17:0 | - | 11.43 | 2.6 ± 0.6 | 1.4 ± 0.8 |
| antéiso-C17:0 | 12.92 | 17.28 | 1.1 ± 0.3 | 11.1 ± 0.4 |
| C18:1 ω9c | - | - | 1.8 ± 1.1 | 1.3 ± 0.8 |
| C18:0 | - | - | 1.3 ± 0.9 | 2.7 ± 1.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Ran, C.; Zhou, N.; Zhao, X.; Liu, X.; Jiang, C.; Zhao, Y.; Lv, Y. Neobacillus terrisolis sp. nov. and Neobacillus solisequens sp. nov. Isolated from Soil. Microorganisms 2025, 13, 2437. https://doi.org/10.3390/microorganisms13112437
Wu H, Ran C, Zhou N, Zhao X, Liu X, Jiang C, Zhao Y, Lv Y. Neobacillus terrisolis sp. nov. and Neobacillus solisequens sp. nov. Isolated from Soil. Microorganisms. 2025; 13(11):2437. https://doi.org/10.3390/microorganisms13112437
Chicago/Turabian StyleWu, Haoyu, Congguo Ran, Nan Zhou, Xize Zhao, Xingyu Liu, Chengying Jiang, Yinghao Zhao, and Ying Lv. 2025. "Neobacillus terrisolis sp. nov. and Neobacillus solisequens sp. nov. Isolated from Soil" Microorganisms 13, no. 11: 2437. https://doi.org/10.3390/microorganisms13112437
APA StyleWu, H., Ran, C., Zhou, N., Zhao, X., Liu, X., Jiang, C., Zhao, Y., & Lv, Y. (2025). Neobacillus terrisolis sp. nov. and Neobacillus solisequens sp. nov. Isolated from Soil. Microorganisms, 13(11), 2437. https://doi.org/10.3390/microorganisms13112437






