Genomic Surveillance and Resistance Profiling of Multidrug-Resistant Acinetobacter baumannii Clinical Isolates: Clonal Diversity and Virulence Insights
Abstract
1. Introduction
- Characterize clonal diversity and resistance profiles at the hospital scale;
- Uncover potential transmission clusters and ward-specific genomic signatures;
- Inform surveillance strategies by integrating genomic data with clinical and epidemiological metadata.
2. Materials and Methods
2.1. Bacterial Isolates
2.2. DNA Extraction and Whole-Genome Sequencing (WGS)
2.3. Bioinformatics Analysis
3. Results
3.1. Isolates
3.2. Quality Control and Assembly Process
3.3. MLST and Phylogenetic Trees
3.4. Pan-Genome Analysis
3.5. Gene Resistance Analysis
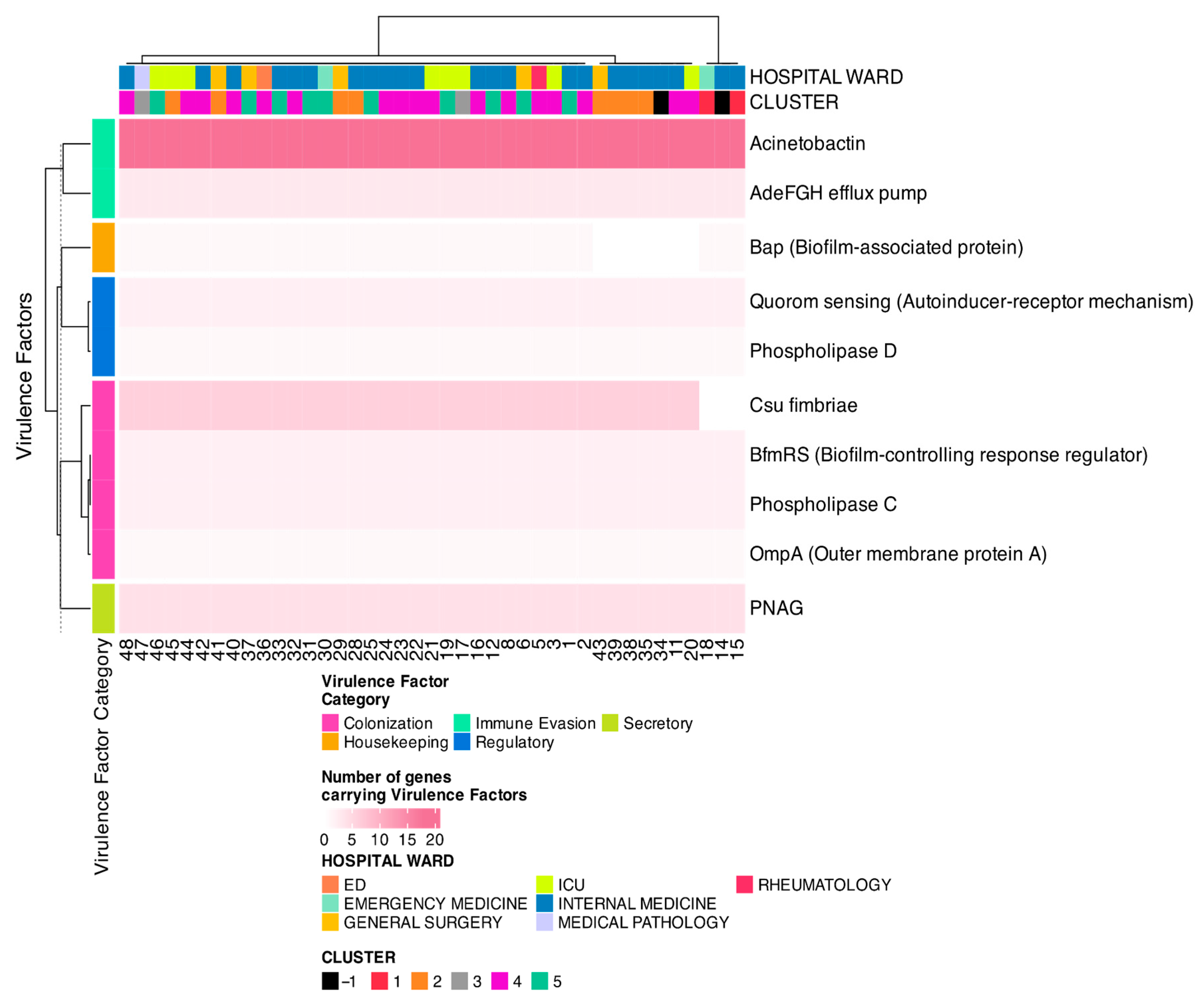
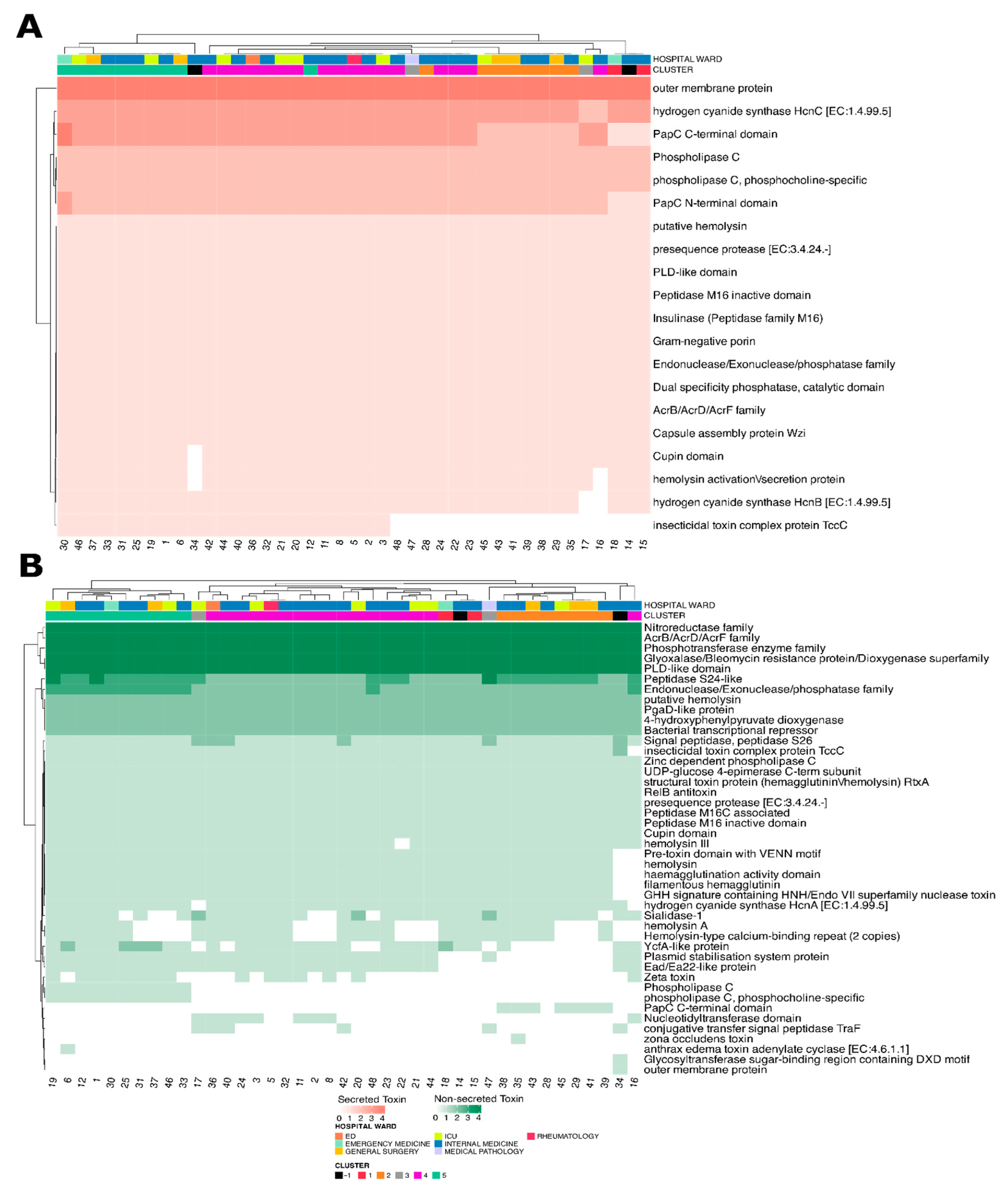
3.6. Virulence Factors and Toxin Analysis
4. Discussion
5. Conclusions
6. Perspectives for Future Studies
- Expanding the number of isolates and including samples from multiple institutions to improve the generalizability of findings.
- Improving detailed clinical metadata such as patient outcomes, antibiotic exposure, or length of hospital stay, which could have enhanced interpretation of resistance and virulence findings.
- Refining clonal classification methods, as the current use of fixed genetic distance thresholds may not fully align with epidemiologically meaningful transmission units.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation/Code | Definition |
| aac(6′)-Ib′ | aminoglycoside N-acetyltransferase AAC(6′)-Ib′ |
| aadA1 | ANT(3″)-Ia family aminoglycoside nucleotidyltransferase AadA1 |
| abaF | fosfomycin efflux MFS transporter AbaF |
| adeC | multidrug efflux RND transporter AdeABC outer membrane channel subunit AdeC |
| AMR | antimicrobial resistance |
| amvA | multidrug efflux MFS transporter AmvA |
| ANI | average nucleotide identity |
| ant(3″)-IIa | aminoglycoside nucleotidyltransferase ANT(3″)-IIa |
| aph(3″)-Ib | aminoglycoside O-phosphotransferase APH(3″)-Ib |
| aph(3′)-Ia | aminoglycoside O-phosphotransferase APH(3′)-Ia |
| aph(6)-Id | aminoglycoside O-phosphotransferase APH(6)-Id |
| armA | 16S rRNA (guanine(1405)-N(7))-methyltransferase ArmA |
| BAL | broncho-alveolar lavage |
| BfmRS | biofilm-controlling two-component regulatory system |
| blaADC-186 | extended-spectrum class C beta-lactamase ADC-186 |
| blaADC-30 | extended-spectrum class C beta-lactamase ADC-30 |
| blaADC-33 | cefepime-hydrolyzing class C beta-lactamase ADC-33 |
| blaADC-73 | extended-spectrum class C beta-lactamase ADC-73 |
| blaOXA-23 | OXA-23 family carbapenem-hydrolyzing class D beta-lactamase OXA-23 |
| blaOXA-66 | OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-66 |
| blaOXA-69 | OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-69 |
| blaOXA-82 | OXA-51 family carbapenem-hydrolyzing class D beta-lactamase OXA-82 |
| catB8 | type B-3 chloramphenicol O-acetyltransferase CatB8 |
| CheckM | tool for assessing genome quality (completeness/contamination) |
| ClusterPicker | software for automatic cluster identification in phylogenetic trees |
| Csu fimbriae | adhesion factor involved in biofilm formation and immune evasion |
| cxpE | chloramphenicol efflux transporter CxpE |
| Element symbol | element name |
| ftsI_A515V | Acinetobacter baumannii carbapenem/sulbactam-durlobactam resistant FtsI |
| GTDB-Tk | genome taxonomy database toolkit for taxonomic classification |
| gyrA_S81L | Acinetobacter baumannii quinolone resistant GyrA |
| ICU | intensive care unit |
| kSNP | tool for SNP-based phylogenetic analysis without a reference genome |
| MALDI-TOF | matrix-assisted laser desorption/ionization–time of flight mass spectrometry |
| MDR | multi-drug resistant |
| MEGAHIT | assembler for large and complex metagenomic datasets |
| MetaVF | tool using VFDB for virulence factor detection |
| MLST | multilocus sequence typing |
| mph(E) | Mph(E) family macrolide 2′-phosphotransferase |
| msr(E) | ABC-F type ribosomal protection protein Msr(E) |
| qacEdelta1 | gene that confers resistance to disinfectants, specifically quaternary ammonium compounds. |
| ST | sequence type: identifier assigned to a specific allelic profile in the MLST scheme. |
| tetA | gene codifying for metal-tetracycline/H(+) antiporter conferring resistance to tetracycline by an active tetracycline efflux. |
| tetB | Tet(B) is a tetracycline efflux protein expressed in many Gram-negative bacteria. It confers resistance to tetracycline, doxycycline, and minocycline, but not tigecycline. |
References
- Asif, M.; Alvi, I.A.; Rehman, S.U. Insight into Acinetobacter baumannii: Pathogenesis, Global Resistance, Mechanisms of Resistance, Treatment Options, and Alternative Modalities. Infect. Drug Resist. 2018, 11, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Linde, J.; Nguyen, N.H.; Nguyen, T.N.M.; Sprague, L.D.; Pletz, M.W.; Neubauer, H. WGS-Based Analysis of Carbapenem-Resistant Acinetobacter baumannii in Vietnam and Molecular Characterization of Antimicrobial Determinants and MLST in Southeast Asia. Antibiotics 2021, 10, 563. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The Population Structure of Acinetobacter baumannii: Expanding Multiresistant Clones from an Ancestral Susceptible Genetic Pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef]
- Mellace, M.; Ceniti, C.; Paonessa, M.; Procopio, A.C.; Tilocca, B. Multidrug-Resistant Acinetobacter baumannii: An Underestimated Pathogen in Veterinary Medicine in Italy. Ger. J. Vet. Res. 2024, 4, 112–126. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hujer, K.M.; Hujer, A.M.; Hulten, E.A.; Bajaksouzian, S.; Adams, J.M.; Donskey, C.J.; Ecker, D.J.; Massire, C.; Eshoo, M.W.; Sampath, R.; et al. Analysis of Antibiotic Resistance Genes in Multidrug-Resistant Acinetobacter sp. Isolates from Military and Civilian Patients Treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 2006, 50, 4114–4123. [Google Scholar] [CrossRef]
- Nemec, A.; Dijkshoorn, L.; Van Der Reijden, T.J.K. Long-Term Predominance of Two Pan-European Clones among Multi-Resistant Acinetobacter baumannii Strains in the Czech Republic. J. Med. Microbiol. 2004, 53, 147–153. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Furuya, E.Y.; Lowy, F.D. Antimicrobial-Resistant Bacteria in the Community Setting. Nat. Rev. Microbiol. 2006, 4, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Cave, R.; Cole, J.; Mkrtchyan, H.V. Surveillance and Prevalence of Antimicrobial Resistant Bacteria from Public Settings within Urban Built Environments: Challenges and Opportunities for Hygiene and Infection Control. Environ. Int. 2021, 157, 106836. [Google Scholar] [CrossRef]
- Almasaudi, S.B. Acinetobacter spp. as Nosocomial Pathogens: Epidemiology and Resistance Features. Saudi J. Biol. Sci. 2018, 25, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Peacock, S.J.; Parkhill, J.; Brown, N.M. Changing the Paradigm for Hospital Outbreak Detection by Leading with Genomic Surveillance of Nosocomial Pathogens. Microbiology 2018, 164, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Aldeyab, M.A.; Harbarth, S.; Vernaz, N.; Kearney, M.P.; Scott, M.G.; Darwish Elhajji, F.W.; Aldiab, M.A.; McElnay, J.C. The Impact of Antibiotic Use on the Incidence and Resistance Pattern of Extended-spectrum Beta-lactamase-producing Bacteria in Primary and Secondary Healthcare Settings. Br. J. Clin. Pharma. 2012, 74, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Karanika, S.; Karantanos, T.; Arvanitis, M.; Grigoras, C.; Mylonakis, E. Fecal Colonization with Extended-Spectrum Beta-Lactamase–Producing Enterobacteriaceae and Risk Factors Among Healthy Individuals: A Systematic Review and Metaanalysis. Clin. Infect. Dis. 2016, 63, 310–318. [Google Scholar] [CrossRef]
- Zeana, C.; Larson, E.; Sahni, J.; Bayuga, S.J.; Wu, F.; Della-Latta, P. The Epidemiology of Multidrug-Resistant Acinetobacter baumannii Does the Community Represent a Reservoir? Infect. Control Hosp. Epidemiol. 2003, 24, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Ciccozzi, M.; Cella, E.; Lai, A.; Florio, L.; Antonelli, F.; Fogolari, M.; Di Matteo, F.M.; Pizzicannella, M.; Colombo, B.; Dicuonzo, G.; et al. Phylogenetic Analysis of Multi-Drug Resistant Klebsiella pneumoniae Strains from Duodenoscope Biofilm: Microbiological Surveillance and Reprocessing Improvements for Infection Prevention. Front. Public Health 2019, 7, 219. [Google Scholar] [CrossRef]
- Angeletti, S.; Cella, E.; Lai, A.; Lo Presti, A.; Antonelli, F.; Conti, A.; Lopalco, M.; Spoto, S.; Zehender, G.; Ciccozzi, M. Whole-Genome Sequencing of Klebsiella pneumoniae MDR Strain Isolated in a Syrian Refugee. Pathog. Glob. Health 2017, 111, 212–215. [Google Scholar] [CrossRef]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the Mechanisms of Acinetobacter baumannii Virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Ristori, M.V.; Scarpa, F.; Sanna, D.; Casu, M.; Petrosillo, N.; Longo, U.G.; Lucia, D.F.; Spoto, S.; Chiantia, R.M.; Caserta, A.; et al. Multidrug-Resistant Klebsiella pneumoniae Strains in a Hospital: Phylogenetic Analysis to Investigate Local Epidemiology. Microorganisms 2024, 12, 2541. [Google Scholar] [CrossRef]
- Angeletti, S.; Dicuonzo, G.; Lo Presti, A.; Cella, E.; Crea, F.; Avola, A.; Vitali, M.A.; Fagioni, M.; De Florio, L. MALDI-TOF Mass Spectrometry and Blakpc Gene Phylogenetic Analysis of an Outbreak of Carbapenem-Resistant K. pneumoniae Strains. New Microbiol. 2015, 38, 541–550. [Google Scholar]
- ISO 20776-1:2019; Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devicesPart 1: Broth micro-dilution reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases. ISO: Geneva, Switzerland, 2019. Available online: https://www.iso.org/standard/70464.html (accessed on 21 May 2025).
- Sserwadda, I.; Mboowa, G. rMAP: The Rapid Microbial Analysis Pipeline for ESKAPE Bacterial Group Whole-Genome Sequence Data. Microb. Genom. 2021, 7, 000583. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Alneberg, J.; Bjarnason, B.S.; De Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning Metagenomic Contigs by Coverage and Composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory Friendly Classification with the Genome Taxonomy Database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Figueras, M.J.; Beaz-Hidalgo, R.; Hossain, M.J.; Liles, M.R. Taxonomic Affiliation of New Genomes Should Be Verified Using Average Nucleotide Identity and Multilocus Phylogenetic Analysis. Genome Announc. 2014, 2, e00927-14. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Prodhan, A. Asadprodhan/Average-Nucleotide-Identity-ANI-Analysis 2025. Available online: https://github.com/asadprodhan/Average-Nucleotide-Identity-ANI-analysis (accessed on 21 May 2025).
- Seemann, T. Tseemann/Mlst 2025. Available online: https://github.com/tseemann/mlst (accessed on 6 June 2025).
- Shakya, M.; Ahmed, S.A.; Davenport, K.W.; Flynn, M.C.; Lo, C.-C.; Chain, P.S.G. Standardized Phylogenetic and Molecular Evolutionary Analysis Applied to Species across the Microbial Tree of Life. Sci. Rep. 2020, 10, 1723. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G.; Nisbet, J. Building Phylogenetic Trees from Genome Sequences With kSNP4. Mol. Biol. Evol. 2023, 40, msad235. [Google Scholar] [CrossRef] [PubMed]
- Ragonnet-Cronin, M.; Hodcroft, E.; Hué, S.; Fearnhill, E.; Delpech, V.; Brown, A.J.L.; Lycett, S. Automated Analysis of Phylogenetic Clusters. BMC Bioinform. 2013, 14, 317. [Google Scholar] [CrossRef] [PubMed]
- FigTree. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 21 May 2024).
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Roary/Contrib/Roary_plots/Roary_plots.Py at Master. Sanger-Pathogens/Roary. Available online: https://github.com/sanger-pathogens/Roary/blob/master/contrib/roary_plots/roary_plots.py (accessed on 6 June 2025).
- Powell, D. Drpowell/FriPan 2024.
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- De Nies, L.; Lopes, S.; Busi, S.B.; Galata, V.; Heintz-Buschart, A.; Laczny, C.C.; May, P.; Wilmes, P. PathoFact: A Pipeline for the Prediction of Virulence Factors and Antimicrobial Resistance Genes in Metagenomic Data. Microbiome 2021, 9, 49. [Google Scholar] [CrossRef]
- Dong, W.; Fan, X.; Guo, Y.; Wang, S.; Jia, S.; Lv, N.; Yuan, T.; Pan, Y.; Xue, Y.; Chen, X.; et al. An Expanded Database and Analytical Toolkit for Identifying Bacterial Virulence Factors and Their Associations with Chronic Diseases. Nat. Commun. 2024, 15, 8084. [Google Scholar] [CrossRef]
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Posit. The Data Science Code Editor. Available online: https://www.posit.co/ (accessed on 6 June 2025).
- Patil, I. Visualizations with Statistical Details: The “ggstatsplot” Approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar] [CrossRef]
- Xu, S.; Dai, Z.; Guo, P.; Fu, X.; Liu, S.; Zhou, L.; Tang, W.; Feng, T.; Chen, M.; Zhan, L.; et al. ggtreeExtra: Compact Visualization of Richly Annotated Phylogenetic Data. Mol. Biol. Evol. 2021, 38, 4039–4042. [Google Scholar] [CrossRef]
- Yu, G. Data Integration, Manipulation and Visualization of Phylogenetic Trees, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2022; ISBN 978-1-00-327924-2. [Google Scholar]
- Wareth, G.; Brandt, C.; Sprague, L.D.; Neubauer, H.; Pletz, M.W. Spatio-Temporal Distribution of Acinetobacter baumannii in Germany—A Comprehensive Systematic Review of Studies on Resistance Development in Humans (2000–2018). Microorganisms 2020, 8, 375. [Google Scholar] [CrossRef]
- Wareth, G.; Abdel-Glil, M.Y.; Schmoock, G.; Steinacker, U.; Kaspar, H.; Neubauer, H.; Sprague, L.D. Draft Genome Sequence of an Acinetobacter baumannii Isolate Recovered from a Horse with Conjunctivitis in Germany. Microbiol. Resour. Announc. 2019, 8, e01128-19. [Google Scholar] [CrossRef]
- Wareth, G.; Linde, J.; Hammer, P.; Nguyen, N.H.; Nguyen, T.N.M.; Splettstoesser, W.D.; Makarewicz, O.; Neubauer, H.; Sprague, L.D.; Pletz, M.W. Phenotypic and WGS-Derived Antimicrobial Resistance Profiles of Clinical and Non-Clinical Acinetobacter baumannii Isolates from Germany and Vietnam. Int. J. Antimicrob. Agents 2020, 56, 106127. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Neubauer, H.; Sprague, L.D. Acinetobacter baumannii–a Neglected Pathogen in Veterinary and Environmental Health in Germany. Vet. Res. Commun. 2019, 43, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Murugaiyan, J.; Walther, B.; Stamm, I.; Abou-Elnaga, Y.; Brueggemann-Schwarze, S.; Vincze, S.; Wieler, L.H.; Lübke-Becker, A.; Semmler, T.; Roesler, U. Species Differentiation within the Staphylococcus Intermedius Group Using a Refined MALDI-TOF MS Database. Clin. Microbiol. Infect. 2014, 20, 1007–1014. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Biswas, I.; Veeraraghavan, B. Accurate Identification of Clinically Important Acinetobacter spp.: An Update. Future Sci. OA 2019, 5, FSO395. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A.F.; Malecki, M.; Otchwemah, R.; Tellez-Castillo, C.J.; Sakka, S.G.; Mattner, F. One-Year Molecular Surveillance of Carbapenem-Susceptible A. Baumannii on a German Intensive Care Unit: Diversity or Clonality. Antimicrob. Resist. Infect. Control. 2018, 7, 145. [Google Scholar] [CrossRef]
- Higgins, P.G.; Schneiders, T.; Hamprecht, A.; Seifert, H. In Vivo Selection of a Missense Mutation in adeR and Conversion of the Novel BlaOXA-164 Gene into BlaOXA-58 in Carbapenem-Resistant Acinetobacter baumannii Isolates from a Hospitalized Patient. Antimicrob. Agents Chemother. 2010, 54, 5021–5027. [Google Scholar] [CrossRef]
- Corbella, X.; Pujol, M.; Ayats, J.; Sendra, M.; Ardanuy, C.; Dominguez, M.A.; Linares, J.; Ariza, J.; Gudiol, F. Relevance of Digestive Tract Colonization in the Epidemiology of Nosocomial Infections Due to Multiresistant Acinetobacter baumannii. Clin. Infect. Dis. 1996, 23, 329–334. [Google Scholar] [CrossRef]
- Ayats, J.; Corbella, X.; Ardanuy, C.; Domínguez, M.A.; Ricart, A.; Ariza, J.; Martin, R.; Liñares, J. Epidemiological Significance of Cutaneous, Pharyngeal, and Digestive Tract Colonization by Multiresistant Acinetobacter baumannii in ICU Patients. J. Hosp. Infect. 1997, 37, 287–295. [Google Scholar] [CrossRef]
- Odih, E.E.; Irek, E.O.; Obadare, T.O.; Oaikhena, A.O.; Afolayan, A.O.; Underwood, A.; Adenekan, A.T.; Ogunleye, V.O.; Argimon, S.; Dalsgaard, A.; et al. Rectal Colonization and Nosocomial Transmission of Carbapenem-Resistant Acinetobacter baumannii in an Intensive Care Unit, Southwest Nigeria. Front. Med. 2022, 9, 846051. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Wareth, G.; Linde, J.; Hammer, P.; Splettstoesser, W.D.; Pletz, M.W.; Neubauer, H.; Sprague, L.D. Molecular Characterization of German Acinetobacter baumannii Isolates and Multilocus Sequence Typing (MLST) Analysis Based on WGS Reveals Novel STs. Pathogens 2021, 10, 690. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Nigro, S.J. Emergence, Molecular Mechanisms and Global Spread of Carbapenem-Resistant Acinetobacter baumannii. Microb. Genom. 2019, 5, e000306. [Google Scholar] [CrossRef]
- Reis, A.C.; Cunha, M.V. The Open Pan-Genome Architecture and Virulence Landscape of Mycobacterium Bovis. Microb. Genom. 2021, 7, 000664. [Google Scholar] [CrossRef] [PubMed]
- Medini, D.; Donati, C.; Tettelin, H.; Masignani, V.; Rappuoli, R. The Microbial Pan-Genome. Curr. Opin. Genet. Dev. 2005, 15, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Vernikos, G.; Medini, D.; Riley, D.R.; Tettelin, H. Ten Years of Pan-Genome Analyses. Curr. Opin. Microbiol. 2015, 23, 148–154. [Google Scholar] [CrossRef]
- Chan, A.P.; Sutton, G.; DePew, J.; Krishnakumar, R.; Choi, Y.; Huang, X.-Z.; Beck, E.; Harkins, D.M.; Kim, M.; Lesho, E.P.; et al. A Novel Method of Consensus Pan-Chromosome Assembly and Large-Scale Comparative Analysis Reveal the Highly Flexible Pan-Genome of Acinetobacter baumannii. Genome Biol. 2015, 16, 143. [Google Scholar] [CrossRef]
- O’Brien, B.; Yushchenko, A.; Suh, J.; Jung, D.; Cai, Z.; Nguyen, N.S.; Semret, M.; Dufour, S.; Fanning, S.; Ronholm, J. Subtle Genomic Differences in Klebsiella pneumoniae Sensu Stricto Isolates Indicate Host Adaptation. One Health 2025, 20, 100970. [Google Scholar] [CrossRef] [PubMed]
- Karampatakis, T.; Tsergouli, K.; Behzadi, P. Pan-Genome Plasticity and Virulence Factors: A Natural Treasure Trove for Acinetobacter baumannii. Antibiotics 2024, 13, 257. [Google Scholar] [CrossRef] [PubMed]
- Marino, A.; Augello, E.; Stracquadanio, S.; Bellanca, C.M.; Cosentino, F.; Spampinato, S.; Cantarella, G.; Bernardini, R.; Stefani, S.; Cacopardo, B.; et al. Unveiling the Secrets of Acinetobacter baumannii: Resistance, Current Treatments, and Future Innovations. Int. J. Mol. Sci. 2024, 25, 6814. [Google Scholar] [CrossRef]
- Zack, K.M.; Sorenson, T.; Joshi, S.G. Types and Mechanisms of Efflux Pump Systems and the Potential of Efflux Pump Inhibitors in the Restoration of Antimicrobial Susceptibility, with a Special Reference to Acinetobacter baumannii. Pathogens 2024, 13, 197. [Google Scholar] [CrossRef]
- Ahmad, I.; Nadeem, A.; Mushtaq, F.; Zlatkov, N.; Shahzad, M.; Zavialov, A.V.; Wai, S.N.; Uhlin, B.E. Csu Pili Dependent Biofilm Formation and Virulence of Acinetobacter baumannii. NPJ Biofilms Microbiomes 2023, 9, 101. [Google Scholar] [CrossRef]
- Kim, H.-J.; Kim, N.-Y.; Ko, S.-Y.; Park, S.-Y.; Oh, M.-H.; Shin, M.-S.; Lee, Y.-C.; Lee, J.-C. Complementary Regulation of BfmRS Two-Component and AbaIR Quorum Sensing Systems to Express Virulence-Associated Genes in Acinetobacter baumannii. Int. J. Mol. Sci. 2022, 23, 13136. [Google Scholar] [CrossRef]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The Mechanisms of Disease Caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef] [PubMed]
- Fiester, S.E.; Arivett, B.A.; Schmidt, R.E.; Beckett, A.C.; Ticak, T.; Carrier, M.V.; Ghosh, R.; Ohneck, E.J.; Metz, M.L.; Sellin Jeffries, M.K.; et al. Iron-Regulated Phospholipase C Activity Contributes to the Cytolytic Activity and Virulence of Acinetobacter baumannii. PLoS ONE 2016, 11, e0167068. [Google Scholar] [CrossRef]
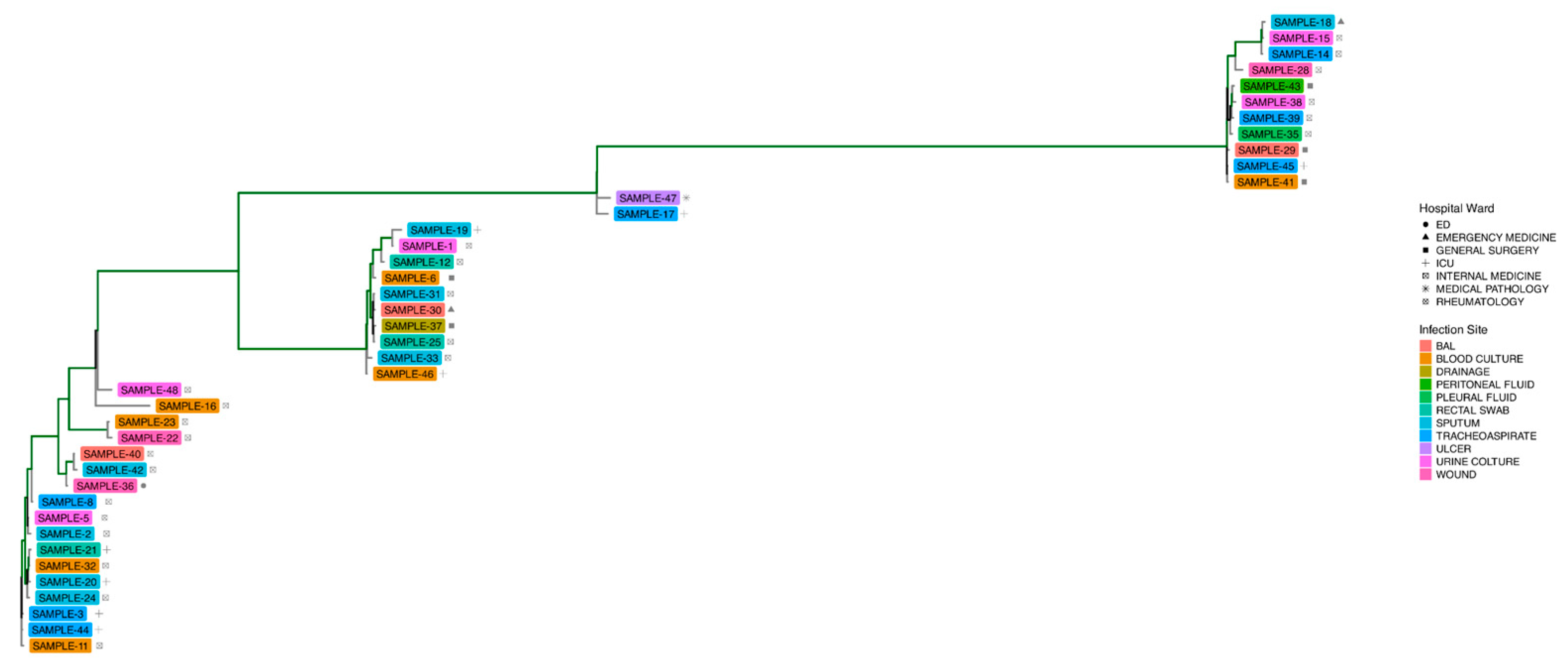
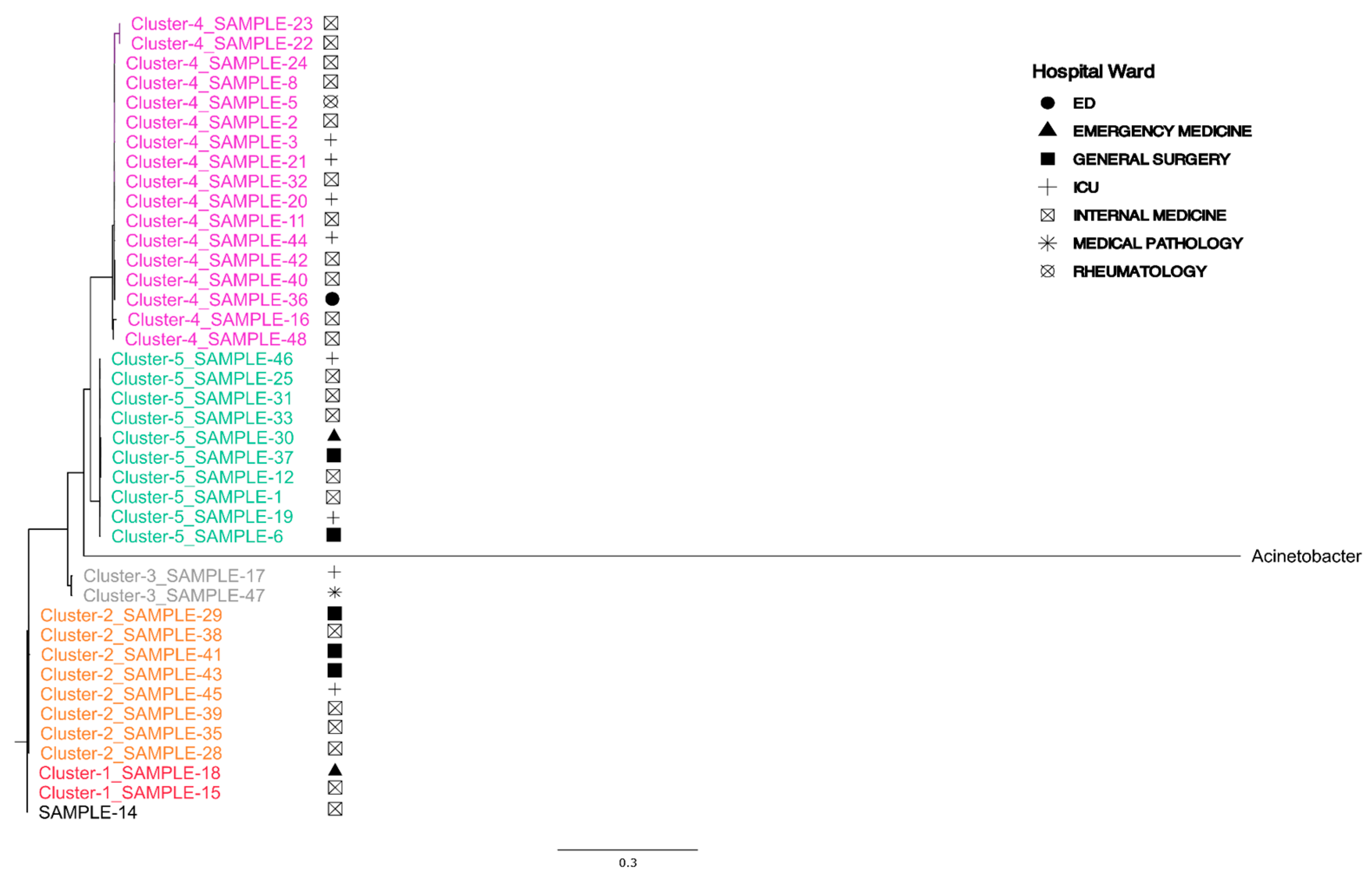
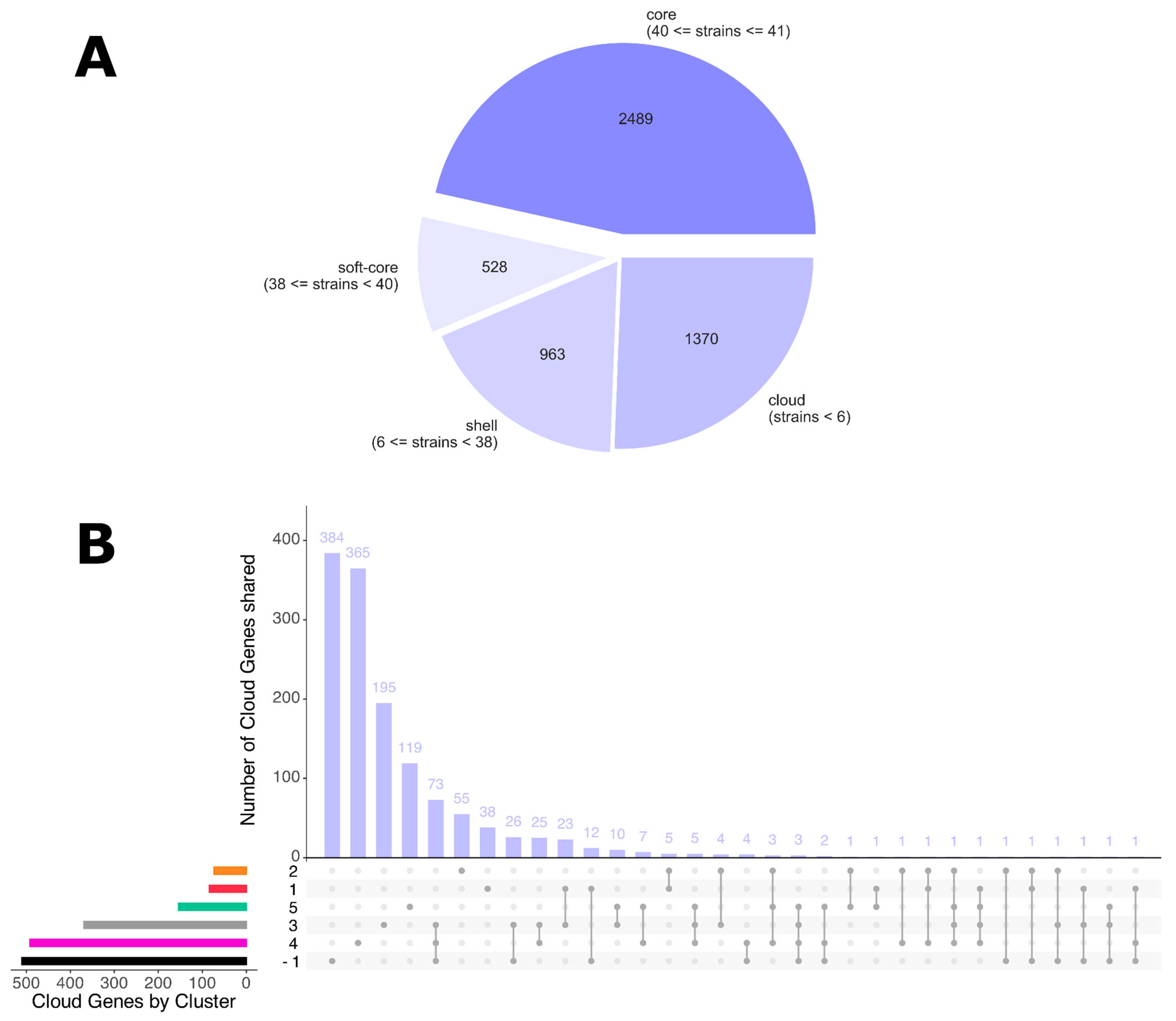
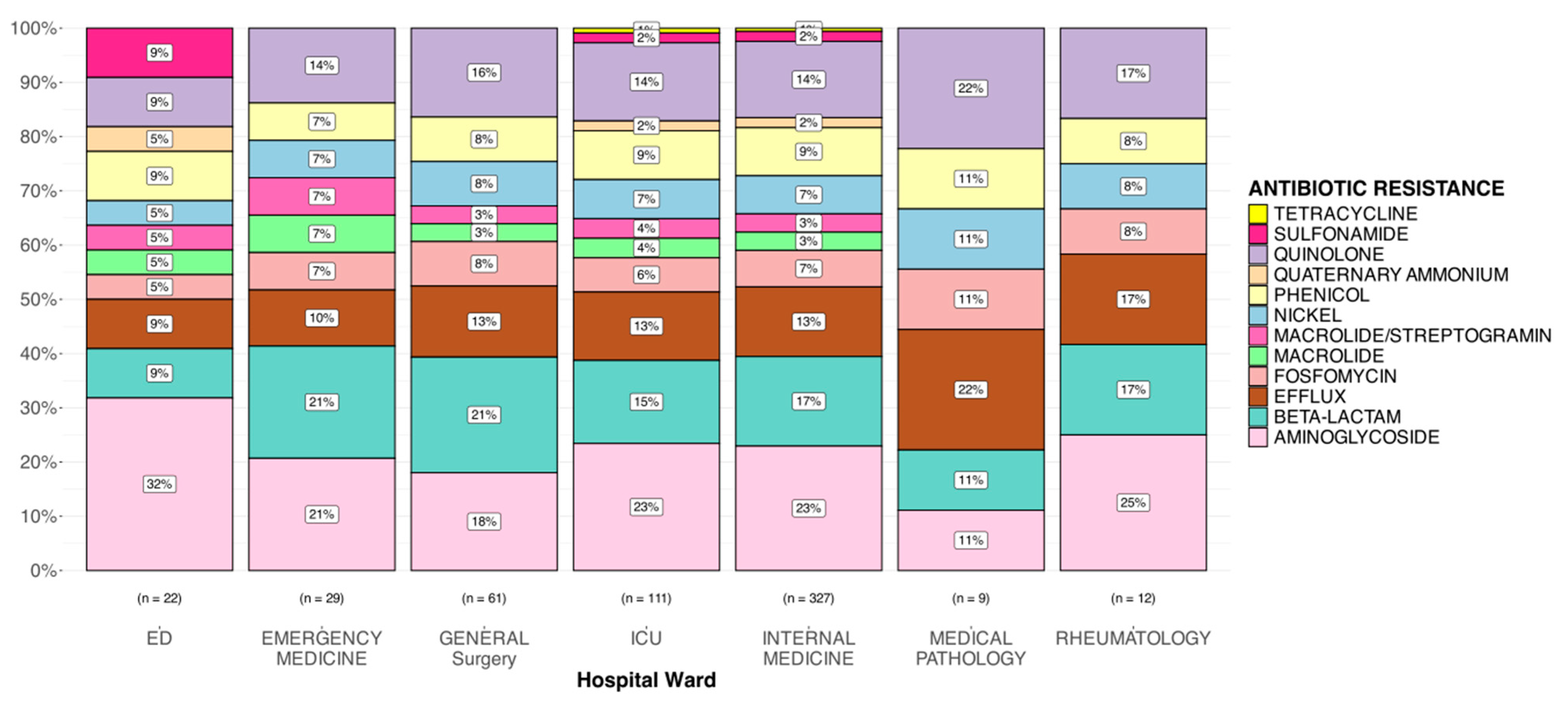

| ID Samples | Hospital Ward | Site of Infection | Date of Isolation |
|---|---|---|---|
| SAMPLE-1 | Internal Medicine | Urine culture | 27 December 2022 |
| SAMPLE-2 | Internal Medicine | Sputum | 8 January 2023 |
| SAMPLE-3 | ICU | Tracheoaspirate | 11 January 2023 |
| SAMPLE-5 | Rheumatology | Urine culture | 15 January 2023 |
| SAMPLE-6 | General Surgery | Blood culture | 17 January 2023 |
| SAMPLE-7 | ICU | Broncho-alveolar lavage (BAL) | 26 January 2023 |
| SAMPLE-8 | Internal Medicine | Tracheoaspirate | 1 February 2023 |
| SAMPLE-11 | Internal Medicine | Blood culture | 16 February 2023 |
| SAMPLE-12 | Internal Medicine | Rectal swab | 22 February 2023 |
| SAMPLE-14 | Internal Medicine | Tracheoaspirate | 24 February 2023 |
| SAMPLE-15 | Internal Medicine | Urine culture | 27 February 2023 |
| SAMPLE-16 | Internal Medicine | Blood culture | 1 March 2023 |
| SAMPLE-17 | ICU | Tracheoaspirate | 5 March 2023 |
| SAMPLE-18 | Emergency Medicine | Sputum | 6 March 2023 |
| SAMPLE-19 | ICU | Sputum | 6 March 2023 |
| SAMPLE-20 | ICU | Sputum | 6 March 2023 |
| SAMPLE-21 | ICU | Rectal swab | 6 March 2023 |
| SAMPLE-22 | Internal Medicine | Wound | 11 March 2023 |
| SAMPLE-23 | Internal Medicine | Blood culture | 13 March 2023 |
| SAMPLE-24 | Internal Medicine | Sputum | 14 March 2023 |
| SAMPLE-25 | Internal Medicine | Rectal swab | 19 March 2023 |
| SAMPLE-26 | Internal Medicine | Rectal swab | 19 March 2023 |
| SAMPLE-27 | Internal Medicine | Blood culture | 21 March 2023 |
| SAMPLE-28 | Internal Medicine | Wound | 22 March 2023 |
| SAMPLE-29 | General Surgery | Bal | 23 March 2023 |
| SAMPLE-31 | Internal Medicine | Sputum | 27 March 2023 |
| SAMPLE-32 | Internal Medicine | Blood culture | 29 March 2023 |
| SAMPLE-33 | Internal Medicine | Sputum | 30 March 2023 |
| SAMPLE-34 | Internal Medicine | Wound | 4 April 2023 |
| SAMPLE-35 | Internal Medicine | Pleural fluid | 5 April 2023 |
| SAMPLE-36 | ED | Wound | 5 April 2023 |
| SAMPLE-37 | General Surgery | Drainage | 7 April 2023 |
| SAMPLE-38 | Internal Medicine | Urine culture | 11 April 2023 |
| SAMPLE-39 | Internal Medicine | Tracheoaspirate | 11 April 2023 |
| SAMPLE-40 | Internal Medicine | Bal | 13 April 2023 |
| SAMPLE-41 | General Surgery | Blood culture | 14 April 2023 |
| SAMPLE-42 | Internal Medicine | Sputum | 22 April 2023 |
| SAMPLE-43 | General Surgery | Peritoneal fluid | 22 April 2023 |
| SAMPLE-44 | ICU | Tracheoaspirate | 23 April 2023 |
| SAMPLE-45 | ICU | Tracheoaspirate | 23 April 2023 |
| SAMPLE-46 | ICU | Blood culture | 28 April 2023 |
| SAMPLE-47 | Medical Pathology | Ulcer | 3 May 2023 |
| SAMPLE-48 | Internal Medicine | Urine culture | 4 May 2023 |
| Site of Infection | Number of Patients | Percentage (%) |
|---|---|---|
| BAL | 3 | 7 |
| Blood culture | 9 | 20 |
| Drainage | 1 | 2 |
| Peritoneal fluid | 1 | 2 |
| Pleural fluid | 1 | 2 |
| Rectal swab | 5 | 11 |
| Sputum | 8 | 18 |
| Tracheoaspirate | 7 | 16 |
| Ulcer | 1 | 2 |
| Urine culture | 5 | 11 |
| Wound | 4 | 9 |
| PHOENIX | SENSITITRE | DISK DIFFUSION | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC (Interpretation) | MIC (Int.) | MIC (Int.) | MIC (Int.) | MIC (Int.) | MIC (Int.) | MIC (Int.) | MIC (Int.) | MIC (Int.) | mm (Int.) | |
| Amikacin | Ciprofloxacin | Gentamicin | Imipenem | Levofloxacin | Meropenem | Tobramycin | Co-trimoxazole | Colistin | Cefiderocol | |
| SAMPLE | AN | CIP | GM | IPM | LVX | MER | NN | SXT | CL | FDC |
| SAMPLE 1 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 0.5 (S) | 23 (S) |
| SAMPLE 2 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 3 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 5 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 23 (S) |
| SAMPLE 6 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 20 (S) |
| SAMPLE 7 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 8 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 23 (S) |
| SAMPLE 11 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 12 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 20 (S) |
| SAMPLE 14 | ≤4 (S) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | >8 (R) | 14 (R) |
| SAMPLE 15 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 8 (R) | 13 (R) |
| SAMPLE 16 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 17 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 18 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 0.5 (S) | 21 (S) |
| SAMPLE 19 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 15 (R) |
| SAMPLE 20 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 20 (S) |
| SAMPLE 21 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 20 (S) |
| SAMPLE 22 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 2 (S) | 22 (S) |
| SAMPLE 23 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 19 (S) |
| SAMPLE 24 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 19 (S) |
| SAMPLE 25 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 26 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 23 (S) |
| SAMPLE 27 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 20 (S) |
| SAMPLE 28 | ≤4 (S) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 23 (S) |
| SAMPLE 29 | ≤4 (S) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 2 (S) | 15 (R) |
| SAMPLE 30 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | ≤0.5 (S) | 20 (S) |
| SAMPLE 31 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | ≤0.5 (S) | 21 (S) |
| SAMPLE 32 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 33 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 23 (S) |
| SAMPLE 34 | ≤4 (S) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 0.5 (S) | 23 (S) |
| SAMPLE 35 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 14 (R) |
| SAMPLE 36 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 21 (S) |
| SAMPLE 37 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 13 (R) |
| SAMPLE 38 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 21 (S) |
| SAMPLE 39 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 15 (R) |
| SAMPLE 40 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 0.5 (S) | 23 (S) |
| SAMPLE 41 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 0.5 (S) | 23 (S) |
| SAMPLE 42 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 20 (S) |
| SAMPLE 43 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
| SAMPLE 44 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 21 (S) |
| SAMPLE 45 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 10 (R) |
| SAMPLE 46 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 2 (S) | 20 (S) |
| SAMPLE 47 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 2 (S) | 20 (S) |
| SAMPLE 48 | >16 (R) | >1 (R) | >4 (R) | >8 (R) | >1 (R) | >16 (R) | >4 (R) | >4/76 (R) | 1 (S) | 22 (S) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ristori, M.V.; Pirona, I.; De Florio, L.; Aita, S.E.; Macari, G.; Spoto, S.; Antonelli Incalzi, R.; Angeletti, S. Genomic Surveillance and Resistance Profiling of Multidrug-Resistant Acinetobacter baumannii Clinical Isolates: Clonal Diversity and Virulence Insights. Microorganisms 2025, 13, 2429. https://doi.org/10.3390/microorganisms13112429
Ristori MV, Pirona I, De Florio L, Aita SE, Macari G, Spoto S, Antonelli Incalzi R, Angeletti S. Genomic Surveillance and Resistance Profiling of Multidrug-Resistant Acinetobacter baumannii Clinical Isolates: Clonal Diversity and Virulence Insights. Microorganisms. 2025; 13(11):2429. https://doi.org/10.3390/microorganisms13112429
Chicago/Turabian StyleRistori, Maria Vittoria, Ilaria Pirona, Lucia De Florio, Sara Elsa Aita, Gabriele Macari, Silvia Spoto, Raffaele Antonelli Incalzi, and Silvia Angeletti. 2025. "Genomic Surveillance and Resistance Profiling of Multidrug-Resistant Acinetobacter baumannii Clinical Isolates: Clonal Diversity and Virulence Insights" Microorganisms 13, no. 11: 2429. https://doi.org/10.3390/microorganisms13112429
APA StyleRistori, M. V., Pirona, I., De Florio, L., Aita, S. E., Macari, G., Spoto, S., Antonelli Incalzi, R., & Angeletti, S. (2025). Genomic Surveillance and Resistance Profiling of Multidrug-Resistant Acinetobacter baumannii Clinical Isolates: Clonal Diversity and Virulence Insights. Microorganisms, 13(11), 2429. https://doi.org/10.3390/microorganisms13112429








