Spatiotemporal Dynamics of Microbial and Fish Communities in the Thracian Sea Revealed by eDNA Metabarcoding
Abstract
1. Introduction
2. Materials and Methods
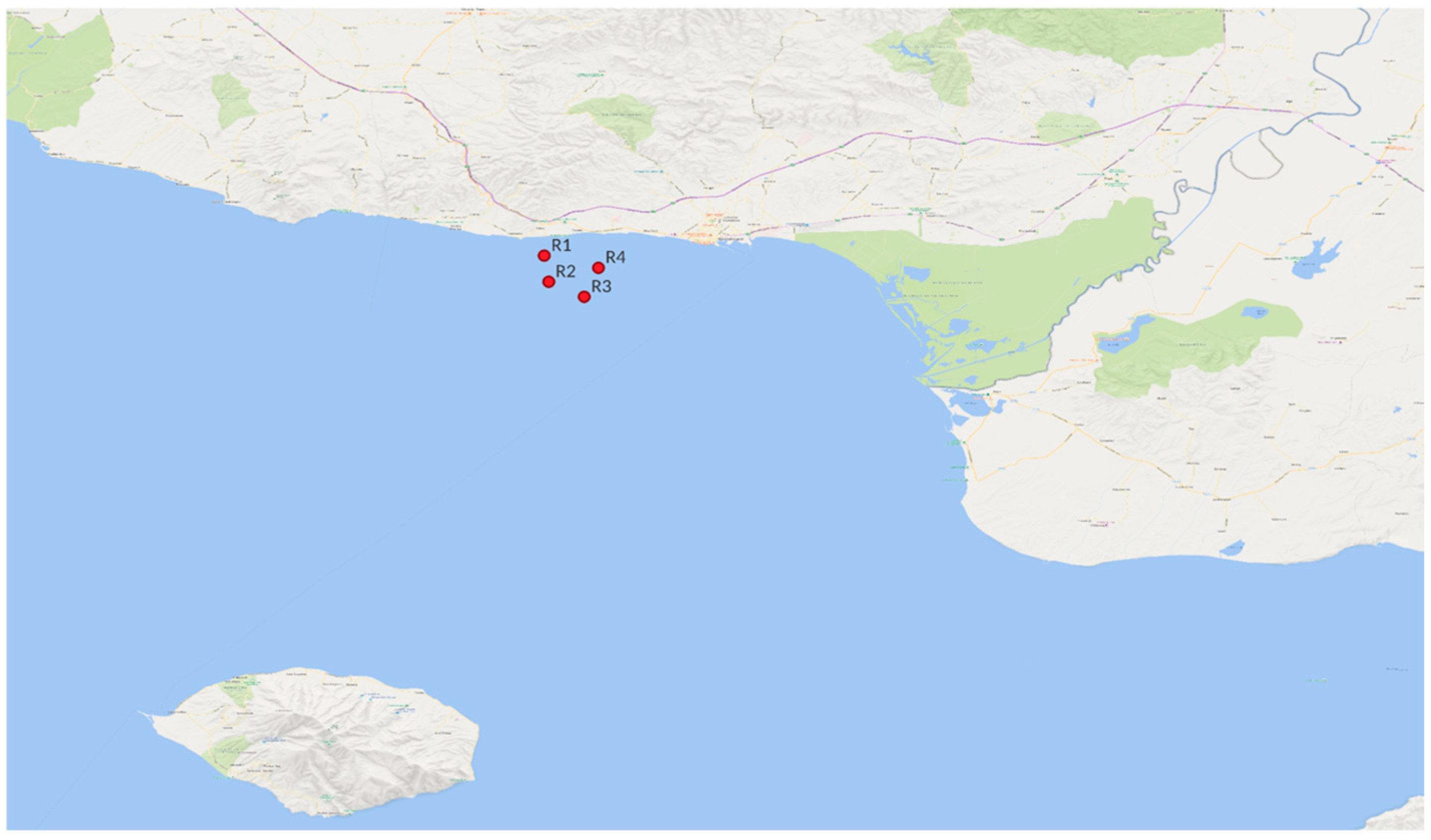
2.1. Study Area and Seawater Sample Collection
2.2. Genetic Marker Amplification
2.3. Library Construction and Sequencing
2.4. NGS Analysis
2.5. Taxonomic Assignment and Diversity Analysis
2.6. Comparative Analysis of Microbiome and Ichthyofauna
3. Results
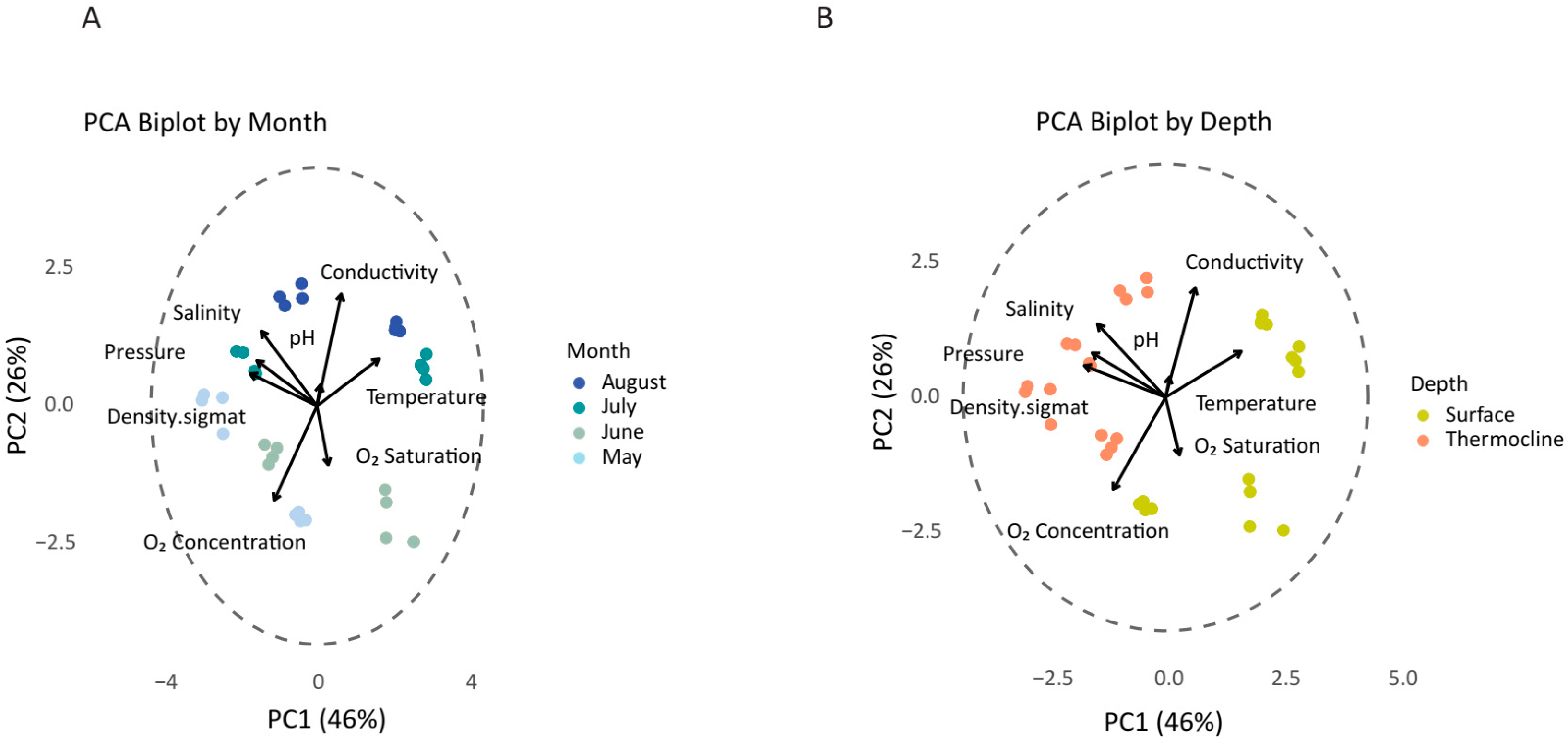
3.1. Environmental Variability
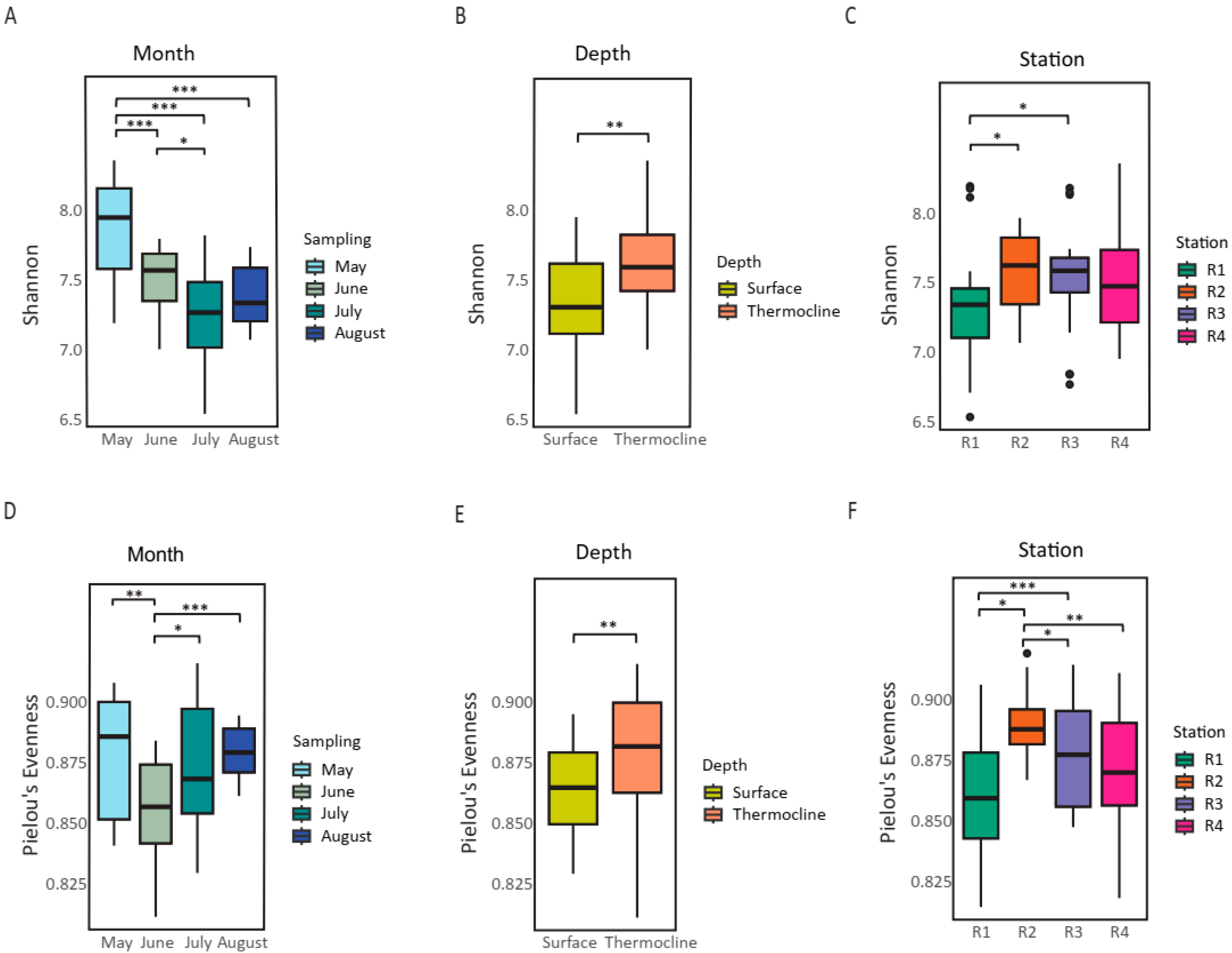
3.2. Temporal and Depth Affect the Microbiome Diversity
3.3. Microbial Community Structure and Core
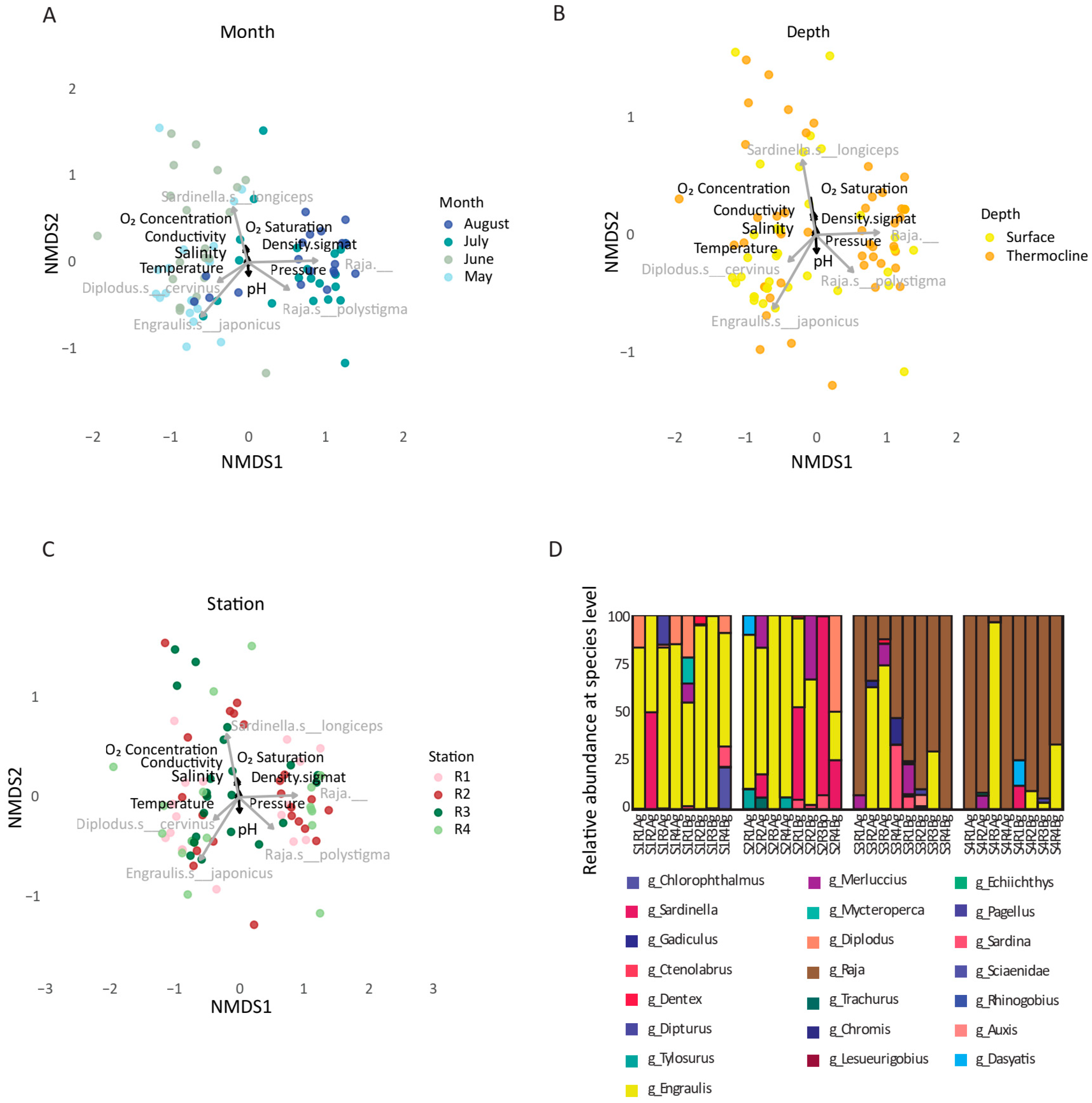
3.4. Temporal Effects Dominate over Spatial Variation in Ichthyofaunal Diversity
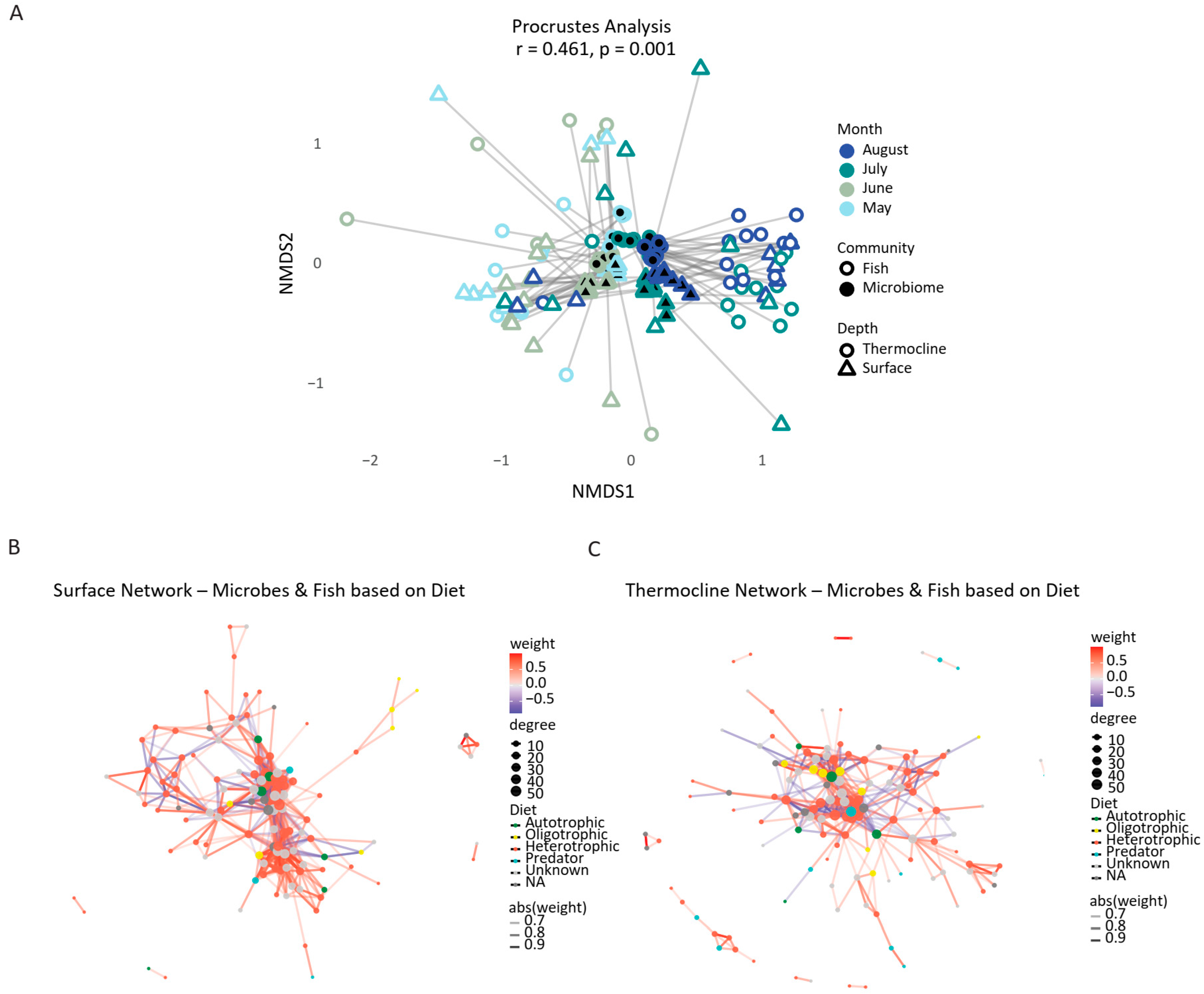
3.5. Linking Microbial and Fish Biodiversity in Coastal Ecosystems
4. Discussion
4.1. Seasonality and Depth as Principal Axes of Microbial Variation and Microbial Core
4.2. Ichthyofaunal Diversity in the Thracian Sea
4.3. Environmental Drivers of Microbial and Ichthyofaunal Communities
4.4. Microbe–Fish Community Coupling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Isari, S.; Psarra, S.; Pitta, P.; Mara, P.; Tomprou, M.O.; Ramfos, A.; Somarakis, S.; Tselepides, A.; Koutsikopoulos, C.; Fragopoulu, N. Differential patterns of mesozooplankters’ distribution in relation to physical and biological variables of the northeastern Aegean Sea (eastern Mediterranean). Mar. Biol. 2007, 151, 1035–1050. [Google Scholar] [CrossRef]
- Anastasiou, S.; Sylaios, G.K.; Tsihrintzis, V.A. Suspended particulate matter estimates using optical and acoustic sensors: Application in Nestos River plume (Thracian Sea, North Aegean Sea). Environ. Monit. Assess. 2015, 187, 392. [Google Scholar] [CrossRef]
- Mertzanis, A.; Papadopoulos, A.; Goudelis, G.; Pantera, A.; Efthimiou, G. Human-induced impact to the environment and changes in the geomorphology: Some examples of inland and coastal environments in Greece. Acad. J. J. Ecol. Nat. Environ. (JENE) 2011, 3, 273–297. [Google Scholar]
- Sylaios, G.; Kamidis, N.; Anastasiou, S.; Tsihrintzis, V.A. Hydrodynamic response of Thassos Passage (N. Aegean Sea) to Nestos River discharge and meteorological forcing. Cont. Shelf Res. 2013, 59, 37–51. [Google Scholar] [CrossRef]
- Mamoutos, I.; Potiris, E.; Androulidakis, Y.; Tragou, E.; Zervakis, V. Evidence for reduced Black Sea water outflow to the North Aegean. Earth Space Sci. 2024, 11, e2024EA003674. [Google Scholar] [CrossRef]
- Potiris, M.; Mamoutos, I.G.; Tragou, E.; Zervakis, V.; Kassis, D.; Ballas, D. Dense water formation variability in the Aegean Sea from 1947 to 2023. Oceans 2024, 5, 611–636. [Google Scholar] [CrossRef]
- Zodiatis, G.; Balopoulos, E. Structure and characteristics of fronts in the North Aegean Sea. Boll. Oceanol. Teor. Appl 1993, 11, 113–124. [Google Scholar]
- Zervakis, V.; Georgopoulos, D. Hydrology and circulation in the North Aegean (eastern Mediterranean) throughout 1997 and 1998. Mediterr. Mar. Sci. 2016, 3, 5–19. [Google Scholar] [CrossRef]
- Siokou-Frangou, I.; Shiganova, T.; Christou, E.D.; Kamburska, L.; Gubanova, A.; Konsulov, A.; Musaeva, E.; Skryabin, V.; Khoroshilov, V. Mesozooplankton communities in the Aegean and Black Seas: A comparative study. Mar. Biol. 2004, 144, 1111–1126. [Google Scholar] [CrossRef]
- Altuğ, G.; Aktan, Y.; Oral, M.; Topaloğlu, B.; Dede, A.; Keskin, Ç.; Işinibilir, M.; Çardak, M.; Çiftçi, P.S. Biodiversity of the northern Aegean Sea and southern part of the Sea of Marmara, Turkey. Mar. Biodivers. Rec. 2011, 4, e65. [Google Scholar] [CrossRef]
- Report of the Mediterranean Regional Workshop to Facilitate the Description of Ecologically or Biologically Significant Marine Areas1. 2014. Available online: https://www.cbd.int/doc/meetings/mar/ebsaws-2014-03/official/ebsaws-2014-03-04-en.pdf (accessed on 30 May 2025).
- Gourgiotis, A.; Koutsi, D.; Krommyda, V.; Stratigea, A. Spatial and Developmental Policy Directions Affecting Marine Spatial Planning in the Northern Aegean Sea, Greece. Oceans 2024, 5, 522–546. [Google Scholar] [CrossRef]
- Trapella, G.; Scicchitano, D.; Foresto, L.; Dell’Acqua, A.N.; Radaelli, E.; Turroni, S.; Rampelli, S.; Corinaldesi, C.; Palladino, G.; Candela, M. Signature of the anthropogenic impacts on the epipelagic microbiome of the North-Western Adriatic Sea (Mediterranean Sea). Front. Mar. Sci. 2024, 11, 1340088. [Google Scholar] [CrossRef]
- Arrigo, K.R. Marine microorganisms and global nutrient cycles. Nature 2005, 437, 349–355, Erratum in Nature 2005, 438, 122. [Google Scholar] [CrossRef]
- Luna, G.M. Diversity of marine microbes in a changing Mediterranean Sea. Rend. Lincei 2015, 26, 49–58. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef] [PubMed]
- Krabberød, A.K.; Deutschmann, I.M.; Bjorbækmo, M.F.; Balagué, V.; Giner, C.R.; Ferrera, I.; Garcés, E.; Massana, R.; Gasol, J.M.; Logares, R. Long-term patterns of an interconnected core marine microbiota. Environ. Microbiome 2022, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Polymenakou, P.N.; Tselepides, A.; Stephanou, E.G.; Bertilsson, S. Carbon speciation and composition of natural microbial communities in polluted and pristine sediments of the Eastern Mediterranean Sea. Mar. Pollut. Bull. 2006, 52, 1396–1405. [Google Scholar] [CrossRef]
- Charalampous, G.; Kormas, K.A.; Antoniou, E.; Kalogerakis, N.; Gontikaki, E. Distinct communities of Bacteria and unicellular eukaryotes in the different water masses of Cretan Passage water column (Eastern Mediterranean Sea). Curr. Microbiol. 2024, 81, 381. [Google Scholar] [CrossRef]
- Ignatiades, L.; Karydis, M.; Vounatsou, P. A possible method for evaluating oligotrophy and eutrophication based on nutrient concentration scales. Mar. Pollut. Bull. 1992, 24, 238–243. [Google Scholar] [CrossRef]
- Hassan, S.; Sabreena; Khurshid, Z.; Bhat, S.A.; Kumar, V.; Ameen, F.; Ganai, B.A. Marine bacteria and omic approaches: A novel and potential repository for bioremediation assessment. J. Appl. Microbiol. 2022, 133, 2299–2313. [Google Scholar] [CrossRef]
- Nagar, D.N.; Mani, K.; Bragança, J.M. Metagenome mining approaches for the discovery of marine microbial natural products. In Marine Bioactive Molecules for Biomedical and Pharmacotherapeutic Applications; Springer: Berlin/Heidelberg, Germany, 2024; pp. 61–83. [Google Scholar]
- Huang, Z.; Fang, F.; Ding, L.; Yu, K.; Zhang, L.; Lu, H. Technological advancements in field investigations of marine microorganisms: From sampling strategies to molecular analyses. J. Mar. Sci. Eng. 2023, 11, 1981. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.; Hao, H. Advances in metagenomics and its application in environmental microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef]
- Miya, M. Environmental DNA metabarcoding: A novel method for biodiversity monitoring of marine fish communities. Annu. Rev. Mar. Sci. 2022, 14, 161–185. [Google Scholar] [CrossRef] [PubMed]
- Rourke, M.L.; Fowler, A.M.; Hughes, J.M.; Broadhurst, M.K.; DiBattista, J.D.; Fielder, S.; Wilkes Walburn, J.; Furlan, E.M. Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 2022, 4, 9–33. [Google Scholar] [CrossRef]
- Ardura, A.; Zaiko, A.; Martinez, J.L.; Samuiloviene, A.; Borrell, Y.; Garcia-Vazquez, E. Environmental DNA evidence of transfer of North Sea molluscs across tropical waters through ballast water. J. Molluscan Stud. 2015, 81, 495–501. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Pääbo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef]
- Lefkaditou, E.; Haralabous, J.; Sarikas, D.; Karamelidou, S.; Kavadas, S. The cephalopods in the small-scale fishery in the eastern Thracian Sea (NE Mediterranean). In Proceedings of the 2004 ICES Annual Science Conference, Vigo, Spain, 22 September 2004. [Google Scholar]
- Fortier, L.; Le Fèvre, J.; Legendre, L. Export of biogenic carbon to fish and to the deep ocean: The role of large planktonic microphages. J. Plankton Res. 1994, 16, 809–839. [Google Scholar] [CrossRef]
- Sini, M.; Katsanevakis, S.; Koukourouvli, N.; Gerovasileiou, V.; Dailianis, T.; Buhl-Mortensen, L.; Damalas, D.; Dendrinos, P.; Dimas, X.; Frantzis, A. Assembling ecological pieces to reconstruct the conservation puzzle of the Aegean Sea. Front. Mar. Sci. 2017, 4, 347. [Google Scholar] [CrossRef]
- Kokkos, N.; Papadopoulou, A.; Zachopoulos, K.; Zoidou, M.; Beguery, L.; Margirier, F.; Sylaios, G. Hydrography and deep chlorophyll maximum patterns of the Athos Basin and the Thracian Sea continental shelf (North Aegean Sea) combining glider and satellite observations. Cont. Shelf Res. 2023, 262, 105029. [Google Scholar] [CrossRef]
- Cucknell, A.; Frantzis, A.; Boisseau, O.; Romagosa, M.; Ryan, C.; Tonay, A.; Alexiadou, P.; Öztürk, A.; Moscrop, A. Harbour porpoises in the Aegean Sea, Eastern Mediterranean: The species’ presence is confirmed. Mar. Biodivers. Rec. 2016, 9, 72. [Google Scholar] [CrossRef]
- Gkanasos, A.; Schismenou, E.; Tsiaras, K.; Somarakis, S.; Giannoulaki, M.; Sofianos, S.; Triantafyllou, G. A three dimensional, full life cycle, anchovy and sardine model for the North Aegean Sea (Eastern Mediterranean): Validation, sensitivity and climatic scenario simulations. Mediterr. Mar. Sci. 2021, 22, 653–668. [Google Scholar] [CrossRef]
- Kallianiotis, A.; Vidoris, P.; Sylaios, G. Fish species assemblages and geographical sub-areas in the North Aegean Sea, Greece. Fish. Res. 2004, 68, 171–187. [Google Scholar] [CrossRef]
- Stergiou, K.; Papaconstantinou, C.; Kleanthous, M.; Fourtouni, A. The relative abundance and distribution of small pelagic fishes in the Aegean Sea. Fresenius Environ. Bull. 1993, 2, 357–362. [Google Scholar]
- Polymenakou, P.N.; Fragkioudaki, G.; Tselepides, A. Bacterial and organic matter distribution in the sediments of the Thracian Sea (NE Aegean Sea). Cont. Shelf Res. 2007, 27, 2187–2197. [Google Scholar] [CrossRef]
- Xanthopoulou, P.; Katsanevakis, S.; Ragkousis, M.; Papadakis, O.; Zotou, M.; Kamidis, N.; Wangensteen, O.S.; Papathanasiou, V.; Karampetsis, D.; Mazaris, A.D. Complementing underwater visual surveys with eDNA metabarcoding to detect Mediterranean non-indigenous fishes. Mediterr. Mar. Sci. 2025, 26, 216–229. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System; Open Source Geospatial Foundation: Beaverton, OR, USA, 2009. [Google Scholar]
- Microsoft Corporation Bing Maps—Hillshaded Base Map. Available online: https://ecn.dynamic.t0.tiles.virtualearth.net/comp/CompositionHandler/{q}?mkt=en-us&it=G,VE,BX,L,LA&shading=hill (accessed on 18 August 2025).
- Becker, R.; Chambers, J.; Wilks, A. The New S Language; Wadsworth & Brooks/Cole: Jamaica, VT, USA, 1988. [Google Scholar]
- Mardia, K.V.; Kent, J.T.; Taylor, C.C. Multivariate Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2024; ISBN 1-118-73802-0. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Slowikowski, K. ggrepel: Automatically Position Non-Overlapping Text Labels with “ggplot2”. 2024. Available online: https://ggrepel.slowkow.com/ (accessed on 30 May 2025).
- Kassambara, A.; Mundt, F. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. Available online: https://cran.r-project.org/web/packages/factoextra/index.html (accessed on 30 May 2025).
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N. Moving pictures of the human microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 2016, 1, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Erratum in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Margalef, R. Information Theory in Ecology; Real Academia de Ciencias y Artes de Barcelona: Barcelona, Spain, 1973. [Google Scholar]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.6-8; R Core Team: Vienna, Austria, 2025.
- Pedersen, T.L. ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. J. Open Source Softw. 2017, 2, 316. [Google Scholar]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Haro-Moreno, J.M.; López-Pérez, M.; de la Torre, J.R.; Picazo, A.; Camacho, A.; Rodriguez-Valera, F. Fine metagenomic profile of the Mediterranean stratified and mixed water columns revealed by assembly and recruitment. Microbiome 2018, 6, 128. [Google Scholar] [CrossRef]
- Winter, C.; Kerros, M.-E.; Weinbauer, M.G. Seasonal changes of bacterial and archaeal communities in the dark ocean: Evidence from the Mediterranean Sea. Limnol. Oceanogr. 2009, 54, 160–170. [Google Scholar] [CrossRef]
- Giovannoni, S.J. SAR11 bacteria: The most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 2017, 9, 231–255. [Google Scholar] [CrossRef]
- Laghdass, M.; Catala, P.; Caparros, J.; Oriol, L.; Lebaron, P.; Obernosterer, I. High contribution of SAR11 to microbial activity in the North West Mediterranean Sea. Microb. Ecol. 2012, 63, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Quéméneur, M.; Bel Hassen, M.; Armougom, F.; Khammeri, Y.; Lajnef, R.; Bellaaj-Zouari, A. Prokaryotic diversity and distribution along physical and nutrient gradients in the Tunisian coastal waters (South Mediterranean Sea). Front. Microbiol. 2020, 11, 593540. [Google Scholar] [CrossRef] [PubMed]
- Feingersch, R.; Suzuki, M.T.; Shmoish, M.; Sharon, I.; Sabehi, G.; Partensky, F.; Béjà, O. Microbial community genomics in eastern Mediterranean Sea surface waters. ISME J. 2010, 4, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Farias, I.P.; Ortí, G.; Sampaio, I.; Schneider, H.; Meyer, A. The cytochrome b gene as a phylogenetic marker: The limits of resolution for analyzing relationships among cichlid fishes. J. Mol. Evol. 2001, 53, 89–103. [Google Scholar] [CrossRef]
- Kochzius, M.; Seidel, C.; Antoniou, A.; Botla, S.K.; Campo, D.; Cariani, A.; Vazquez, E.G.; Hauschild, J.; Hervet, C.; Hjörleifsdottir, S. Identifying fishes through DNA barcodes and microarrays. PLoS ONE 2010, 5, e12620. [Google Scholar] [CrossRef]
- Li, X.; Shen, X.; Chen, X.; Xiang, D.; Murphy, R.W.; Shen, Y. Detection of potential problematic Cytb gene sequences of fishes in GenBank. Front. Genet. 2018, 9, 30. [Google Scholar] [CrossRef]
- Bariche, M.; Sadek, R.; Al-Zein, M.; El-Fadel, M. Diversity of juvenile fish assemblages in the pelagic waters of Lebanon (eastern Mediterranean). Hydrobiologia 2007, 580, 109–115. [Google Scholar] [CrossRef]
- Jemaa, S.; Lteif, M.; Khala, F.; Fakhri, M. The biodiversity and seasonal variation of pelagic fish caught by purse seines in tripoli, Northern Lebanese Coast. Leban. Sci. J. 2021, 22, 082–097. [Google Scholar] [CrossRef]
- Saad, A. Check–list of bony fish collected from the coast of Syria. Turk. J. Fish. Aquat. Sci. 2005, 5, 99–106. [Google Scholar]
- Cicala, D.; Maiello, G.; Fiorentino, F.; Garofalo, G.; Massi, D.; Sbrana, A.; Mariani, S.; D’Alessandro, S.; Stefani, M.; Perrodin, L. Spatial analysis of demersal food webs through integration of eDNA metabarcoding with fishing activities. Front. Mar. Sci. 2024, 10, 1209093, Erratum in Front. Mar. Sci. 2024, 11, 1380910. [Google Scholar] [CrossRef]
- Djurhuus, A.; Closek, C.J.; Kelly, R.P.; Pitz, K.J.; Michisaki, R.P.; Starks, H.A.; Walz, K.R.; Andruszkiewicz, E.A.; Olesin, E.; Hubbard, K. Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 2020, 11, 254. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokamani, M.; Liakopoulos, P.; Tegopoulos, K.; Zigkou, A.-M.; Triantaphyllidis, G.; Kamidis, N.; Grigoriou, M.E.; Sandaltzopoulos, R.; Kolovos, P. Spatiotemporal Dynamics of Microbial and Fish Communities in the Thracian Sea Revealed by eDNA Metabarcoding. Microorganisms 2025, 13, 2373. https://doi.org/10.3390/microorganisms13102373
Tokamani M, Liakopoulos P, Tegopoulos K, Zigkou A-M, Triantaphyllidis G, Kamidis N, Grigoriou ME, Sandaltzopoulos R, Kolovos P. Spatiotemporal Dynamics of Microbial and Fish Communities in the Thracian Sea Revealed by eDNA Metabarcoding. Microorganisms. 2025; 13(10):2373. https://doi.org/10.3390/microorganisms13102373
Chicago/Turabian StyleTokamani, Maria, Panagiotis Liakopoulos, Konstantinos Tegopoulos, Aristea-Marina Zigkou, George Triantaphyllidis, Nikolaos Kamidis, Maria E. Grigoriou, Raphael Sandaltzopoulos, and Petros Kolovos. 2025. "Spatiotemporal Dynamics of Microbial and Fish Communities in the Thracian Sea Revealed by eDNA Metabarcoding" Microorganisms 13, no. 10: 2373. https://doi.org/10.3390/microorganisms13102373
APA StyleTokamani, M., Liakopoulos, P., Tegopoulos, K., Zigkou, A.-M., Triantaphyllidis, G., Kamidis, N., Grigoriou, M. E., Sandaltzopoulos, R., & Kolovos, P. (2025). Spatiotemporal Dynamics of Microbial and Fish Communities in the Thracian Sea Revealed by eDNA Metabarcoding. Microorganisms, 13(10), 2373. https://doi.org/10.3390/microorganisms13102373






