Comparative Meta-Analysis of Long-Read and Short-Read Sequencing for Metagenomic Profiling of the Lower Respiratory Tract Infections
Abstract
1. Introduction
2. Methods
2.1. Search Strategy
2.2. Study Eligibility
2.3. Study Selection
2.4. Data Synthesis
2.5. Quality Assessment
2.6. Outcome Measures
3. Results
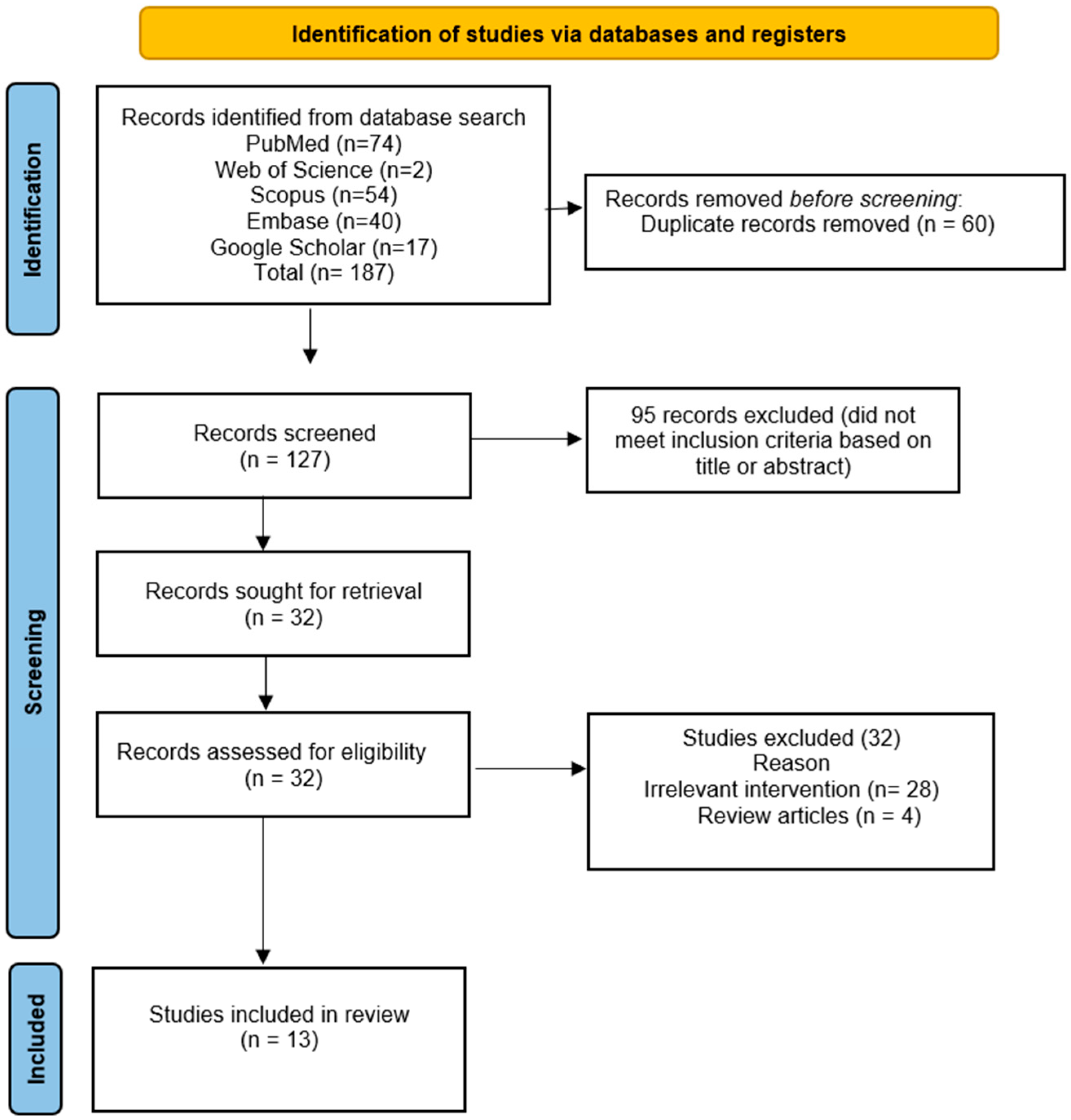
3.1. Included Studies
3.2. Flow Diagram
3.3. Study Characteristics
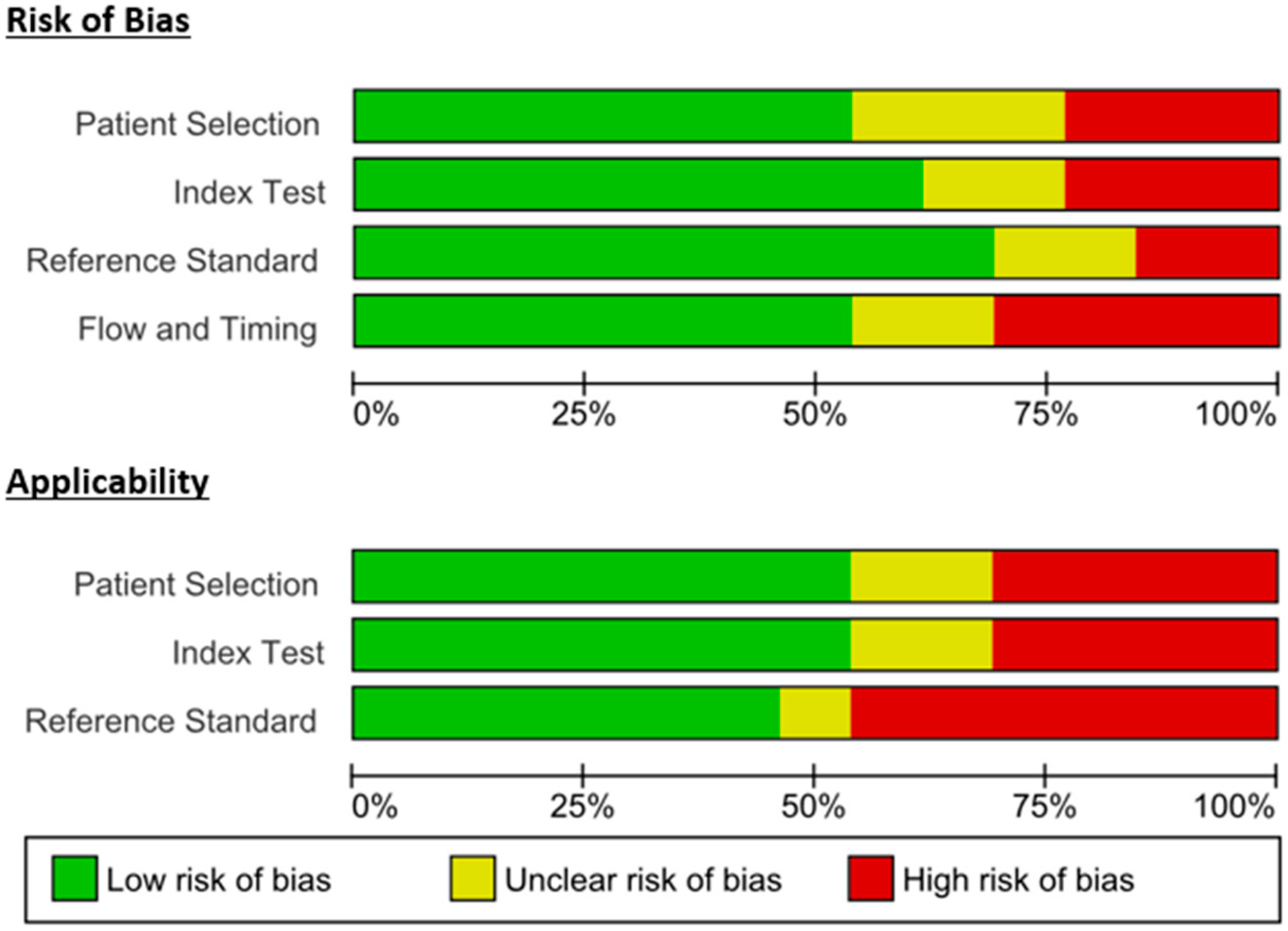
3.4. Methods Quality Assessment
4. Discussion
4.1. Strengths and Limitations
4.2. Implications for Practice and Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Key Variable | Sub Terms | Search Options | PubMed | Web of Science | Scopus | Embase |
|---|---|---|---|---|---|---|
| 1. Metagenomics | 1.1 Metagenomics | Mesh | 12,372 | |||
| 1.2 Metagenomics | TI/AB | 16,240 | 2093 | 12,244 | 12,823 | |
| 1.3 Metagenomic sequencing | TI/AB | 5293 | 1562 | 15,275 | 6274 | |
| ((“Metagenomics” [Mesh]) OR (Metagenomics [Title/Abstract])) OR (Metagenomic sequencing [Title/Abstract]) | 26,072 | 3655 | 25,114 | 18315 | ||
| 2. Short vs. Long | 2.1 Illumina | TI/AB | 32,276 | 1302 | 33,937 | 59,185 |
| 2.2 short-read | TI/AB | 4550 | 585 | 6303 | 5530 | |
| 2.3 Nanopore sequencing | TI/AB | 3349 | 1688 | 7794 | 3521 | |
| 2.4 long-read | TI/AB | 7104 | 8321 | 8776 | 8535 | |
| 2.5 PacBio | TI/AB | 4621 | 345 | 4326 | 4803 | |
| (PacBio [Title/Abstract]) OR ((((Illumina [Title/Abstract]) OR (short-read [Title/Abstract])) OR (Nanopore sequencing [Title/Abstract])) OR (long-read [Title/Abstract])) | 42,425 | 11,292 | 50,409 | 73,056 | ||
| 3. LRTI | 3.1 Respiratory Tract Infections | Mesh | 694,847 | |||
| 3.2 Respiratory Tract Infections | TI/AB | 21,010 | 7704 | 41,928 | 20,238 | |
| 3.3 Lower respiratory tract | TI/AB | 16,308 | 3054 | 25,280 | 23,303 | |
| 3.4 pneumonia | TI/AB | 171,735 | 78,891 | 158,958 | 259,012 | |
| 3.5 bronchoalveolar lavage | TI/AB | 37,229 | 5049 | 35,492 | 55,243 | |
| 3.6 LRTI | TI/AB | 2033 | 59 | 1885 | 3392 | |
| 3.7 lower respiratory tract infection | TI/AB | 4250 | 19,257 | 17,447 | 6093 | |
| (lower respiratory tract infection [Title/Abstract]) OR ((((((“Respiratory Tract Infections” [Mesh]) OR (Respiratory Tract Infections [Title/Abstract])) OR (Lower respiratory tract [Title/Abstract])) OR (pneumonia [Title/Abstract])) OR (bronchoalveolar lavage [Title/Abstract])) OR (LRTI [Title/Abstract])) | 820,678 | 105,613 | 231,479 | 333,441 | ||
| ((((“Metagenomics” [Mesh]) OR (Metagenomics [Title/Abstract])) OR (Metagenomic sequencing [Title/Abstract])) AND ((((Illumina [Title/Abstract]) OR (short-read [Title/Abstract])) OR (Nanopore sequencing [Title/Abstract])) OR (long-read [Title/Abstract]))) AND ((lower respiratory tract infection [Title/Abstract]) OR ((((((“Respiratory Tract Infections” [Mesh]) OR (Respiratory Tract Infections [Title/Abstract])) OR (Lower respiratory tract [Title/Abstract])) OR (pneumonia [Title/Abstract])) OR (bronchoalveolar lavage [Title/Abstract])) OR (LRTI [Title/Abstract]))) | 74 | 2 | 54 | 40 | ||
References
- Bender, R.G.; Sirota, S.B.; Swetschinski, L.R.; Dominguez, R.M.V.; Novotney, A.; Wool, E.E.; Ikuta, K.S.; Vongpradith, A.; Rogowski, E.L.B.; Doxey, M.; et al. Global, regional, and national incidence and mortality burden of non-COVID-19 lower respiratory infections and aetiologies, 1990-2021: A systematic analysis from the Global Burden of Disease Study 2021. Lancet Infect. Dis. 2024, 24, 974–1002. [Google Scholar] [CrossRef]
- van Heijl, I.; Schweitzer, V.A.; Zhang, L.; van der Linden, P.D.; van Werkhoven, C.H.; Postma, D.F. Inappropriate Use of Antimicrobials for Lower Respiratory Tract Infections in Elderly Patients: Patient- and Community-Related Implications and Possible Interventions. Drugs Aging 2018, 35, 389–398. [Google Scholar] [CrossRef]
- Bustos, I.G.; Martinez-Lemus, L.F.; Reyes, L.F.; Martin-Loeches, I. Transforming Microbiological Diagnostics in Nosocomial Lower Respiratory Tract Infections: Innovations Shaping the Future. Diagnostics 2025, 15, 265. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, G.; Lau, H.C.; Yu, J. Metagenomic Sequencing for Microbial DNA in Human Samples: Emerging Technological Advances. Int. J. Mol. Sci. 2022, 23, 2181. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, T.; Shen, M.; Zhang, Y.; Chen, W.; Chen, H.; Wang, Y.; Liu, J.; Tao, J.; He, L.; et al. Utilizing metagenomic next-generation sequencing for diagnosis and lung microbiome probing of pediatric pneumonia through bronchoalveolar lavage fluid in pediatric intensive care unit: Results from a large real-world cohort. Front. Cell. Infect. Microbiol. 2023, 13, 1200806. [Google Scholar] [CrossRef]
- Latorre-Pérez, A.; Villalba-Bermell, P.; Pascual, J.; Vilanova, C. Assembly methods for nanopore-based metagenomic sequencing: A comparative study. Sci. Rep. 2020, 10, 13588. [Google Scholar] [CrossRef] [PubMed]
- Bejaoui, S.; Nielsen, S.H.; Rasmussen, A.; Coia, J.E.; Andersen, D.T.; Pedersen, T.B.; Møller, M.V.; Kusk Nielsen, M.T.; Frees, D.; Persson, S. Comparison of Illumina and Oxford Nanopore sequencing data quality for Clostridioides difficile genome analysis and their application for epidemiological surveillance. BMC Genom. 2025, 26, 92. [Google Scholar] [CrossRef]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; AbuOun, M.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 2019, 5, e000294. [Google Scholar] [CrossRef] [PubMed]
- Maghini Dylan, G.; Kiguchi, Y.; Darling Aaron, E.; Monahan Leigh, G.; Halpern Aaron, L.; Burke Catherine, M.; Jaeger, E.; Statham, A.; Truong, T.; Ying, K.; et al. Illumina complete long read assay yields contiguous bacterial genomes from human gut metagenomes. mSystems 2025, 10, e01531-24. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Greenman, N.; Hassouneh, S.A.; Abdelli, L.S.; Johnston, C.; Azarian, T. Improving Bacterial Metagenomic Research through Long-Read Sequencing. Microorganisms 2024, 12, 935. [Google Scholar] [CrossRef]
- Oehler, J.B.; Wright, H.; Stark, Z.; Mallett, A.J.; Schmitz, U. The application of long-read sequencing in clinical settings. Human. Genom. 2023, 17, 73. [Google Scholar] [CrossRef]
- Yan, M.; Shang, L.; Wang, Y.; Wang, C.; Cao, B. Metagenomic next-generation sequencing on treatment strategies and prognosis of patients with lower respiratory tract infections: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2025, 65, 107440. [Google Scholar] [CrossRef]
- Guo, Q.; Xiao, Y.; Zhang, S. Metagenomic next generation sequencing of bronchoalveolar lavage samples for the diagnosis of lower respiratory tract infections: A systematic review and meta-analysis. Heliyon 2024, 10, e23188. [Google Scholar] [CrossRef]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, P.; Lin, Y.; Chen, H.; Yang, L.; Li, J.; Zhang, T.; Chen, Q.; Li, Z.; Du, X.; et al. Metagenomic Diagnosis for a Culture-Negative Sample From a Patient With Severe Pneumonia by Nanopore and Next-Generation Sequencing. Front. Cell. Infect. Microbiol. 2020, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Capraru, I.D.; Romanescu, M.; Anghel, F.M.; Oancea, C.; Marian, C.; Sirbu, I.O.; Chis, A.R.; Ciordas, P.D. Identification of Genomic Variants of SARS-CoV-2 Using Nanopore Sequencing. Medicina 2022, 58, 1841. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, L.; Zhu, C.; Jin, J.; Song, C.; Dong, H.; Li, Z.; Wang, Z.; Chen, Y.; Yang, Z.; et al. Clinical value of metagenomic next-generation sequencing by Illumina and Nanopore for the detection of pathogens in bronchoalveolar lavage fluid in suspected community-acquired pneumonia patients. Front. Cell. Infect. Microbiol. 2022, 12, 1021320, Erratum in Front. Cell. Infect. Microbiol. 2024, 14, 1468511. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhu, C.; Yan, T.; Hu, Y.; Zhou, J.; Li, Y.; Du, F.; Zhou, J. Illumina and Nanopore sequencing in culture-negative samples from suspected lower respiratory tract infection patients. Front. Cell. Infect. Microbiol. 2024, 14, 1230650. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, H.; Mao, L.; Yu, H.; Yu, X.; Sun, Z.; Qian, X.; Cheng, S.; Chen, S.; Chen, J.; et al. Rapid genomic characterization of SARS-CoV-2 viruses from clinical specimens using nanopore sequencing. Sci. Rep. 2020, 10, 17492. [Google Scholar] [CrossRef]
- Carbo, E.C.; Mourik, K.; Boers, S.A.; Munnink, B.O.; Nieuwenhuijse, D.; Jonges, M.; Welkers, M.R.A.; Matamoros, S.; van Harinxma thoe Slooten, J.; Kraakman, M.E.M.; et al. A comparison of five Illumina, Ion Torrent, and nanopore sequencing technology-based approaches for whole genome sequencing of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 701–713. [Google Scholar] [CrossRef]
- Hahn, A.; Sanyal, A.; Perez, G.F.; Colberg-Poley, A.M.; Campos, J.; Rose, M.C.; Pérez-Losada, M. Different next generation sequencing platforms produce different microbial profiles and diversity in cystic fibrosis sputum. J. Microbiol. Methods 2016, 130, 95–99. [Google Scholar] [CrossRef]
- Zhang, X.; Ye, J.; Wang, L.; Zhang, L.; Wang, L.; Jin, H. Nanopore Sequencing Technology: A Reliable Method for Pathogen Diagnosis in Elderly Patients with Community-Acquired Pneumonia. Infect. Drug Resist. 2024, 17, 3659–3667. [Google Scholar] [CrossRef]
- Heikema, A.P.; Horst-Kreft, D.; Boers, S.A.; Jansen, R.; Hiltemann, S.D.; de Koning, W.; Kraaij, R.; de Ridder, M.A.; van Houten, C.B.; Bont, L.J. Comparison of illumina versus nanopore 16S rRNA gene sequencing of the human nasal microbiota. Genes 2020, 11, 1105. [Google Scholar] [CrossRef]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J.; et al. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2019, 58, e00963-19. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Horiba, K.; Haruta, K.; Takeuchi, S.; Suzuki, T.; Torii, Y.; Kawabe, S.; Wada, S.; Ikeyama, T.; Ito, Y.; et al. Utility of nanopore sequencing for detecting pathogens in bronchoalveolar lavage fluid from pediatric patients with respiratory failure. J. Clin. Virol. Plus 2023, 3, 100154. [Google Scholar] [CrossRef]
- Jabeen Maisha, F.; Sanderson Nicholas, D.; Foster, D.; Crook Derrick, W.; Cane Jennifer, L.; Borg, C.; Connolly, C.; Thulborn, S.; Pavord Ian, D.; Klenerman, P.; et al. Identifying Bacterial Airways Infection in Stable Severe Asthma Using Oxford Nanopore Sequencing Technologies. Microbiol. Spectr. 2022, 10, e02279-21. [Google Scholar] [CrossRef] [PubMed]
- Serpa, P.H.; Deng, X.; Abdelghany, M.; Crawford, E.; Malcolm, K.; Caldera, S.; Fung, M.; McGeever, A.; Kalantar, K.L.; Lyden, A.; et al. Metagenomic prediction of antimicrobial resistance in critically ill patients with lower respiratory tract infections. Genome Med. 2022, 14, 74. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Wang, G.-H.; Xu, X.-Y.; Nong, X.-H.; Wang, J.; Amin, M.; Qi, S.-H. Exploring fungal diversity in deep-sea sediments from Okinawa Trough using high-throughput Illumina sequencing. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2016, 116, 99–105. [Google Scholar] [CrossRef]
- Horiba, K.; Torii, Y.; Aizawa, Y.; Yamaguchi, M.; Haruta, K.; Okumura, T.; Suzuki, T.; Kawano, Y.; Kawada, J.I.; Hara, S.; et al. Performance of Nanopore and Illumina Metagenomic Sequencing for Pathogen Detection and Transcriptome Analysis in Infantile Central Nervous System Infections. Open Forum Infect. Dis. 2022, 9, ofac504. [Google Scholar] [CrossRef]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef]
- Zheng, P.; Zhou, C.; Ding, Y.; Liu, B.; Lu, L.; Zhu, F.; Duan, S. Nanopore sequencing technology and its applications. MedComm 2023, 4, e316. [Google Scholar] [CrossRef]
- Gu, W.; Deng, X.; Lee, M.; Sucu, Y.D.; Arevalo, S.; Stryke, D.; Federman, S.; Gopez, A.; Reyes, K.; Zorn, K.; et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 2021, 27, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Luan, T.; Commichaux, S.; Hoffmann, M.; Jayeola, V.; Jang, J.H.; Pop, M.; Rand, H.; Luo, Y. Benchmarking short and long read polishing tools for nanopore assemblies: Achieving near-perfect genomes for outbreak isolates. BMC Genom. 2024, 25, 679. [Google Scholar] [CrossRef]
- Deremarque, T.; Gozlan, R.E.; Ravaozafindrasoa, R.; Mucci, G.; Delalex, L.; Foissy, J.-M.; Cagnant, M.; Clair, M.; Givens, J.; Justy, F.; et al. How does the efficiency of Nanopore compare to Illumina sequencing in tracking the spread of aquatic invasive species? Water Biol. Secur. 2025, 100423. [Google Scholar] [CrossRef]
- Petersen, L.M.; Martin, I.W.; Moschetti, W.E.; Kershaw, C.M.; Tsongalis, G.J. Third-Generation Sequencing in the Clinical Laboratory: Exploring the Advantages and Challenges of Nanopore Sequencing. J. Clin. Microbiol. 2019, 58. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, K.; Johnson, B.; Klimowich, K.; Chiotos, K.; Jensen, E.A.; Planet, P.; Phinizy, P.; Piccione, J. Comparison of tracheal aspirate and bronchoalveolar lavage samples in the microbiological diagnosis of lower respiratory tract infection in pediatric patients. Pediatr. Pulmonol. 2022, 57, 2405–2410. [Google Scholar] [CrossRef] [PubMed]
- Veselovsky, V.; Romanov, M.; Zoruk, P.; Larin, A.; Babenko, V.; Morozov, M.; Strokach, A.; Zakharevich, N.; Khamidova, S.; Danilova, A.; et al. Comparative evaluation of sequencing platforms: Pacific Biosciences, Oxford Nanopore Technologies, and Illumina for 16S rRNA-based soil microbiome profiling. Front. Microbiol. 2025, 16, 1633360. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Feng, S.; Yu, X.; Zhao, J.; Wan, Y.; Yao, Z.; Li, D. Diagnostic value of metagenomic next-generation sequencing combined by medical thoracoscopy surgery among infectious pleural effusion patients. BMC Infect. Dis. 2025, 25, 407. [Google Scholar] [CrossRef] [PubMed]

| Authors | Year | Study Design | Participants | Age | Males | Comparator | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| Zhang et al. [21] | 2022 | Prospective | 66 | 68 (58, 72) | 63.6% | Illumina, Nanopore | Illumina: 46.7%, Nanopore: 40% | NA |
| Ma et al. [22] | 2024 | Retrospective | 38 | 55 (36, 63) | 63.20% | Illumina, Nanopore | Illumina: 80.6%, Nanopore: 93.5% | Illumina: 42.9%, Nanopore: 28.6% |
| Zhang et al. [26] | 2024 | Prospective | 29 | 67 (65, 73.5) | NA | Illumina, Nanopore | Nanopore, 82.3%, Illumina, 88.2% | Nanopore, 75%, Illumina, 50% |
| Heikema et al. [27] | 2020 | Cross-sectional | 10 adults and 49 children | NA | NA | Illumina, Nanopore | NA | NA |
| Carbo et al. [24] | 2023 | Retrospective | 24 | NA | NA | Illumina, Ion Torrent, and Nanopore | NA | NA |
| Capraru et al. [20] | 2022 | Cross-sectional analytical | 103 samples | NA | NA | Ion torrent, Nanopore | NA | NA |
| Wang et al. [19] | 2020 | Case study | 63-year-old male | - | - | Nanopore and BGISEQ-500 | - | - |
| Lewandowski et al. [28] | 2019 | Methodological validation study | 50 samples | NA | NA | Illumina and nanopore | Nanopore: 83%, Illumina: NA | Nanopore: 100%, Illumina: NA |
| Yamaguchi et al. [29] | 2023 | Prospective | 31 samples | NA | NA | Illumina and nanopore | NA | NA |
| Hahn et al. [25] | 2016 | Cross-sectional | 12 samples | NA | NA | PacBio, Illumina | NA | NA |
| Jabeen et al. [30] | 2022 | Prospective | 23 | 67 (10) | 57% | Illumina, Nanopore | NA | NA |
| Serpa et al. [31] | 2022 | Retrospective | 88 | NA | NA | Illumina, Nanopore | Illumina: Gram-positive: 70%, Gram-negative: 100%, Nanopore: 100% | Illumina: Gram-positive: 95%, Gram-negative: 64%, Nanopore: NA |
| Li et al. [23] | 2020 | Cross-sectional diagnostic accuracy study | 29 clinical SARS-CoV-2 specimens | NA | NA | Illumina, Nanopore | NA | NA |
| Authors | Platform | Concordance/Agreement (%) | Positivity Rate | Turn-Around Time | Genome Coverage (%) | Read Metrics | Other Outcomes | Conclusion |
|---|---|---|---|---|---|---|---|---|
| Zhang et al. [21] | Illumina | 56.1% | NA | 20 (19–21) | NA | NA | AUC Bacteria: 0.73, Fungi: 0.73 | Nanopore detected more taxa overall than Illumina. |
| Nanopore | 57.6% | NA | 14 (11–15) | NA | NA | AUC Bacteria: 0.60, Fungi: 0.81 | ||
| Ma et al. [22] | Illumina | 63.9% | Bacteria: 71.4%, Fungi: 50% | NA | NA | NA | 61.1% detected (with antibiotics) and 46.2% detected (without antibiotics) | Nanopore sequencing showed higher sensitivity and better concordance than Illumina, particularly for detecting Mycobacterium. |
| Nanopore | 83.3% | Bacteria: 78.6%, Fungi: 62.5% | NA | NA | NA | 77.8% detected (with antibiotics) and 76.9% detected (without antibiotics) | ||
| Zhang et al. [26] | Illumina | NA | 51.7% | 24 | NA | NA | NA | Nanopore exhibited relatively better consistency. |
| Nanopore | NA | 48.3% | 8 | NA | NA | Required shorter time | ||
| Heikema et al. [27] | Illumina | 69.1% with nanopore | 91% | NA | NA | 131,024 | ISI: 2.7, Mean genera detected (≥1%): 4.4 | Both comparable but nanopore is not that effective with genus Corynebacterium |
| Nanopore | - | 78% | NA | NA | 21,907 | ISI: 2.2, Genera detected: 4.5 | ||
| Carbo et al. [24] | Illumina | NA | NA | 3 days | 99.8% | Depth: 860 | NA | Illumina has higher accuracy but longer time. |
| Nanopore | NA | NA | <24 h | 81.2% | Depth: >2000 | NA | ||
| Capraru et al. [20] | Ion torrent | Clade: 90.90% | NA | NA | NA | 190 base pair | NA | Nanopore is faster with deeper coverage; Ion Torrent higher alignment rates |
| Nanopore | NA | NA | >250× | 519.17 bases | NA | |||
| Wang et al. [19] | BGISEQ-500 | 100% | - | NA | 100% | 129,512,318 | NA | Both rapidly and reliably identified the causative pathogen. |
| Nanopore | 100% | - | 12.14 h | 45% | 34,831 | NA | ||
| Lewandowski et al. [28] | Illumina | 100% with nanopore | NA | NA | 26.6% | NA | NA | Nanopore is comparable to Illumina in sequencing influenza viruses. |
| Nanopore | - | NA | NA | ≥99.3% per segment | 3.8 × 105 reads | Limit of Detection: 102–103 copies/mL | ||
| Yamaguchi et al. [29] | Illumina | Reference | NA | NA | NA | 2,155,152, 264,467,762 bases | NA | In a comparison of 7 BALF samples, nanopore sequencing detected the same RNA viruses as Illumina. |
| Nanopore | 71.4% | 41.7% | NA | 81.38% | 220,600, 699,203,556 bases | NA | ||
| Hahn et al. [25] | Illumina | NA | 49.4% | NA | NA | 479,220 reads Per-sample | MiSeq sequencing of the 16S rRNA V4 region provided higher alpha-diversity estimates | PacBio identified Burkholderia while MiSeq detected more Escherichia. |
| PacBio | NA | 99.3% | NA | NA | 122,526 reads Per-sample | |||
| Jabeen et al. [30] | Illumina | NA | NA | NA | NA | 172 base pair | Nanopore sequencing achieved near-complete genome coverage and depth at all read–depth thresholds compared with MiSeq | |
| Nanopore | NA | NA | NA | 24.2–94.2 | 2013 base pair | |||
| Serpa et al. [31] | Illumina | 100% of AMR loci identified by Illumina | NA | NA | NA | 6.9 × 107 reads per sample | NA | Illumina and nanopore has similar sensitivity |
| Nanopore | 81% of culture-confirmed bacterial pathogens | NA | NA | NA | 1.19 × 106 total reads per sample | NA | ||
| Li et al. [23] | Illumina | NA | NA | NA | NA | NA | NA | Nanopore detected whole genomes from samples diluted up to 100,000× (undetectable by qRT-PCR), with ≥97.6% completeness at >250× depth |
| Nanopore | 100% with Illumina | NA | NA | 98.08–100% | NA | NA | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorenzin, G.; Carlin, M. Comparative Meta-Analysis of Long-Read and Short-Read Sequencing for Metagenomic Profiling of the Lower Respiratory Tract Infections. Microorganisms 2025, 13, 2366. https://doi.org/10.3390/microorganisms13102366
Lorenzin G, Carlin M. Comparative Meta-Analysis of Long-Read and Short-Read Sequencing for Metagenomic Profiling of the Lower Respiratory Tract Infections. Microorganisms. 2025; 13(10):2366. https://doi.org/10.3390/microorganisms13102366
Chicago/Turabian StyleLorenzin, Giovanni, and Maddalena Carlin. 2025. "Comparative Meta-Analysis of Long-Read and Short-Read Sequencing for Metagenomic Profiling of the Lower Respiratory Tract Infections" Microorganisms 13, no. 10: 2366. https://doi.org/10.3390/microorganisms13102366
APA StyleLorenzin, G., & Carlin, M. (2025). Comparative Meta-Analysis of Long-Read and Short-Read Sequencing for Metagenomic Profiling of the Lower Respiratory Tract Infections. Microorganisms, 13(10), 2366. https://doi.org/10.3390/microorganisms13102366






