FucR Functions as a Transcriptional Regulator for L-Fucose Utilization in Campylobacter jejuni
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Construction and Complementation of Mutants
2.3. Confirmation of Polycistronic Transcript
2.4. Semi-Quantitative Reverse Transcription PCR
2.5. Transcriptomic Analysis of Fucose Induction in NCTC 11168 and Cj0480c Mutant
2.6. Construction and Production of 6x His-Tagged Cj0480c
2.7. Electrophoretic Mobility Shift Assay (EMSA)
3. Results
3.1. Genetic Organization and Co-Transcription of PR2
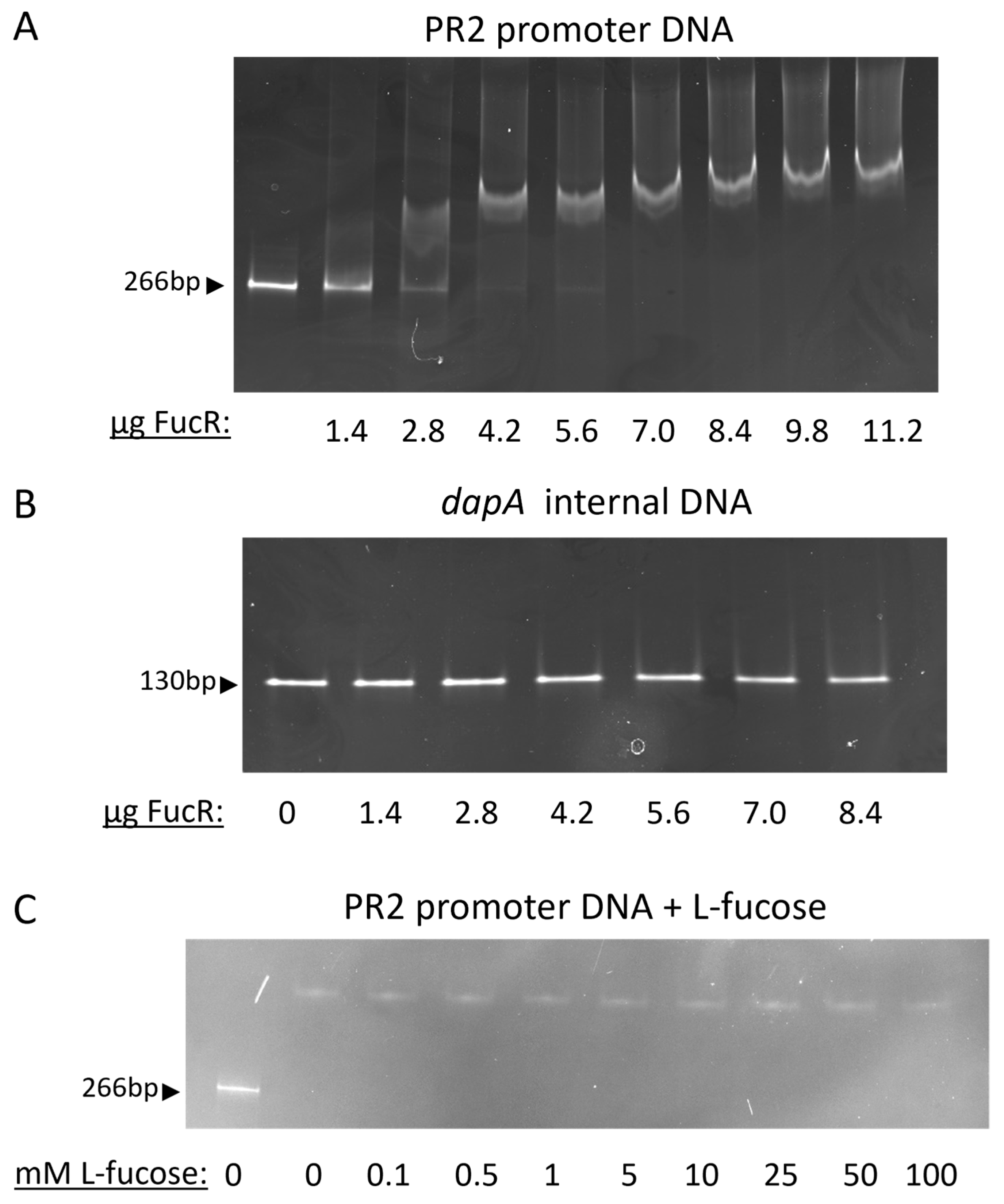
3.2. PR2 Harbors a Transcriptional Regulator That Senses and Responds to Fucose
3.3. Transcriptome Analysis of the FucR Mutant and Wild-Type Strain Grown with Fucose
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Humphrey, T.; O’Brien, S.; Madsen, M. Campylobacters as zoonotic pathogens: A food production perspective. Int. J. Food Microbiol. 2007, 117, 237–257. [Google Scholar] [CrossRef]
- McCarthy, N.; Giesecke, J. Incidence of Guillain-Barre syndrome following infection with Campylobacter jejuni. Am. J. Epidemiol. 2001, 153, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, G.I.; Blaser, M.J. Campylobacter and Helicobacter. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Olson, C.K.; Ethelberg, S.; van Pelt, W.; Tauxe, R.V. Epidemiology of infections in industrialized nations. In Campylobacter, 3rd ed.; Wiley: Hoboken, NJ, USA, 2008; pp. 163–189. [Google Scholar]
- van Gerwe, T.; Miflin, J.K.; Templeton, J.M.; Bouma, A.; Wagenaar, J.A.; Jacobs-Reitsma, W.F.; Stegeman, A.; Klinkenberg, D. Quantifying transmission of Campylobacter jejuni in commercial broiler flocks. Appl. Environ. Microbiol. 2009, 75, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Velayudhan, J.; Kelly, D.J. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: An essential role for phosphoenolpyruvate carboxykinase. Microbiology 2002, 148, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Stahl, M.; Butcher, J.; Stintzi, A. Nutrient acquisition and metabolism by Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2012, 2, 5. [Google Scholar] [CrossRef]
- Stahl, M.; Friis, L.M.; Nothaft, H.; Liu, X.; Li, J.; Szymanski, C.M.; Stintzi, A. L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. USA 2011, 108, 7194–7199. [Google Scholar] [CrossRef]
- Muraoka, W.T.; Zhang, Q. Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 2011, 193, 1065–1075. [Google Scholar] [CrossRef]
- Allen, K.J.; Griffiths, M.W. Effect of environmental and chemotactic stimuli on the activity of the Campylobacter jejuni flaA sigma(28) promoter. FEMS Microbiol. Lett. 2001, 205, 43–48. [Google Scholar] [CrossRef]
- Middendorf, P.S.; Zomer, A.L.; Bergval, I.L.; Jacobs-Reitsma, W.F.; den Besten, H.M.W.; Abee, T. Host associations of Campylobacter jejuni and Campylobacter coli isolates carrying the L-fucose or d-glucose utilization cluster. Int. J. Food Microbiol. 2024, 425, 110855. [Google Scholar] [CrossRef]
- Zautner, A.E.; Ohk, C.; Tareen, A.M.; Lugert, R.; Gross, U. Epidemiological association of Campylobacter jejuni groups with pathogenicity-associated genetic markers. BMC Microbiol. 2012, 12, 171. [Google Scholar] [CrossRef]
- Luijkx, Y.; Bleumink, N.M.C.; Jiang, J.; Overkleeft, H.S.; Wosten, M.; Strijbis, K.; Wennekes, T. Bacteroides fragilis fucosidases facilitate growth and invasion of Campylobacter jejuni in the presence of mucins. Cell. Microbiol. 2020, 22, e13252. [Google Scholar] [CrossRef]
- Middendorf, P.S.; Wijnands, L.M.; Boeren, S.; Zomer, A.L.; Jacobs-Reitsma, W.F.; den Besten, H.M.W.; Abee, T. Activation of the l-fucose utilization cluster in Campylobacter jejuni induces proteomic changes and enhances Caco-2 cell invasion and fibronectin binding. Heliyon 2024, 10, e34996. [Google Scholar] [CrossRef] [PubMed]
- Alemka, A.; Corcionivoschi, N.; Bourke, B. Defense and adaptation: The complex inter-relationship between Campylobacter jejuni and mucus. Front. Cell. Infect. Microbiol. 2012, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S. Characterization of l-2-keto-3-deoxyfuconate aldolases in a nonphosphorylating l-fucose metabolism pathway in anaerobic bacteria. J. Biol. Chem. 2020, 295, 1338–1349. [Google Scholar] [CrossRef] [PubMed]
- Garber, J.M.; Nothaft, H.; Pluvinage, B.; Stahl, M.; Bian, X.; Porfirio, S.; Enriquez, A.; Butcher, J.; Huang, H.; Glushka, J.; et al. The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus. Commun. Biol. 2020, 3, 2. [Google Scholar] [CrossRef]
- Zhou, B.; Garber, J.M.; Vlach, J.; Azadi, P.; Ng, K.K.S.; Escalante-Semerena, J.C.; Szymanski, C.M. Campylobacter jejuni uses energy taxis and a dehydrogenase enzyme for l-fucose chemotaxis. mBio 2023, 14, e0273223. [Google Scholar] [CrossRef]
- Hartley, L.E.; Kaakoush, N.O.; Ford, J.L.; Korolik, V.; Mendz, G.L. Characterisation of Campylobacter jejuni genes potentially involved in phosphonate degradation. Gut Pathog. 2009, 1, 13. [Google Scholar] [CrossRef]
- Hwang, S.; Kim, M.; Ryu, S.; Jeon, B. Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PLoS ONE 2011, 6, e22300. [Google Scholar] [CrossRef]
- Hwang, S.; Jeon, B.; Yun, J.; Ryu, S. Roles of RpoN in the resistance of Campylobacter jejuni under various stress conditions. BMC Microbiol. 2011, 11, 207. [Google Scholar] [CrossRef]
- Hwang, S.; Zhang, Q.; Ryu, S.; Jeon, B. Transcriptional regulation of the CmeABC multidrug efflux pump and the KatA catalase by CosR in Campylobacter jejuni. J. Bacteriol. 2012, 194, 6883. [Google Scholar] [CrossRef]
- Monk, C.E.; Pearson, B.M.; Mulholland, F.; Smith, H.K.; Poole, R.K. Oxygen- and NssR-dependent globin expression and enhanced iron acquisition in the response of Campylobacter to nitrosative stress. J. Biol. Chem. 2008, 283, 28413–28425. [Google Scholar] [CrossRef]
- Cameron, A.; Frirdich, E.; Huynh, S.; Parker, C.T.; Gaynor, E.C. Hyperosmotic stress response of Campylobacter jejuni. J. Bacteriol. 2012, 194, 6116–6130. [Google Scholar] [CrossRef]
- Xia, Q.; Muraoka, W.T.; Shen, Z.; Sahin, O.; Wang, H.; Wu, Z.; Liu, P.; Zhang, Q. Adaptive mechanisms of Campylobacter jejuni to erythromycin treatment. BMC Microbiol. 2013, 13, 133. [Google Scholar] [CrossRef]
- Gangaiah, D.; Liu, Z.; Arcos, J.; Kassem, I.I.; Sanad, Y.; Torrelles, J.B.; Rajashekara, G. Polyphosphate kinase 2: A novel determinant of stress responses and pathogenesis in Campylobacter jejuni. PLoS ONE 2010, 5, e12142. [Google Scholar] [CrossRef]
- Lin, J.; Akiba, M.; Sahin, O.; Zhang, Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 2005, 49, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Sahin, O.; Michel, L.O.; Zhang, Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 2003, 71, 4250–4259. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Lin, E.C. The nucleotide sequence of Escherichia coli genes for L-fucose dissimilation. Nucleic Acids Res. 1989, 17, 4883–4884. [Google Scholar] [CrossRef] [PubMed]
- Bartkus, J.M.; Mortlock, R.P. Isolation of a mutation resulting in constitutive synthesis of L-fucose catabolic enzymes. J. Bacteriol. 1986, 165, 710–714. [Google Scholar] [CrossRef]
- Hooper, L.V.; Xu, J.; Falk, P.G.; Midtvedt, T.; Gordon, J.I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 1999, 96, 9833–9838. [Google Scholar] [CrossRef]
- Molina-Henares, A.J.; Krell, T.; Eugenia, G.M.; Segura, A.; Ramos, J.L. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 2006, 30, 157–186. [Google Scholar] [CrossRef]
- Krell, T.; Molina-Henares, A.J.; Ramos, J.L. The IclR family of transcriptional activators and repressors can be defined by a single profile. Protein Sci. 2006, 15, 1207–1213. [Google Scholar] [CrossRef]
- Parkhill, J.; Wren, B.W.; Mungall, K.; Ketley, J.M.; Churcher, C.; Basham, D.; Chillingworth, T.; Davies, R.M.; Feltwell, T.; Holroyd, S.; et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000, 403, 665–668. [Google Scholar] [CrossRef]
- Wang, Y.; Taylor, D.E. Chloramphenicol resistance in Campylobacter coli: Nucleotide sequence, expression, and cloning vector construction. Gene 1990, 94, 23–28. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, A.H.; Wood, A.C.; Henderson, J.; Wooldridge, K.; Ketley, J. Genetic manipulation of enteric Campylobacter species. In Methods in Microbiology; Academic Press Ltd.: Cambridge, MA, USA, 1998; pp. 407–419. [Google Scholar]
- Karlyshev, A.V.; Wren, B.W. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl. Environ. Microbiol. 2005, 71, 4004–4013. [Google Scholar] [CrossRef] [PubMed]
- Holm, S. A simple sequentially rejective Bonferroni test procedure. Scand. J. Statist. 1979, 6, 65–70. [Google Scholar]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Stowell, J.A.W.; Tang, T.T.L.; Seidel, M.; Passmore, L.A. Gel-Based Analysis of Protein-Nucleic Acid Interactions. Methods Mol. Biol. 2021, 2263, 321–339. [Google Scholar] [CrossRef]
- Gundogdu, O.; Bentley, S.D.; Holden, M.T.; Parkhill, J.; Dorrell, N.; Wren, B.W. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genom. 2007, 8, 162. [Google Scholar] [CrossRef]
- Page, W.J.; Huyer, G.; Huyer, M.; Worobec, E.A. Characterization of the porins of Campylobacter jejuni and Campylobacter coli and implications for antibiotic susceptibility. Antimicrob. Agents Chemother. 1989, 33, 297–303. [Google Scholar] [CrossRef]
- Guccione, E.; Leon-Kempis, M.R.; Pearson, B.M.; Hitchin, E.; Mulholland, F.; van Diemen, P.M.; Stevens, M.P.; Kelly, D.J. Amino acid-dependent growth of Campylobacter jejuni: Key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 2008, 69, 77–93. [Google Scholar] [CrossRef]
- Chaudhuri, R.R.; Yu, L.; Kanji, A.; Perkins, T.T.; Gardner, P.P.; Choudhary, J.; Maskell, D.J.; Grant, A.J. Quantitative RNA-seq analysis of the Campylobacter jejuni transcriptome. Microbiology 2011, 157, 2922–2932. [Google Scholar] [CrossRef]
- Reid, A.N.; Pandey, R.; Palyada, K.; Naikare, H.; Stintzi, A. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl. Environ. Microbiol. 2008, 74, 1583–1597. [Google Scholar] [CrossRef]
- Hofreuter, D.; Novik, V.; Galan, J.E. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 2008, 4, 425–433. [Google Scholar] [CrossRef]
- Fearnley, C.; Manning, G.; Bagnall, M.; Javed, M.A.; Wassenaar, T.M.; Newell, D.G. Identification of hyperinvasive Campylobacter jejuni strains isolated from poultry and human clinical sources. J. Med. Microbiol. 2008, 57, 570–580. [Google Scholar] [CrossRef]
- de Haan, C.P.; Llarena, A.K.; Revez, J.; Hanninen, M.L. Association of Campylobacter jejuni metabolic traits with multilocus sequence types. Appl. Environ. Microbiol. 2012, 78, 5550–5554. [Google Scholar] [CrossRef]
- Middendorf, P.S.; Jacobs-Reitsma, W.F.; Zomer, A.L.; den Besten, H.M.W.; Abee, T. Comparative analysis of L-fucose utilization and its impact on growth and survival of Campylobacter isolates. Front. Microbiol. 2022, 13, 872207. [Google Scholar] [CrossRef] [PubMed]
- Dedieu, L.; Pages, J.M.; Bolla, J.M. Environmental regulation of Campylobacter jejuni major outer membrane protein porin expression in Escherichia coli monitored by using green fluorescent protein. Appl. Environ. Microbiol. 2002, 68, 4209–4215. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, R.C.; Neal-McKinney, J.M.; Dhillon, A.S.; Miller, W.G.; Konkel, M.E. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 2009, 77, 2399–2407. [Google Scholar] [CrossRef]
- Jeon, B.; Zhang, Q. Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J. Antimicrob. Chemother. 2009, 63, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Matin, A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol. Microbiol. 1991, 5, 3–10. [Google Scholar] [CrossRef]
- Dubey, A.K.; Baker, C.S.; Suzuki, K.; Jones, A.D.; Pandit, P.; Romeo, T.; Babitzke, P. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 2003, 185, 4450–4460. [Google Scholar] [CrossRef]
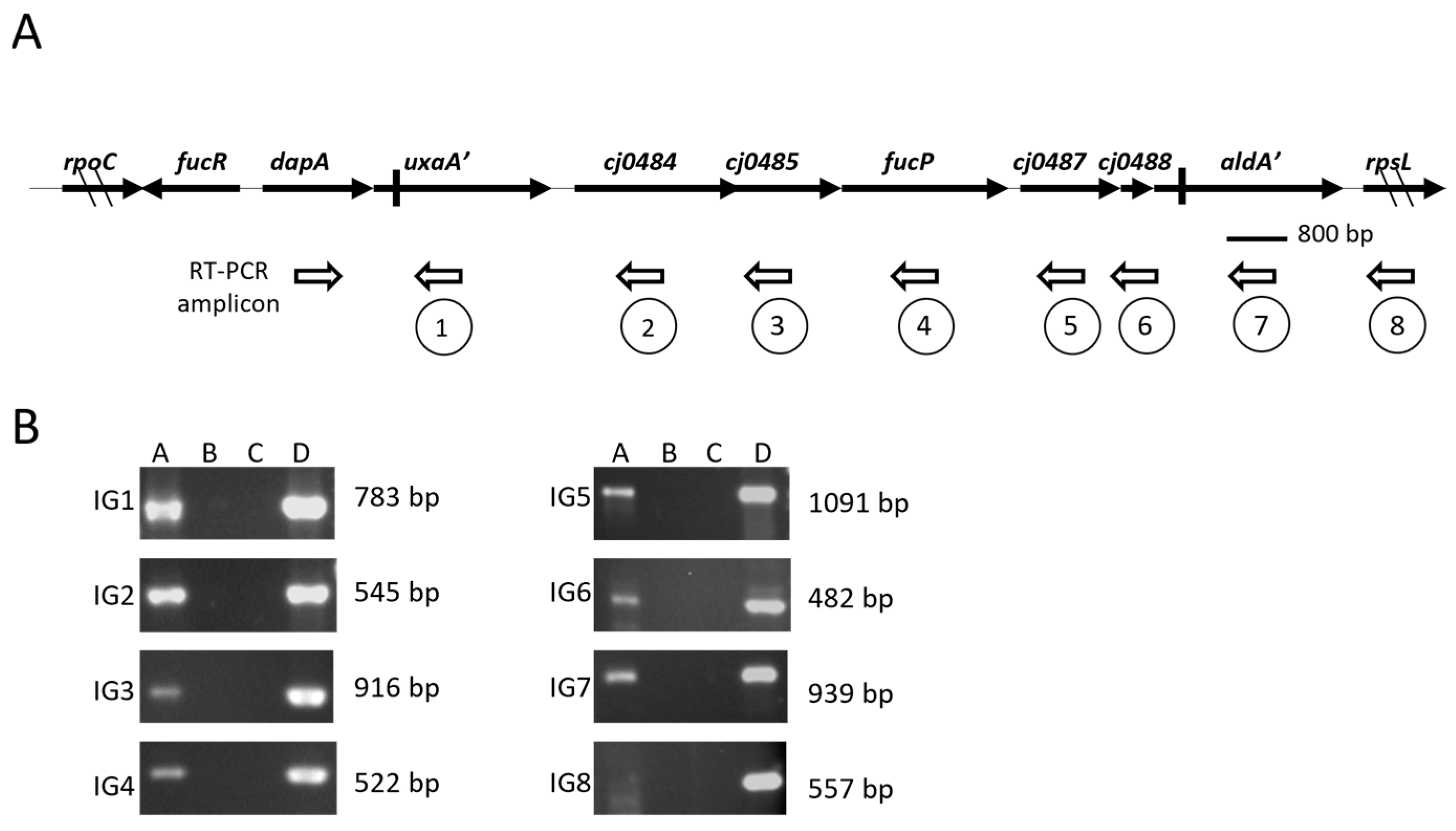

| Bacterial Strain or Plasmid | Description or Relevant Genotype | Source or Reference |
|---|---|---|
| E. coli strain | ||
| DH5α Origami B(DE3) | Plasmid propagation strain Protein expression strain | Invitrogen Sigma-Aldrich |
| C. jejuni strains | ||
| NCTC 11168 | Wild-type C. jejuni | [34] |
| CjWM116a | NCTC 11168 derivative, Δcj0480c::cat; Cmr | This study |
| CjWM233b | CjWM116a derivative, Δcj0480c::cat, ITS1::cj0480c; Cmr, Kanr | This study |
| Plasmids | ||
| pGEM-T | Cloning vector | Promega |
| pUOA18 pTrcHis | E. coli-C. jejuni shuttle vector E. coli expression vector | [35] ThermoFisher |
| pcj0480c | pGEM-T::cj0480c | This study |
| pWM15 | ΔfucP::cat; Cmr suicide vector to construct CjWM114a | This study |
| pRRK | pRR::aphA3 | [9] |
| pRRKWM14 pTrc-cj0480c | pRRK::cj0480c pTrc::cj0480c-6xHis | This study This study |
| Primer Name | Primer Sequence (5′ → 3′) |
|---|---|
| Primers to construct mutant and complement strains | |
| cj0480c_L | CGCTCAAAGCTTGAGAATCC |
| cj0480c_R | GTTTATCGCGGACAAGGTGT |
| icj0480c_L | CGCGGATCCCCAAAGAGTTCCAGCAGGAA |
| icj0480c_R | CGCGGATCCTCCTCAAATTGAATGTATGGCT |
| KIcj0480c_L | CGCCCTAGGAAGCTGTTAACTTGGTAAAAATTCG |
| KIcj0480c_R | CGCCCTAGGAGCCCTTCGGGGCTATATTA |
| cj0480cBamHI | GGATCGATGGGGATCCATGCATCAGCCCAC |
| cj0480cEcoRI | GCCAAGCTTCGAATTCTTAATACAGTGTATCTAAATC |
| fucRpro_L | GCTGATGCATATTGTCTTTATCC |
| fucRpro_R | GTAAGTAAAGCCGGTAAAGTTCC |
| dapAintl_L | ATGAAAAAGAATTTATTCGC |
| dapAintl_R | ACGCTCAAAGCTTG |
| Primers to amplify intergenic sequences (IG) | |
| IG1_L | GCATAGGTGGAGTTTTTCCAG |
| IG1_R | AAAAACATTGGCATTGCTCC |
| IG2_L | TAGCCCAGGAAATATGGCAG |
| IG2_R | AAAGGCAAAATACGCCAAGA |
| IG3_L | TCGCCTTGCCAATATTTACC |
| IG3_R | CTTAGCAAAAGCACAAGCCC |
| IG4_L | TGACCAAAGAATGTTTGCCTT |
| IG4_R | AAGTCCATAGCTTACTCCCCA |
| IG5_L | GGCTTTTAGCGCAGTTTTTG |
| IG5_R | TGCGAAAAACCATCAGGAAT |
| IG6_L | CAGACATGGCTAAAATGGCA |
| IG6_R | TTCGGCCGTATAATCCATATAA |
| IG7_L | GACGATGAAAAATTAGAGCA |
| IG7_R | TTTCCACCATTTTTGTGTGC |
| IG8_L | CAAGGTTTTCATGCAGGCTT |
| IG8_R | ACCCTAGTGCAAACTCCCCT |
| Primers for RT-PCR | |
| qfucP_L | GGCTTTTAGCGCAGTTTTTG |
| qfucP_RT | CAATGCGCCCTAGCATAAAT |
| cjr01_L | TCCCAGTTCGGATTGTTCTC |
| cjr01_RT | GTACAAGACCCGGGAACGTA |
| pdapA_R | GCAAAGCAAAACCCATAGGA |
| tdapA_L | GCATAGGTGGAGTTTTTCGAG |
| Primers to confirm microarray | |
| cj0917c_F | GCGTTTTATCCGTCCAGGTA |
| cj0917c_R | TTGCCAAAGTTGGAGCTTCT |
| cj0927_F | GCAGGAACAGAAAGCAGAGG |
| cj0927_R | TCAAAAGGGAGTTTTCCAGG |
| cj1548c_F | AGAAGGCTCAAGCGTAGCAG |
| cj1548c_R | ACACCCATAGCCAAAGCATC |
| cj0762c_F | AAGCCCTAATTTCAGCCGTT |
| cj0762c_R | GTTTTTCAAAGGCTTGACGC |
| cj0087_F | TGGGGAATTGGAAATCTCTG |
| cj0087_R | CAAAGCCCTAACAAAGCGAG |
| cj0437_F | AATTGGATCAGGTGGAGCAG |
| cj0437_R | CCACCCTCTGCCATACAAGT |
| cj1604_F | CAAGATGCAAAAACTTGCGA |
| cj1604_R | CCTTCATCCAAAGACGCTGT |
| cj0169_F | TTCAAATGGGGGCGTATTTA |
| cj0169_R | TACTTTGACATGAACCGCCA |
| cj0933c_F | CAAAACACGGTACCACAACG |
| cj0933c_R | CAATTTGGCGGTAGATCCAT |
| cj1259_F | GCAGAGCAAGGTGCAGATTT |
| cj1259_R | AGCAGCAGCACCGTAAAGAT |
| cj1339c_F | GCAGGCTCAGGTTTTTCAAG |
| cj1339c_R | CGGCTGCAAAGTCTACATCA |
| Wild-Type + Fucose | fucR Mutant | ||||
|---|---|---|---|---|---|
| Gene ¶ | Function | Microarray log2 Fold-Change (q-Value) * | qRT-PCR log2 Fold-Change (p-Value) * | Microarray log2 Fold-Change (q-Value) * | qRT-PCR log2 Fold-Change (p-Value) * |
| dapA | putative dihydrodipicolinate synthase | 3.799 (1.75 × 10−10) | 3.781 (1.28 × 10−7) | ||
| cj0488 | conserved hypothetical protein Cj0488 | 3.673 (4.58 × 10−8) | 3.600 (1.28 × 10−7) | ||
| uxaA’ | putative altronate hydrolase C-terminus | 3.644 (4.09 × 10−10) | 3.892 (2.06 × 10−6) | ||
| cj0487 | putative amidohydrolase | 3.638 (1.28 × 10−8) | 3.846 (1.83 × 10−7) | ||
| ald’ | putative aldehyde dehydrogenase N-terminus | 3.629 (1.75 × 10−10) | 3.664 (8.21 × 10−7) | ||
| uxaA’ | putative altronate hydrolase N-terminus | 3.573 (3.11 × 10−8) | 3.184 (2.06 × 10−6) | ||
| cj0486 | putative sugar transporter | 3.387 (5.18 × 10−8) | 3.606 (2.06 × 10−6) | ||
| cj0485 | short chain dehydrogenase | 3.339 (1.75 × 10−10) | 3.254 (4.63 × 10−6) | ||
| ald’ | putative aldehyde dehydrogenase C-terminus | 2.581 (7.66 × 10−6) | 2.593 (1.83 × 10−7) | ||
| cj0484 | putative MFS (Major Facilitator Superfamily) transport protein | 1.847 (4.48 × 10−4) | 3.961 (0.001) | 2.039 (5.23 × 10−7) | 5.388 (0.000) |
| cj1064 | pseudo | 1.408 (3.36 × 10−5) | 1.551 (2.34 × 10−3) | ||
| aspB | aspartate aminotransferase | 1.347 (1.05 × 10−5) | −0.089 (0.620) | 1.271 (3.59 × 10−2) | −0.046 (0.822) |
| porA | major outer membrane protein | 0.769 (2.39 × 10−4) | −0.052 (0.553) | 0.784 (9.61 × 10−3) | −0.069 (0.473) |
| rrc | non-haem iron protein | 0.701 (3.21 × 10−3) | |||
| aspA | aspartate ammonia-lyase | 0.637 (3.73 × 10−3) | 0.298 (0.324) | 0.067 (0.649) | |
| rplF | 50S ribosomal protein L6 | 0.611 (1.22 × 10−2) | |||
| sdhA | succinate dehydrogenase flavoprotein subunit | 0.587 (3.21 × 10−3) | 0.466 (0.233) | 0.636 (0.077) | |
| rplE | 50S ribosomal protein L5 | 0.552 (3.11 × 10−2) | |||
| rplX | 50S ribosomal protein L24 | 0.535 (2.90 × 10−2) | |||
| cj1534c | putative bacterioferritin | 0.533 (1.66 × 10−2) | |||
| sdhB | putative succinate dehydrogenase iron-sulfur protein | 0.524 (2.90 × 10−2) | |||
| ndk | nucleoside diphosphate kinase | 0.486 (2.92 × 10−2) | |||
| rpsF | 30S ribosomal protein S6 | 0.476 (2.78 × 10−2) | |||
| cj0416 | hypothetical protein | −0.427 (4.01 × 10−2) | |||
| putA | putative proline dehydrogenase | −0.463 (3.05 × 10−2) | |||
| prsA | ribose-phosphate pyrophosphokinase | −0.472 (3.40 × 10−2) | |||
| cstA | putative integral membrane protein (CstA homolog) | −0.473 (3.29 × 10−2) | −0.199 (0.003) | −0.040 (0.391) | |
| cj1111c | putative MarC family integral membrane protein | −0.520 (4.77 × 10−2) | |||
| cj1548c | putative NADP-dependent alcohol dehydrogenase | −0.523 (2.78 × 10−2) | −0.319 (0.034) | ||
| aptA | adenine phosphoribosyltransferase | −0.821 (8.81 × 10−3) | −0.028 (0.837) | −0.423 (0.08) | |
| glnA | glutamine synthetase | −0.956 (3.33 × 10−2) | |||
| cj0037c | putative cytochrome C | −0.532 (8.82 × 10−3) | |||
| rpmF | 50S ribosomal protein L32 | 0.495 (1.28 × 10−2) | |||
| flgH | flagellar basal body L-ring protein | −1.07 (2.46 × 10−2) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muraoka, W.T.; Lizer, N.; Liu, P.; Shen, Z.; Xia, Q.; Ilgu, M.; Zhang, Q. FucR Functions as a Transcriptional Regulator for L-Fucose Utilization in Campylobacter jejuni. Microorganisms 2025, 13, 2364. https://doi.org/10.3390/microorganisms13102364
Muraoka WT, Lizer N, Liu P, Shen Z, Xia Q, Ilgu M, Zhang Q. FucR Functions as a Transcriptional Regulator for L-Fucose Utilization in Campylobacter jejuni. Microorganisms. 2025; 13(10):2364. https://doi.org/10.3390/microorganisms13102364
Chicago/Turabian StyleMuraoka, Wayne T., Nicholas Lizer, Peng Liu, Zhangqi Shen, Qingqing Xia, Muslum Ilgu, and Qijing Zhang. 2025. "FucR Functions as a Transcriptional Regulator for L-Fucose Utilization in Campylobacter jejuni" Microorganisms 13, no. 10: 2364. https://doi.org/10.3390/microorganisms13102364
APA StyleMuraoka, W. T., Lizer, N., Liu, P., Shen, Z., Xia, Q., Ilgu, M., & Zhang, Q. (2025). FucR Functions as a Transcriptional Regulator for L-Fucose Utilization in Campylobacter jejuni. Microorganisms, 13(10), 2364. https://doi.org/10.3390/microorganisms13102364






