Integrated Pathogen–Host Analysis of Citrobacter braakii SCGY-1L: Genomic Determinants and Host Transcriptional Dynamics During Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation
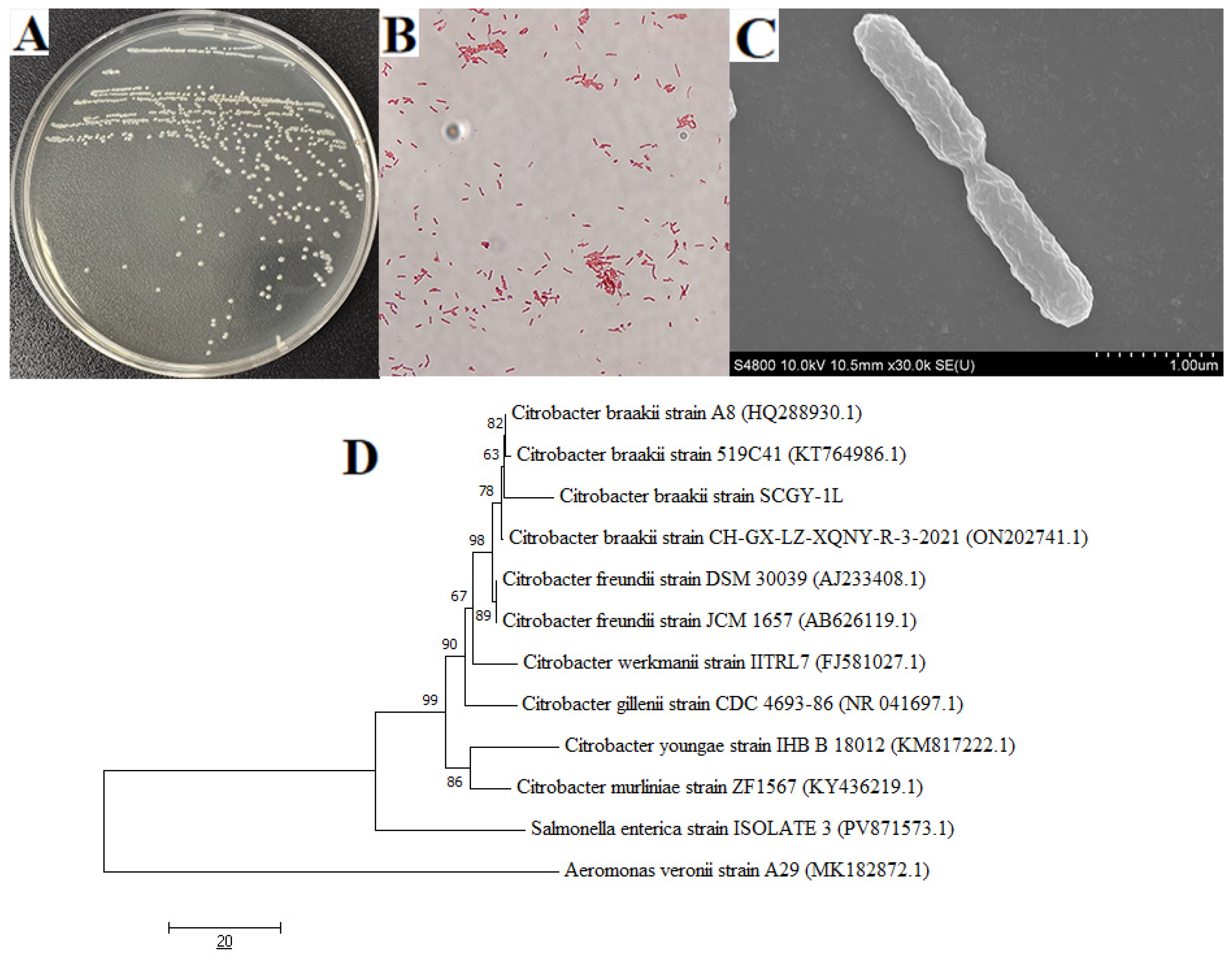
2.2. Bacterial Identification and Phylogenetic Analysis
2.3. Physiological and Biochemical Characteristics, and Antibiotic Susceptibility
2.4. Bacterial Challenge and Histopathological Analysis
2.5. Bacterial Library Construction and Genome Sequencing
2.6. Genome Assembly and Gene Annotation
2.7. Construction of Genome Circle Diagrams and Gene Functional Analysis
2.8. Transcriptomic Analysis of Hosts Infected by Bacteria
2.9. Functional Annotation of Differentially Expressed Genes
2.10. Real-Time Quantitative PCR
3. Results
3.1. Isolation and Identification of C. braakii SCGY-1L Strain
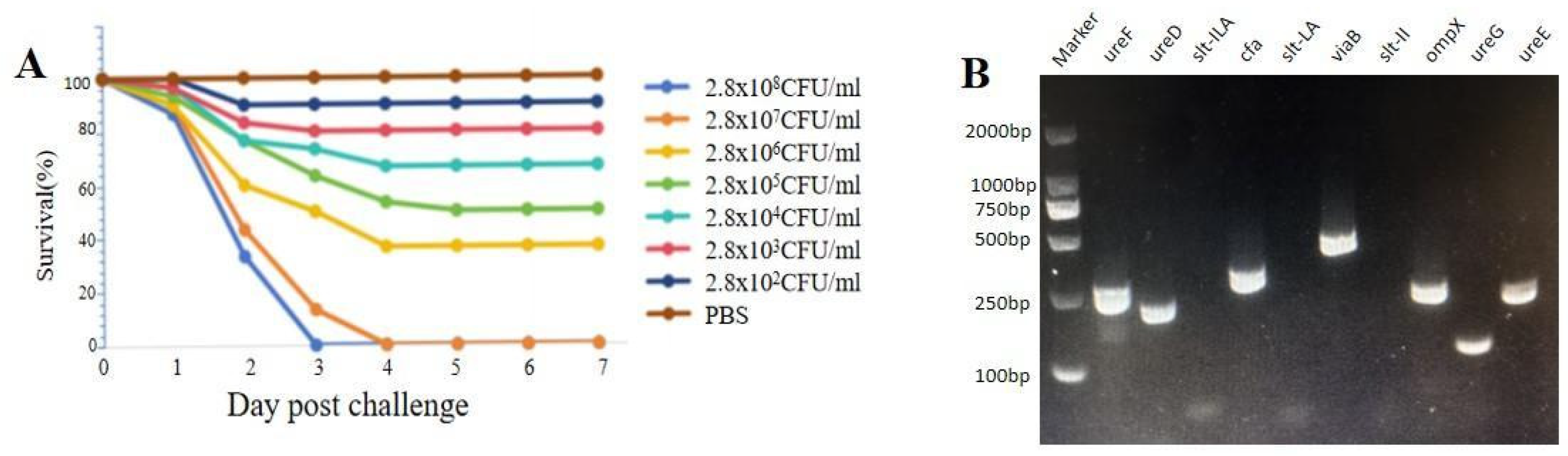
3.2. Pathogenicity Analysis and Antibiotic Susceptibility Test
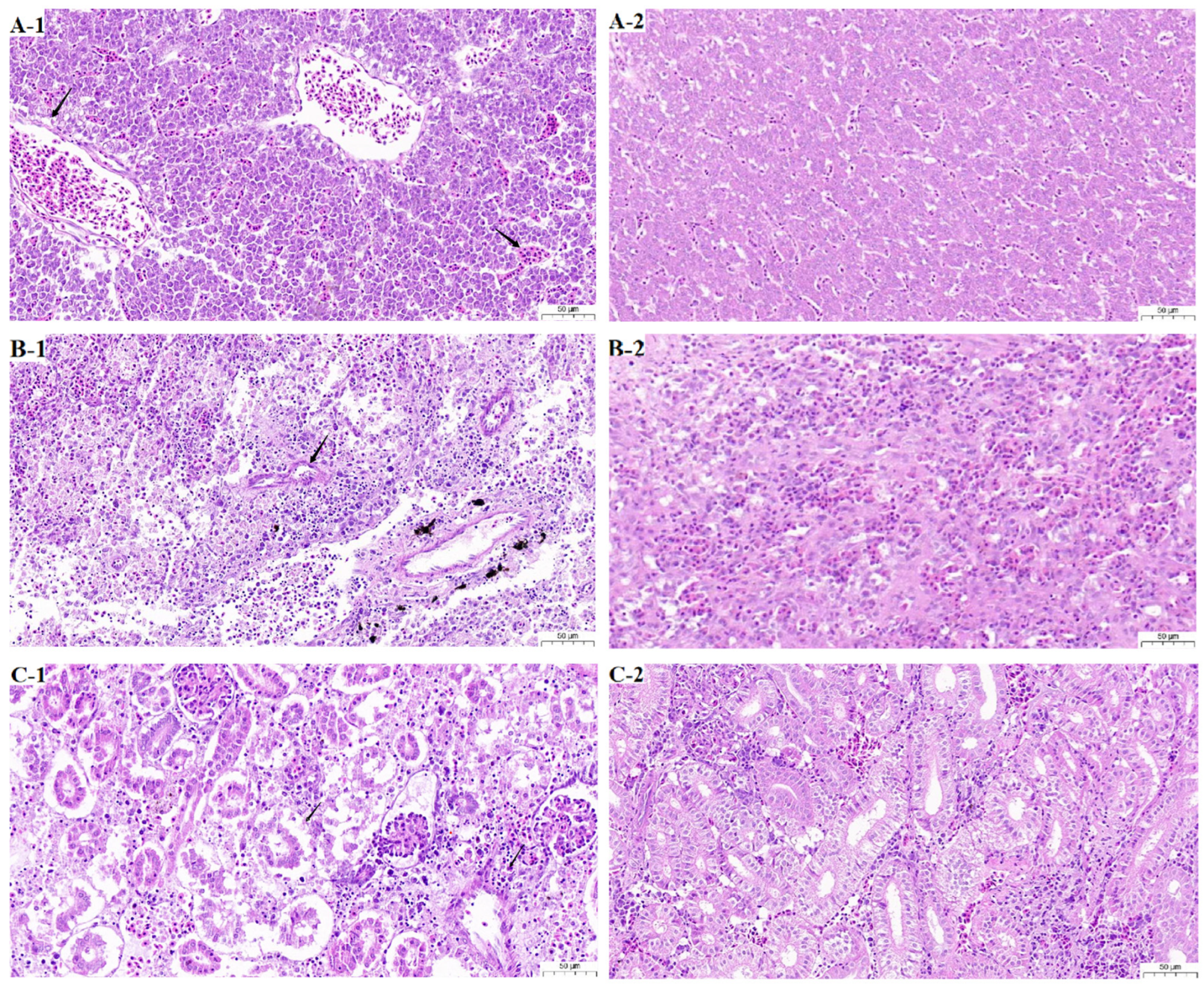
3.3. Histopathology
3.4. Data Statistics and Genome Assessment
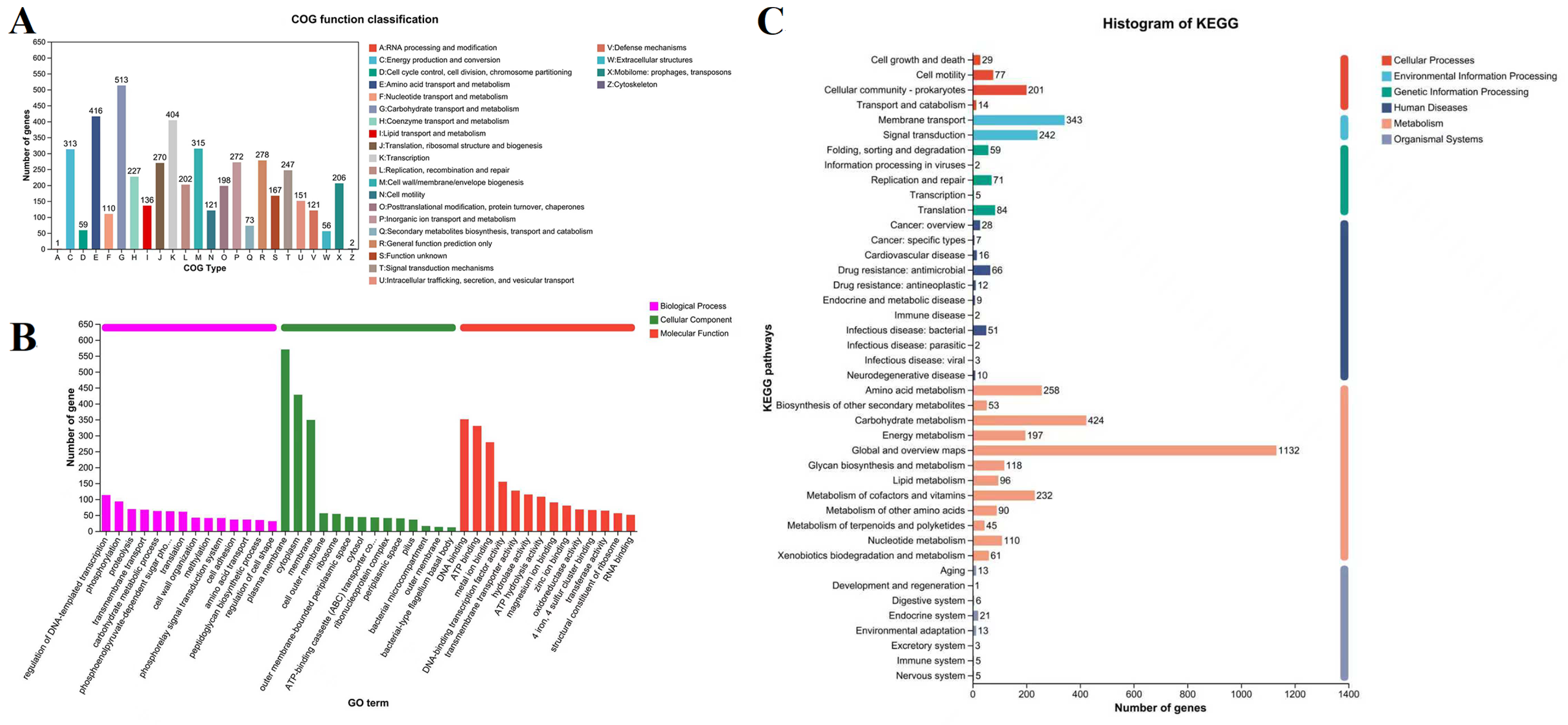
3.5. Bacterial Gene Annotation
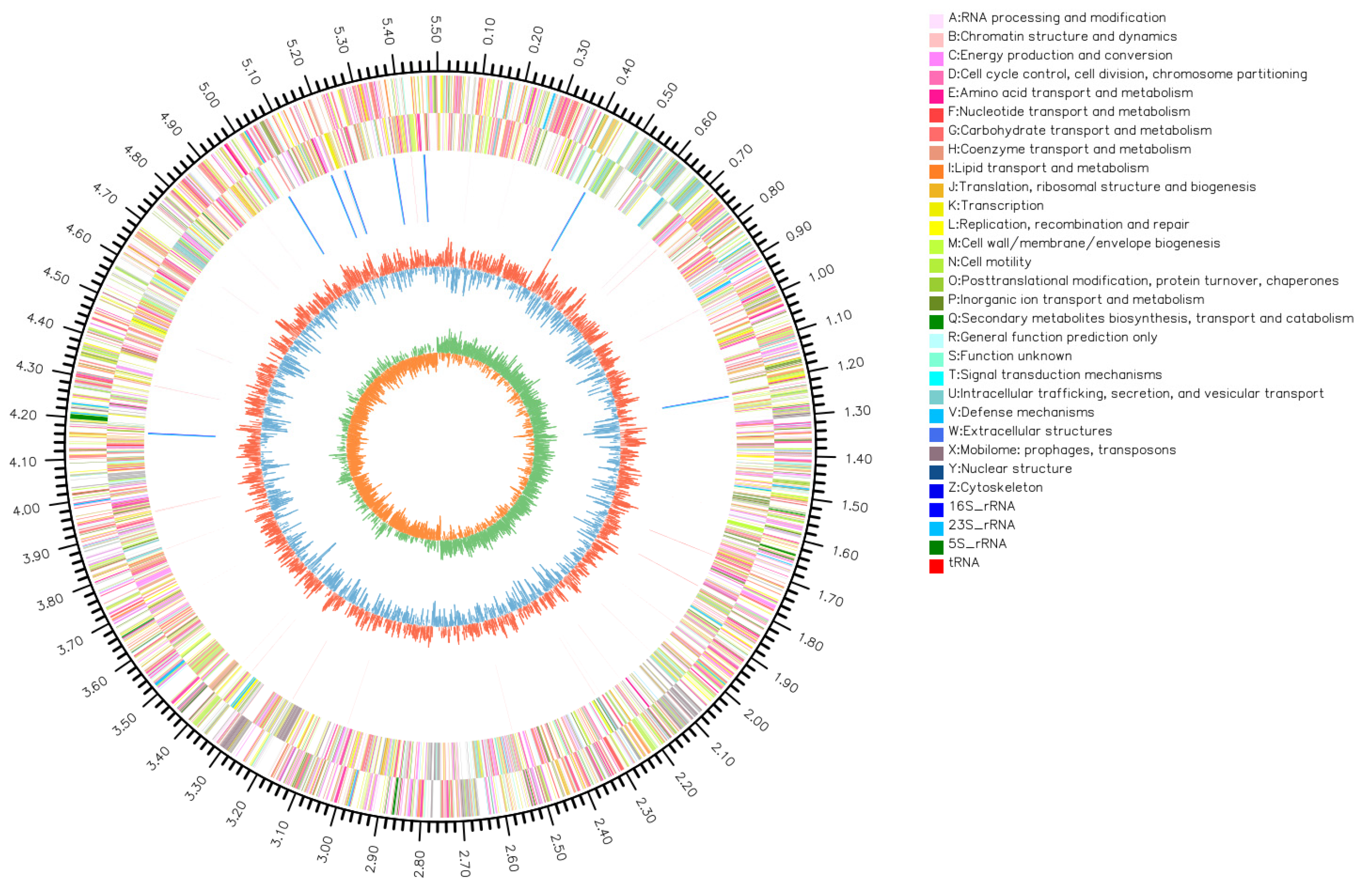
3.6. Bacterial Genomic Circos Plot Analysis
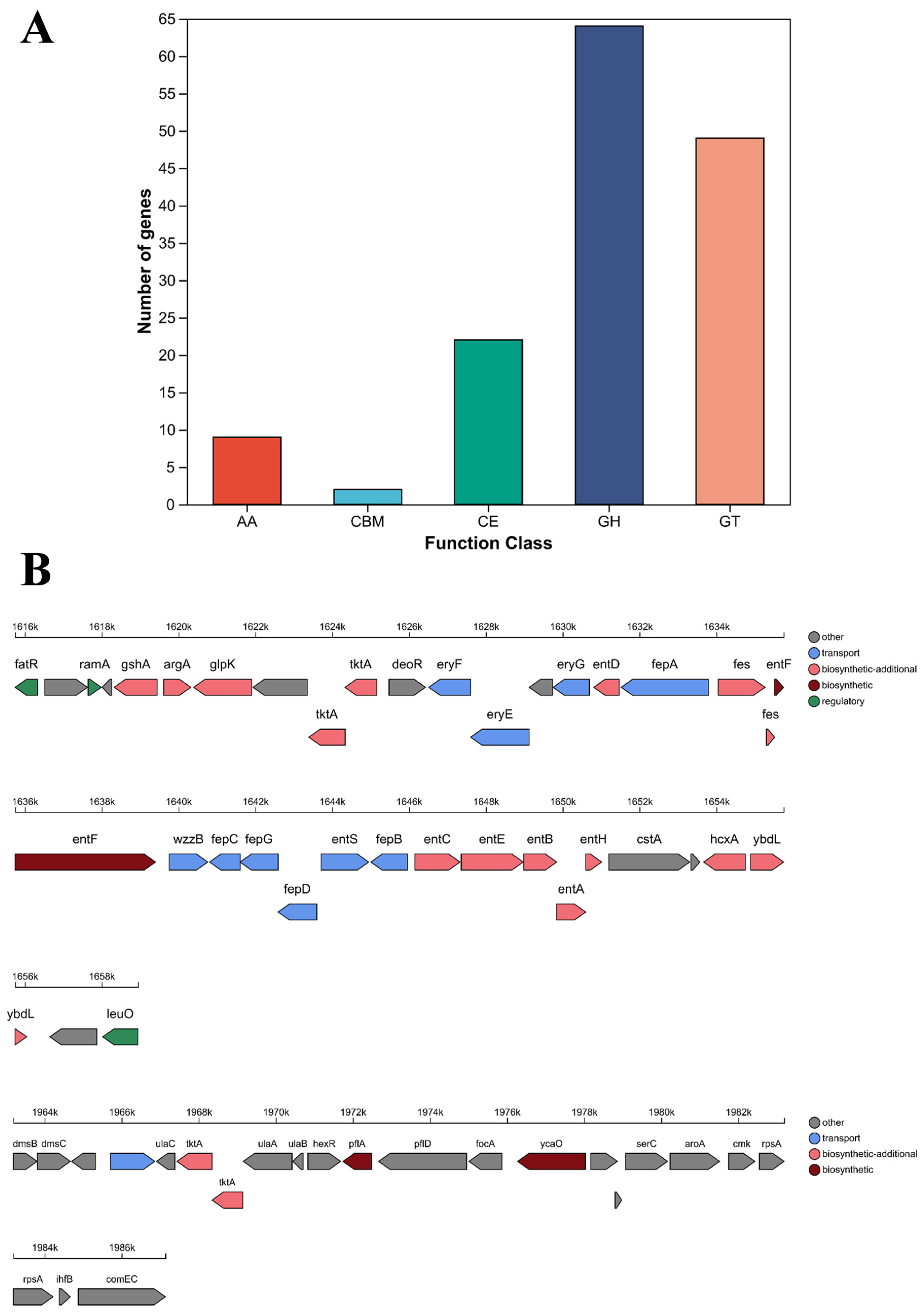
3.7. Carbohydrate-Active Enzymes Analysis
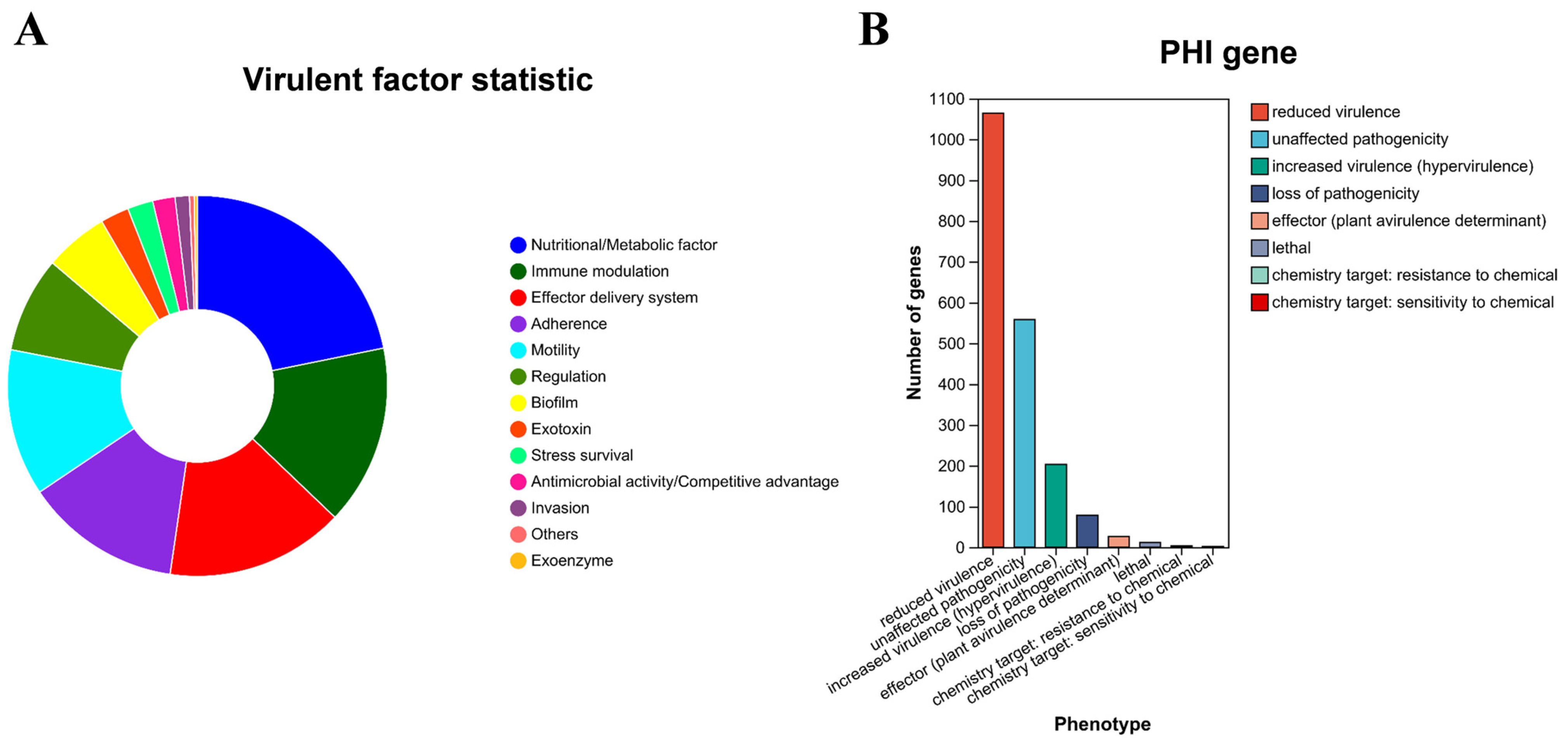
3.8. Pathogenicity-Associated Gene Analysis
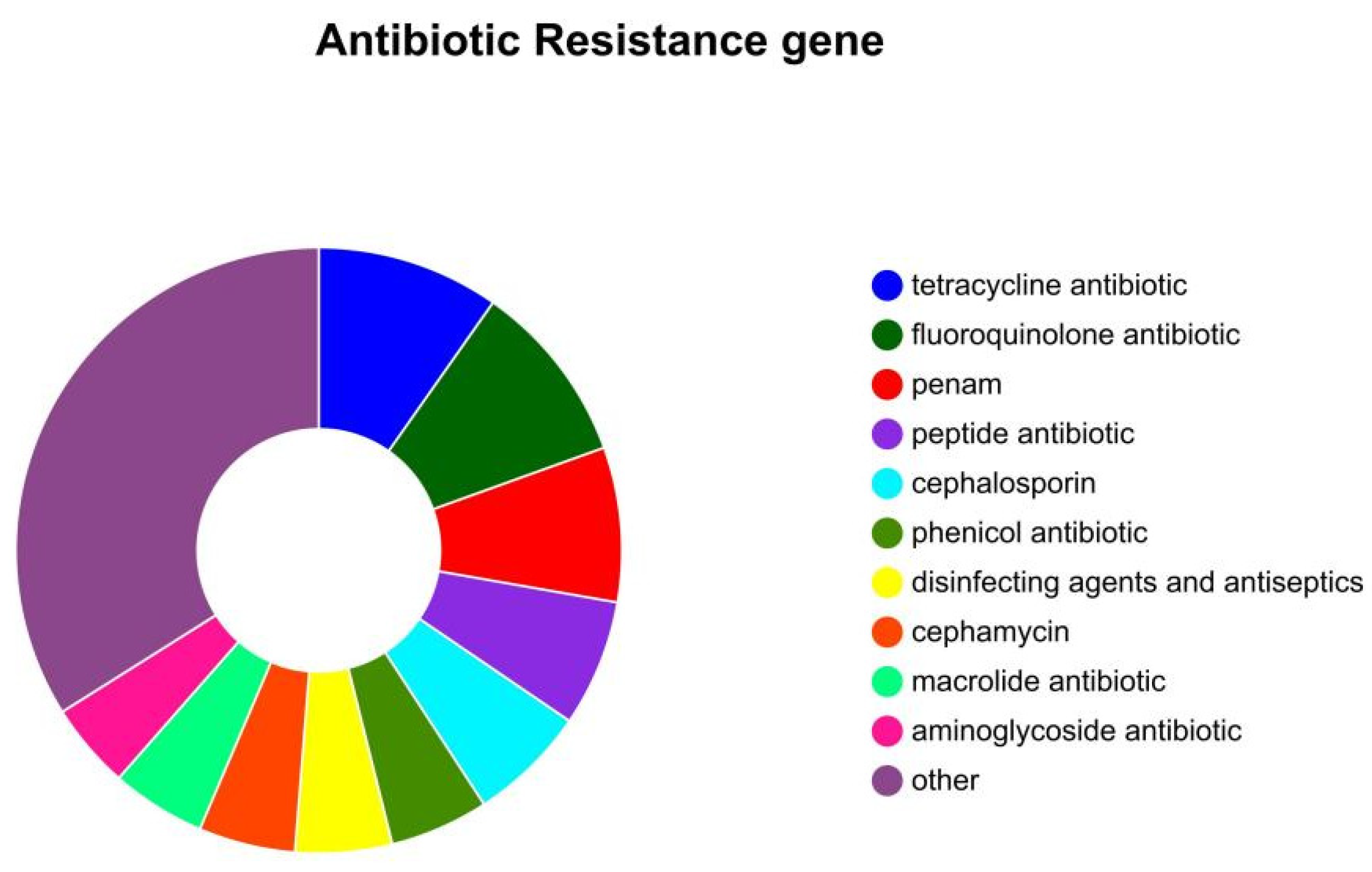
3.9. Antibiotic Resistance Gene Analysis
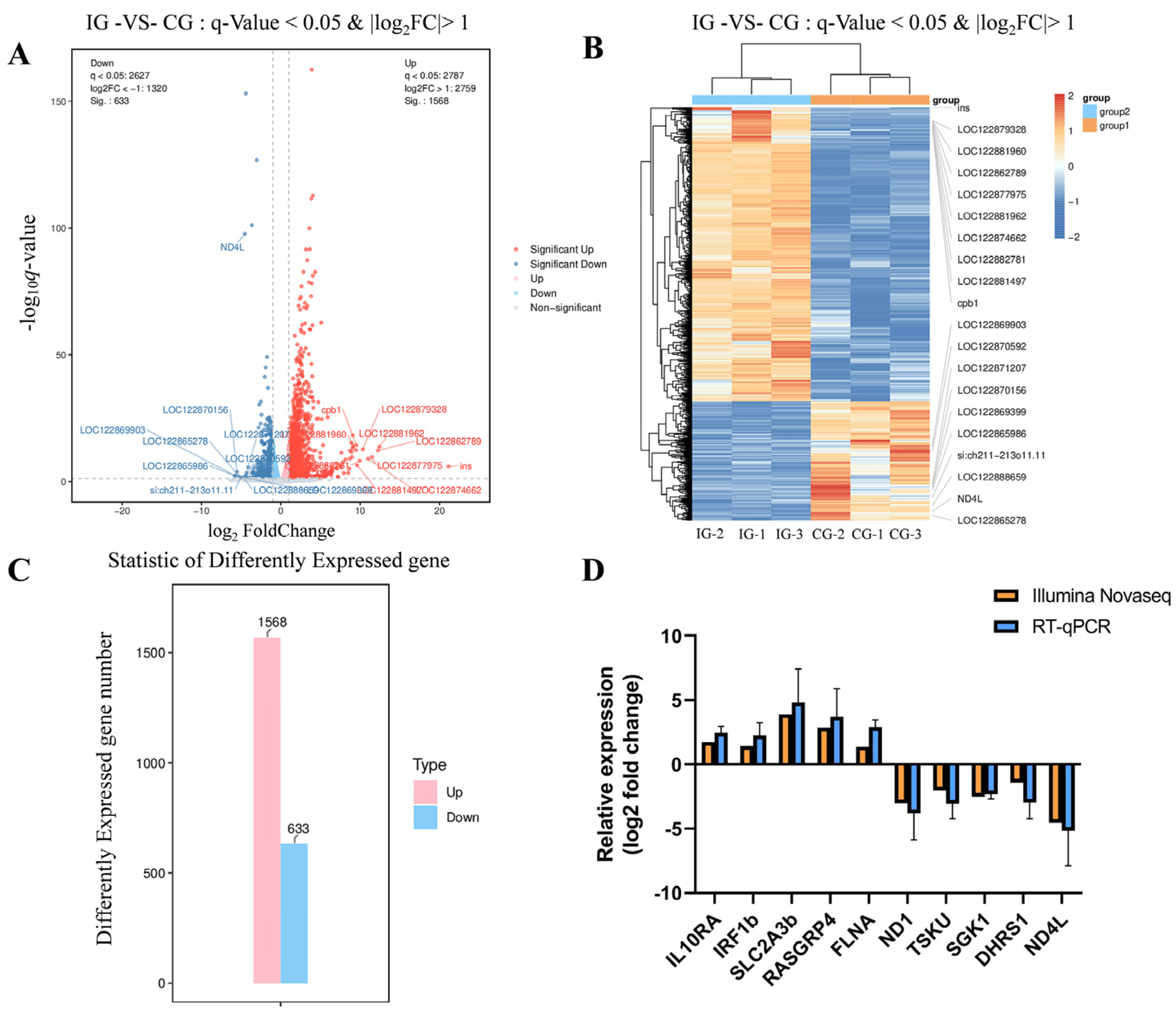
3.10. Transcriptome Sequencing Data Statistics and Identification of Differentially Expressed Genes
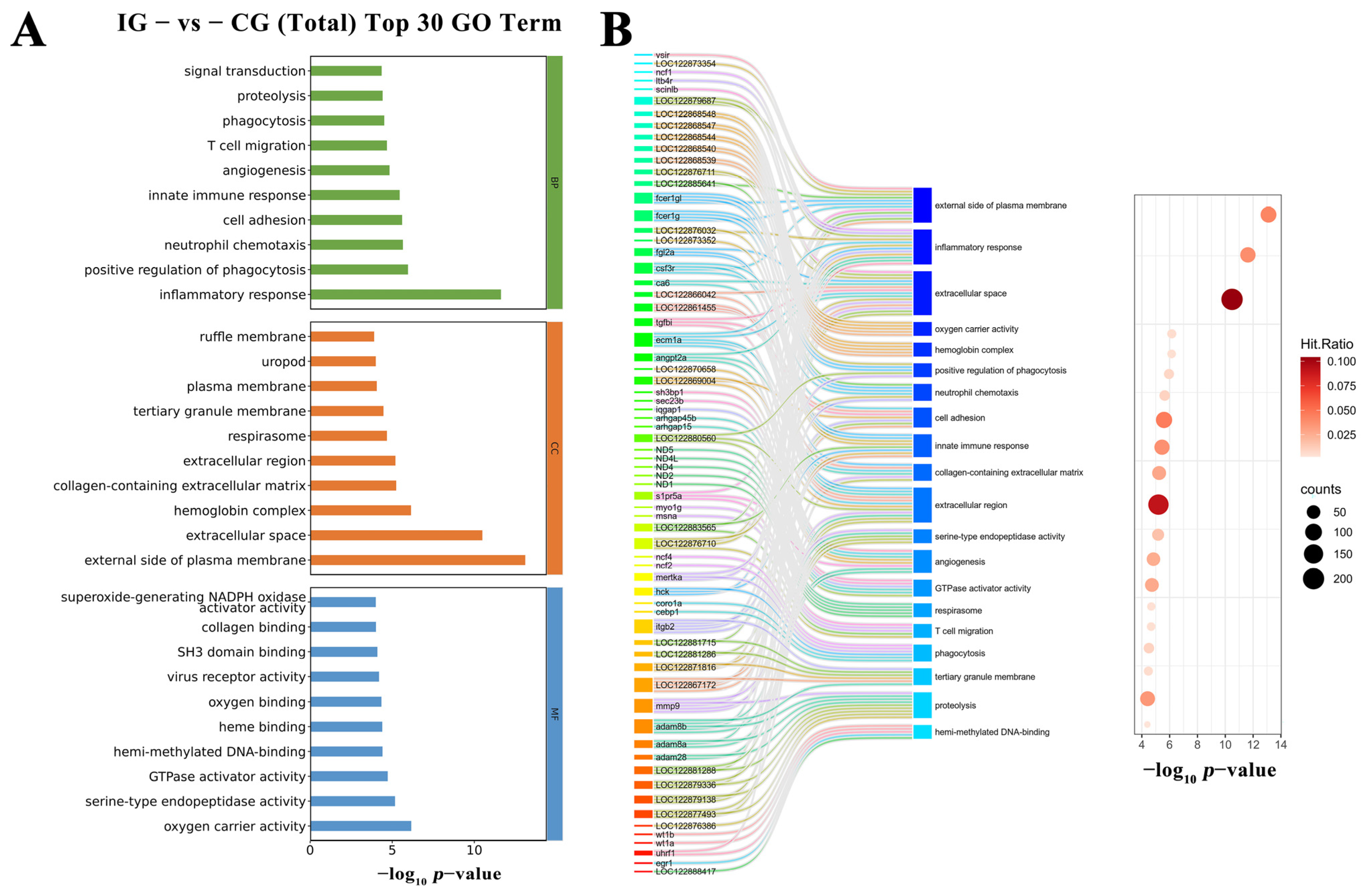
3.11. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yao, Y.; Falgenhauer, L.; Falgenhauer, J.; Hauri, A.M.; Heinmüller, P.; Domann, E.; Chakraborty, T.; Imirzalioglu, C. Carbapenem-Resistant Citrobacter spp. as an Emerging Concern in the Hospital-Setting: Results from a Genome-Based Regional Surveillance Study. Front. Cell Infect. Microbiol. 2021, 11, 744431. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, L.; Zhou, H.; Yuan, M.; Hu, D.; Wang, Y.; Sun, H.; Xu, J.; Lan, R. Antimicrobial Resistance and Molecular Characterization of Citrobacter spp. Causing Extraintestinal Infections. Front. Cell Infect. Microbiol. 2021, 11, 737636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qiao, D.; Wang, H.; Zhao, X.; Jiang, X.; Zhu, L.; Zhang, J.; Li, L.; Kong, X.; Pei, C. Mixed Infection in Common Carp (Cyprinus carpio) Caused by Aeromonas veronii, Aeromonas hydrophila, Plesiomonas shigelloides, and Citrobacter freundii. Animals 2025, 15, 805. [Google Scholar] [CrossRef] [PubMed]
- Pastuszka, A.; Guz, L.; Puk, K.; Pietras-Ożga, D. Occurrence of virulence factors and antimicrobial susceptibility of Citrobacter freundii isolated from diseased ornamental fish in Poland. J. Vet. Res. 2025, 69, 17–26. [Google Scholar] [CrossRef]
- Nawaz, M.; Khan, A.A.; Khan, S.; Sung, K.; Steele, R. Isolation and characterization of tetracycline-resistant Citrobacter spp. from catfish. Food Microbiol. 2008, 25, 85–91. [Google Scholar] [CrossRef]
- Vega-Manriquez, D.X.; Davila-Arrellano, R.P.; Eslava-Campos, C.A.; Salazar Jimenez, E.; Negrete-Philippe, A.C.; Raigoza-Figueras, R.; Muñoz-Tenería, F.A. Identification of bacteria present in ulcerative stomatitis lesions of captive sea turtles Chelonia mydas. Vet. Res. Commun. 2018, 42, 251–254. [Google Scholar] [CrossRef]
- Huang, C.; Feng, C.; Liu, X.; Zhao, R.; Wang, Z.; Xi, H.; Ou, H.; Han, W.; Guo, Z.; Gu, J.; et al. The Bacteriophage vB_CbrM_HP1 Protects Crucian Carp Against Citrobacter braakii Infection. Front. Vet. Sci. 2022, 9, 888561. [Google Scholar] [CrossRef]
- Pasquali, F.; Crippa, C.; Lucchi, A.; Francati, S.; Dindo, M.L.; Manfreda, G. Citrobacter braakii Isolated from Salami and Soft Cheese: An Emerging Food Safety Hazard? Foods 2025, 14, 1887. [Google Scholar] [CrossRef]
- Dong, X.; Lv, M.; Zeng, M.; Chen, X.; Wang, J.; Liang, X.F. Genome-Wide Identification, Characterization of the ORA (Olfactory Receptor Class A) Gene Family, and Potential Roles in Bile Acid and Pheromone Recognition in Mandarin Fish (Siniperca chuatsi). Cells 2025, 14, 189. [Google Scholar] [CrossRef]
- Zhu, C.; Li, D.; Chen, W.; Ban, S.; Liu, T.; Wen, H.; Jiang, M. Effects of dietary host-associated Lactococcus lactis on growth performance, disease resistance, intestinal morphology and intestinal microbiota of mandarin fish (Siniperca chuatsi). Aquaculture 2021, 540, 736702. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, X.; Wu, H.; Gao, J.; Wu, J.; Xiong, Z.; Feng, Z.; Xie, M.; Li, S.; Xie, Z.; et al. The effect of short-term artificial feed domestication on the expression of oxidative-stress-related genes and antioxidant capacity in the liver and Gill tissues of Mandarin Fish (Siniperca chuatsi). Genes 2024, 15, 487. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, D.; Dong, J.; Yan, Y.; Liu, S.; Ye, X.; He, J.; Sun, C. Genomic Characterization and Pathogenicity of a Novel Birnavirus Strain Isolated from Mandarin Fish (Siniperca chuatsi). Genes 2025, 16, 629. [Google Scholar] [CrossRef]
- Zhou, D.Y.; Chen, J.M.; Sun, Z.N.; Zhang, H.Q.; Yuan, M.Z.; Wang, X.B. Transcriptional landscape of brain adaptation to artificial feed domestication in mandarin fish (Siniperca chuatsi) revealed by weighted gene co-expression network analysis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 56, 101543. [Google Scholar] [CrossRef]
- He, S.; You, J.J.; Liang, X.F.; Zhang, Z.L.; Zhang, Y.P. Transcriptome sequencing and metabolome analysis of food habits domestication from live prey fish to artificial diets in mandarin fish (Siniperca chuatsi). BMC Genom. 2021, 22, 129. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Hao, Y.; Zhang, L.; Gao, X.; Xu, Y.; Wang, J.J.; Hanafiah, F.; Khor, W.; Sun, Y.; Wu, C. Profiling the gut structure and microbiota, and identifying two dominant bacteria belonging to the weissella genus in mandarin fish (Siniperca chuatsi) fed an artificial diet. Front. Microbiol. 2024, 15, 1486501. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.L.; Li, L.; Miao, Y.L.; Liang, H.; Zou, J.M.; You, J.J.; Liang, X.F.; He, S. Effects and regulatory pathway of proopinmelanocortin on feeding habit domestication in mandarin fish. Gene 2023, 878, 147581. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.Y.; Ding, Z.L.; Zhou, A.N.; Zhu, C.B.; Yang, S.; Fei, H. Bacterial diseases in Siniperca chuatsi: Status and therapeutic strategies. Vet. Res. Commun. 2024, 48, 3579–3592. [Google Scholar] [CrossRef]
- Rossi, F.; Santonicola, S.; Amadoro, C.; Marino, L.; Colavita, G. Recent Records on Bacterial Opportunistic Infections via the Dietary Route. Microorganisms 2023, 12, 69. [Google Scholar] [CrossRef]
- He, X.; Wu, J.; Tan, X.; Xu, S.; Kong, W.; Liu, X. Development of Duplex Loop-Mediated Isothermal Amplification with Hydroxynaphthol Blue for Detection of Infectious Spleen and Kidney Necrosis Virus and Aeromonas hydrophila in Chinese Perch (Siniperca chuatsi). Microorganisms 2025, 13, 586. [Google Scholar] [CrossRef]
- Gao, J.H.; Zhao, J.L.; Yao, X.L.; Tola, T.; Zheng, J.; Xue, W.B.; Wang, D.W.; Xing, Y. Identification of antimicrobial peptide genes from transcriptomes in Mandarin fish (Siniperca chuatsi) and their response to infection with Aeromonas hydrophila. Fish Shellfish Immunol. 2024, 144, 109247. [Google Scholar] [CrossRef]
- Zhou, W.D.; Zhang, Y.L.; Wen, Y.; Ji, W.; Zhou, Y.; Ji, Y.C.; Liu, X.L.; Wang, W.M.; Asim, M.; Liang, X.F.; et al. Analysis of the transcriptomic profilings of Mandarin fish (Siniperca chuatsi) infected with Flavobacterium columnare with an emphasis on immune responses. Fish Shellfish Immunol. 2015, 43, 111–119. [Google Scholar] [CrossRef]
- Tian, J.Y.; Qi, Z.T.; Wu, N.; Chang, M.X.; Nie, P. Complementary DNA sequences of the constant regions of T-cell antigen receptors α, β and γ in mandarin fish, Siniperca chuatsi Basilewsky, and their transcriptional changes after stimulation with Flavobacterium columnare. J. Fish Dis. 2014, 37, 89–101. [Google Scholar] [CrossRef]
- Wu, Y.L.; Miao, P.F.; Yu, H.; Tan, S.W.; Yang, H.; Peng, Z.Q.; Yang, Y. Isolation, identification and drug susceptibility test of pathogenic Edwardsiella tarda in Siniperca chuatsi. J. South. Agric. 2018, 49, 794–799. [Google Scholar] [CrossRef]
- Zhu, C.B.; Shen, Y.T.; Ren, C.H.; Yang, S.; Fei, H. A novel formula of herbal extracts regulates growth performance, antioxidant capacity, intestinal microbiota and resistance against Aeromonas veronii in largemouth bass (Micropterus salmoides). Aquaculture 2024, 583, 740614. [Google Scholar] [CrossRef]
- Luo, X.; Fu, X.Z.; Liao, G.L.; Chang, O.Q.; Huang, Z.B.; Li, N.Q. Isolation, pathogenicity and characterization of a novel bacterial pathogen Streptococcus uberis from diseased mandarin fish Siniperca chuatsi. Microb. Pathog. 2017, 107, 380–389. [Google Scholar] [CrossRef]
- Weisburg, W.; Barns, S.; Pelletier, D.; Lane, D. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.; Vignaud, M.L.; Cadel-Six, S. Whole-Genome Sequences of Two Salmonella enterica Serovar Dublin Strains That Harbor the viaA, viaB, and ompB Loci of the Vi Antigen. Microbiol. Resour. Announc. 2019, 8, e00028-19. [Google Scholar] [CrossRef] [PubMed]
- Avijit, R.; Amer, F.; Barthe, G.; Brlansky, R.H. A multiplex polymerase chain reaction method for reliable, sensitive and simultaneous detection of multiple viruses in citrus trees. J. Virol. Methods 2005, 129, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Qin, L.Y.; Hao, S.; Lan, R.T.; Xu, B.H.; Guo, Y.M.; Jiang, R.P.; Sun, H.; Chen, X.P.; Lv, X.C.; et al. Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp. Pathogens 2020, 9, 195. [Google Scholar] [CrossRef]
- Jiang, G.L.; Guo, C.Y.; Kuai, X.; Ji, L.L.; Zhao, J.; Zhou, Y.X.; Wang, X.L.; Zhang, M.H.; Zhou, Z.M.; Li, H. Foodborne diarrhea outbreaks linked to Citrobacter species in East China 2022–2023. Food Microbiol. 2025, 132, 104839. [Google Scholar] [CrossRef]
- Tran, T.D.; Lee, S.; Hnasko, R.; McGarvey, J.A. Complete genome sequence of Citrobacter braakii ASE1 generated by PacBio sequencing. Microbiol. Resour. Announc. 2024, 13, e0100023. [Google Scholar] [CrossRef]
- Schneider, D.; Ganbarzade, A.; Post, S.; Zühlke, D.; Hinzke, T.; Hollensteiner, J.; Poehlein, A.; Riedel, K.; Daniel, R. Complete Genome Sequence of Citrobacter braakii GW-Imi-1b1, Isolated from Hospital Wastewater in Greifswald, Germany. Microbiol. Resour. Announc. 2023, 12, e0014323. [Google Scholar] [CrossRef] [PubMed]
- Oyeka, M.; Antony, S. Citrobacter braakii Bacteremia: Case Report and Review of the Literature. Infect. Disord. Drug Targets 2017, 17, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.Q.; Yu, X.M.; Zhang, F.L.; Ma, J.; Ji, P. Whole-genome sequencing of coexisting blaNDM-1, blaKPC-2, and mcr-9.1 in a clinical carbapenem-resistant Citrobacter braakii isolate. J. Glob. Antimicrob. Resist. 2025, 44, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zou, H.; Xia, H.; Zhou, Z.; Zhao, L.; Meng, M.; Li, Q.; Guan, Y.; Li, X. Genomic insight into transmission mechanisms of carbapenem-producing Citrobacter spp. isolates between the WWTP and connecting rivers. Ecotoxicol. Environ. Saf. 2023, 262, 115150. [Google Scholar] [CrossRef]
- Mohiuddin, S.G.; Kavousi, P.; Figueroa, D.; Ghosh, S.; Orman, M.A. The diverse phenotypic and mutational landscape induced by fluoroquinolone treatment. mSystems 2025, 10, e0071325. [Google Scholar] [CrossRef]
- Ding, W.D.; Zhang, X.H.; Zhao, X.M.; Jing, W.; Cao, Z.M.; Li, J.; Huang, Y.; You, X.X.; Wang, M.; Shi, Q.; et al. A Chromosome-Level Genome Assembly of the Mandarin Fish (Siniperca chuatsi). Front. Genet. 2021, 12, 671650. [Google Scholar] [CrossRef]
- Li, M.; Wang, J.; Deng, H.; Li, L.; Huang, X.; Chen, D.; Ouyang, P.; Geng, Y.; Yang, S.; Yin, L.; et al. The Damage of the Crayfish (Procambarus Clarkii) Digestive Organs Caused by Citrobacter Freundii Is Associated with the Disturbance of Intestinal Microbiota and Disruption of Intestinal-Liver Axis Homeostasis. Front. Cell Infect. Microbiol. 2022, 5, 940576. [Google Scholar] [CrossRef]
- Denzer, L.; Schroten, H.; Schwerk, C. From Gene to Protein—How Bacterial Virulence Factors Manipulate Host Gene Expression During Infection. Int. J. Mol. Sci. 2020, 21, 3730. [Google Scholar] [CrossRef]
- Mnich, M.E.; Dalen, R.V.; Sorge, N.M.V. C-Type Lectin Receptors in Host Defense Against Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2020, 10, 309. [Google Scholar] [CrossRef]
- Wong, J.; Chen, Y.H.; Gan, Y.H. Host cytosolic glutathione sensing by a membrane histidine kinase activates the type VI secretion system in an intracellular bacterium. Cell Host Microbe 2015, 18, 38–48. [Google Scholar] [CrossRef]
- Doebel, T.; Voisin, B.; Nagao, K. Langerhans cells—The macrophage in dendritic cell clothing. Trends Immunol. 2017, 38, 817–828. [Google Scholar] [CrossRef]
- Deckers, J.; Hammad, H.; Hoste, E. Langerhans cells: Sensing the environment in health and disease. Front. Immunol. 2018, 9, 93. [Google Scholar] [CrossRef]
- Yang, K.; Park, C.G.; Cheong, C.; Bulgheresi, S.; Zhang, S.; Zhang, P.; He, Y.; Jiang, L.; Huang, H.; Ding, H.; et al. Host Langerin (CD207) is a receptor for Yersinia pestis phagocytosis and promotes dissemination. Immunol. Cell Biol. 2015, 93, 815–824. [Google Scholar] [CrossRef]

| Target Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Reference or Genebank Accession No. |
|---|---|---|---|
| cfa | GCGGTTACTGGAAAGATG | CGGCGATACTGAAATAGG | CP137123.1 |
| viaB | TGTCGAGCAGATGGATGAGCAT | ACGGCTGAAGGTTACGGACCGA | CP183865.1 |
| ompX | CTACGAATACGGCTCTGC | ATCGGTTCCTTCACTTACAC | CP016762 |
| ureD | GCCAGATGTCACGCATAACG | GGCTGCCACTGCTGATAGAA | CP128207.1 |
| ureE | TAACAGGCTTTGGCGAGTAGGA | CGCCTTGACCACGCTCACT | CP069787.1 |
| ureF | ATGCCGCAGAGTTGGCTGTC | GGAGATTGGCTGGGTGAAAA | CP174029.1 |
| ureG | AGGTTATCGCCACCGCTTTC | GGTTGCCCGCATACTGCT | CP137175.1 |
| slt-II | CCGGATCCATGAAGTGTATATTATTTAAATGG | CCCGAATTCTCAGTCATTATTAAACTGCAC | [27] |
| slt-LA | TTTACGATAGACTTCTCGAC | CACATATAAATTATTTCGCTC | [28] |
| slt-ILA | CCCGGATCCATGAAGTGTATATTATTTAAATGG | CCCGAATTCTTATTTACCCGTTGTATATAAAAA | [28] |
| Characteristic | C. braakii SCGY-1L | Characteristic | SCGY-1L |
|---|---|---|---|
| Glucose gas | + | Sorbose | + |
| Citrate | + | Mannitol | + |
| Arginine | + | Raffinose | + |
| Alanine | + | KCN | + |
| Aesculin | − | MR | + |
| V-P test | − | H2S | + |
| Lactose | + | Nitrate reduction | + |
| Sucrose | + | ornithine decarboxylase | + |
| Indole | - | lysine decarboxylase | − |
| Maltose | + | Urea | − |
| Antibiotic | Concentration (μg/piece) | Test Diameter of the Inhibition Zone (mm) | Sensitivity |
|---|---|---|---|
| Amikacin | 30 | 18 | I |
| Gentamicin | 10 | 15 | I |
| Kanamycin | 30 | 20 | S |
| Neomycin | 30 | 16 | I |
| Erythromycin | 15 | 0 | R |
| Medemycin | 30 | 0 | R |
| Norfloxacin | 10 | 25 | S |
| Ofloxacin | 5 | 24 | S |
| Ciprofloxacin | 5 | 28 | S |
| Polymyxin B | 300 | 14 | S |
| Clindamycin | 2 | 0 | R |
| Furazolidone | 300 | 18 | S |
| Tetracycline | 30 | 20 | S |
| Doxycycline | 30 | 18 | S |
| Minocycline | 30 | 14 | I |
| Penicillin | 10 | 16 | R |
| Oxacillin | 1 | 0 | R |
| Ampicillin | 10 | 15 | I |
| Carbenicillin | 100 | 27 | S |
| Piperacillin | 100 | 22 | S |
| Cephalexin | 30 | 21 | S |
| Cefazolin | 30 | 26 | S |
| Cefradine | 30 | 18 | I |
| Cefuroxim | 30 | 22 | S |
| Ceftazidime | 30 | 21 | S |
| Ceftriaxone | 30 | 28 | S |
| Cefoperazone | 75 | 25 | S |
| Vancomycin | 30 | 0 | R |
| Paediatric Compound Sulfamethoxazole Tablets | 23.75/1.25 | 24 | S |
| Chloramphenicol | 30 | 27 | S |
| Florfenicol | 30 | 22 | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhou, T.; Gu, S.; Yao, J.; Liu, S.; Mao, J. Integrated Pathogen–Host Analysis of Citrobacter braakii SCGY-1L: Genomic Determinants and Host Transcriptional Dynamics During Infection. Microorganisms 2025, 13, 2310. https://doi.org/10.3390/microorganisms13102310
Wang Z, Zhou T, Gu S, Yao J, Liu S, Mao J. Integrated Pathogen–Host Analysis of Citrobacter braakii SCGY-1L: Genomic Determinants and Host Transcriptional Dynamics During Infection. Microorganisms. 2025; 13(10):2310. https://doi.org/10.3390/microorganisms13102310
Chicago/Turabian StyleWang, Zhixiu, Tingting Zhou, Shaoxuan Gu, Jiaqi Yao, Suli Liu, and Jiaming Mao. 2025. "Integrated Pathogen–Host Analysis of Citrobacter braakii SCGY-1L: Genomic Determinants and Host Transcriptional Dynamics During Infection" Microorganisms 13, no. 10: 2310. https://doi.org/10.3390/microorganisms13102310
APA StyleWang, Z., Zhou, T., Gu, S., Yao, J., Liu, S., & Mao, J. (2025). Integrated Pathogen–Host Analysis of Citrobacter braakii SCGY-1L: Genomic Determinants and Host Transcriptional Dynamics During Infection. Microorganisms, 13(10), 2310. https://doi.org/10.3390/microorganisms13102310




