Insights into Cold-Season Adaptation of Mongolian Wild Asses Revealed by Gut Microbiome Metagenomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Metagenomic Sequencing and Annotation
2.3. trnL-Based Plant Composition Sequencing and Annotation
2.4. Data Analysis
3. Results
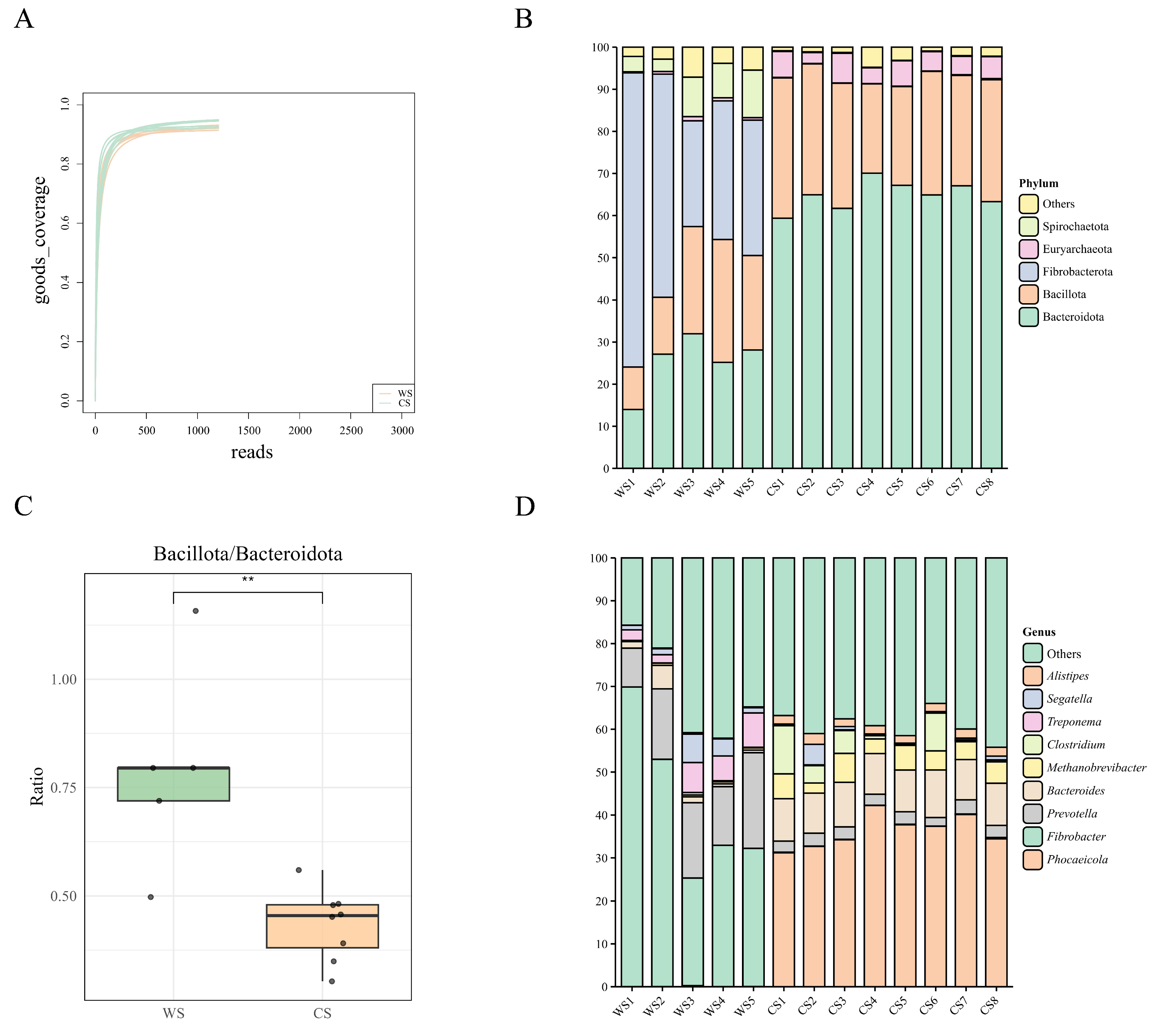
3.1. Gut Microbiota Composition and Abundance Analysis
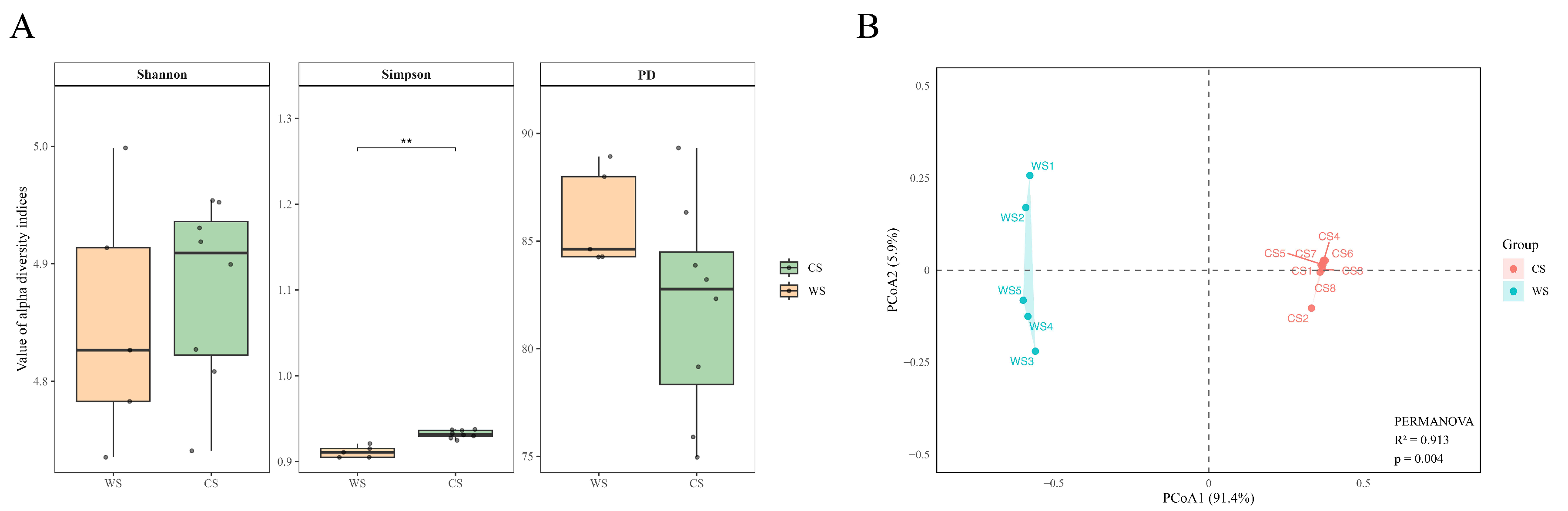
3.2. Gut Microbial Community Structure
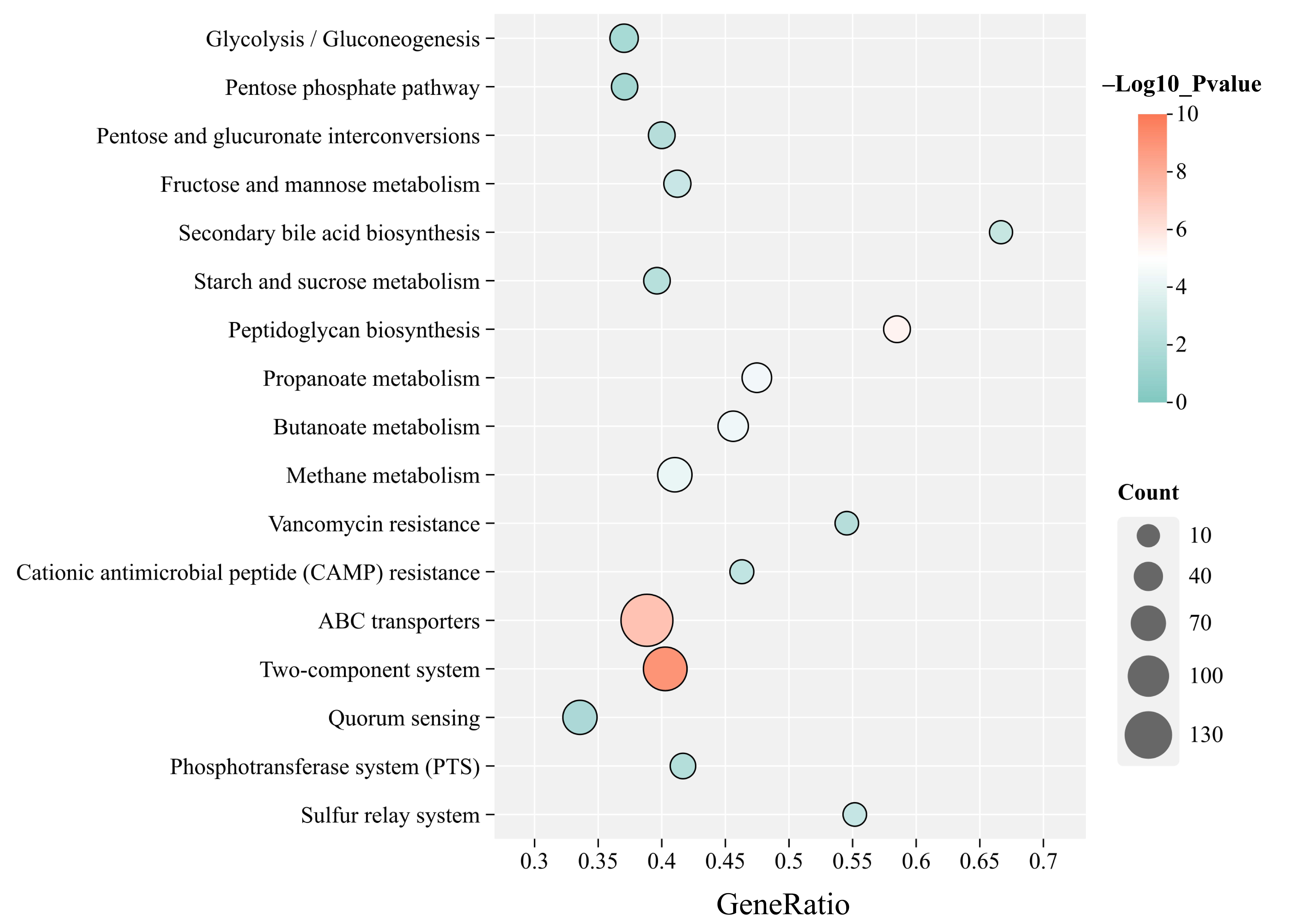
3.3. KEGG Functional Analysis
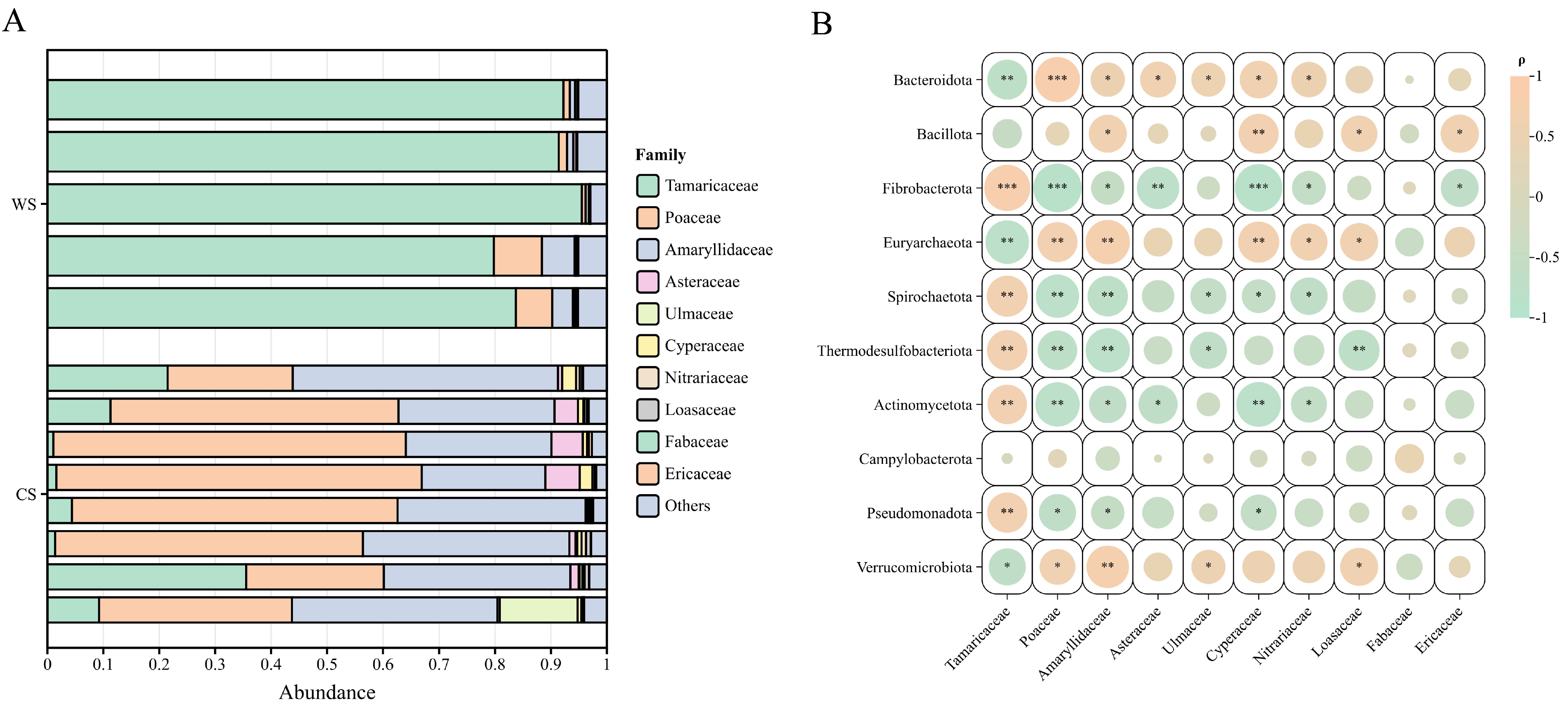
3.4. trnL-Based Plant Composition
4. Discussion
4.1. Cold Adaptation of Gut Microbiota Composition and Structure
4.2. Cold-Season Functional Adaptations of Gut Microbiota
4.3. trnL-Based Plant Composition and Seasonal Feeding
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, S.; Hu, Y.; Shao, C.; Jiang, L.; Zhang, Y.; Su, Z.; Wu, B.; Li, J. Vulnerability Assessment of Suitable Habitats for Equus hemionus in the Kalamaili National Park under Climate Change. Acta Theriol. Sin. 2024, 44, 287–296. [Google Scholar] [CrossRef]
- Ding, J.; Zhuo, Y.; Xu, W.; Kessler, M.; Wang, M.; Yang, W. Synergistic Effects of Climate and Land Use Change on Khulan (Equus hemionus hemionus) Habitat in China. Glob. Ecol. Conserv. 2024, 54, e03181. [Google Scholar] [CrossRef]
- Gao, S.; Davaasuren, D.; Li, J.; Shao, C.; Zhang, Y.; Wang, X.; Li, J. Projected Impacts of Climate Change and Border Fencing on the Distribution of Khulan (Equus hemionus hemionus). Glob. Ecol. Conserv. 2025, 60, e03593. [Google Scholar] [CrossRef]
- Buuveibaatar, B.; Mueller, T.; Strindberg, S.; Leimgruber, P.; Kaczensky, P.; Fuller, T.K. Human Activities Negatively Impact Distribution of Ungulates in the Mongolian Gobi. Biol. Conserv. 2016, 203, 168–175. [Google Scholar] [CrossRef]
- Kaczensky, P.; Salemgareyev, A.; Linnell, J.D.C.; Zuther, S.; Walzer, C.; Huber, N.; Petit, T. Post-Release Movement Behaviour and Survival of Kulan Reintroduced to the Steppes and Deserts of Central Kazakhstan. Front. Conserv. Sci. 2021, 2, 703358. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The Microbiota in Adaptive Immune Homeostasis and Disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Risely, A.; Müller-Klein, N.; Schmid, D.W.; Wilhelm, K.; Clutton-Brock, T.H.; Manser, M.B.; Sommer, S. Climate Change Drives Loss of Bacterial Gut Mutualists at the Expense of Host Survival in Wild Meerkats. Glob. Change Biol. 2023, 29, 5816–5828. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, C.; Stojanović, O.; Colin, D.J.; Suarez-Zamorano, N.; Tarallo, V.; Veyrat-Durebex, C.; Rigo, D.; Fabbiano, S.; Stevanović, A.; Hagemann, S.; et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell 2015, 163, 1360–1374. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. The Hologenome Concept of Evolution after 10 Years. Microbiome 2018, 6, 78. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial Degradation of Complex Carbohydrates in the Gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef]
- Li, B.; Liang, C.; Xu, B.; Song, P.; Liu, D.; Zhang, J.; Gu, H.; Jiang, F.; Gao, H.; Cai, Z.; et al. Extreme Winter Environment Dominates Gut Microbiota and Metabolome of White-Lipped Deer. Microbiol. Res. 2025, 297, 128182. [Google Scholar] [CrossRef]
- Yen, S.; Johnson, J.S. Metagenomics: A Path to Understanding the Gut Microbiome. Mamm Genome 2021, 32, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Palau-Rodriguez, M.; Tulipani, S.; Isabel Queipo-Ortuño, M.; Urpi-Sarda, M.; Tinahones, F.J.; Andres-Lacueva, C. Metabolomic Insights into the Intricate Gut Microbial–Host Interaction in the Development of Obesity and Type 2 Diabetes. Front. Microbiol. 2015, 6, 1151. [Google Scholar] [CrossRef]
- Qin, X.; Xi, L.; Zhao, L.; Han, J.; Qu, H.; Xu, Y.; Weng, W. Exploring the Distinctive Characteristics of Gut Microbiota across Different Horse Breeds and Ages Using Metataxonomics. Front. Cell. Infect. Microbiol. 2025, 15, 839. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and Limitations of the Chloroplast Trn L (UAA) Intron for Plant DNA Barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef]
- Valentini, A.; Miquel, C.; Nawaz, M.A.; Bellemain, E.; Coissac, E.; Pompanon, F.; Gielly, L.; Cruaud, C.; Nascetti, G.; Wincker, P.; et al. New Perspectives in Diet Analysis Based on DNA Barcoding and Parallel Pyrosequencing: The trnL Approach. Mol. Ecol. Resour. 2009, 9, 51–60. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Ma, S.; Zhang, Y.; Yang, X.; Zhang, H.; Han, Y.; Liu, Y.; Gao, F.; Yuan, Z. Seasonal Variation in Gut Microbiota of the Wild Daurian Ground Squirrel (Spermophilus dauricus): Metagenomic Insights into Seasonal Breeding. Animals 2023, 13, 2235. [Google Scholar] [CrossRef] [PubMed]
- Bo, T.-B.; Zhang, X.-Y.; Wen, J.; Deng, K.; Qin, X.-W.; Wang, D.-H. The Microbiota–Gut–Brain Interaction in Regulating Host Metabolic Adaptation to Cold in Male Brandt’s Voles (Lasiopodomys brandtii). ISME J. 2019, 13, 3037–3053. [Google Scholar] [CrossRef]
- Fan, Q.; Cui, X.; Wang, Z.; Chang, S.; Wanapat, M.; Yan, T.; Hou, F. Rumen Microbiota of Tibetan Sheep (Ovis aries) Adaptation to Extremely Cold Season on the Qinghai-Tibetan Plateau. Front. Vet. Sci. 2021, 8, 673822. [Google Scholar] [CrossRef]
- Belzer, C.; de Vos, W.M. Microbes inside—From Diversity to Function: The Case of Akkermansia. ISME J. 2012, 6, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, T.J.; Duddleston, K.N.; Buck, C.L. Effects of Season and Host Physiological State on the Diversity, Density, and Activity of the Arctic Ground Squirrel Cecal Microbiota. Appl. Environ. Microbiol. 2014, 80, 5611–5622. [Google Scholar] [CrossRef]
- Foley, M.H.; Cockburn, D.W.; Koropatkin, N.M. The Sus Operon: A Model System for Starch Uptake by the Human Gut Bacteroidetes. Cell. Mol. Life Sci. 2016, 73, 2603–2617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Sukhchuluun, G.; Bo, T.-B.; Chi, Q.-S.; Yang, J.-J.; Chen, B.; Zhang, L.; Wang, D.-H. Huddling Remodels Gut Microbiota to Reduce Energy Requirements in a Small Mammal Species during Cold Exposure. Microbiome 2018, 6, 103. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Yin, J.; Li, Y.; Han, H.; Chen, S.; Gao, J.; Liu, G.; Wu, X.; Deng, J.; Yu, Q.; Huang, X.; et al. Melatonin Reprogramming of Gut Microbiota Improves Lipid Dysmetabolism in High-Fat Diet-Fed Mice. J. Pineal Res. 2018, 65, e12524. [Google Scholar] [CrossRef] [PubMed]
- Telesford, K.M.; Yan, W.; Ochoa-Reparaz, J.; Pant, A.; Kircher, C.; Christy, M.A.; Begum-Haque, S.; Kasper, D.L.; Kasper, L.H. A Commensal Symbiotic Factor Derived from Bacteroides Fragilis Promotes Human CD39+Foxp3+ T Cells and Treg Function. Gut Microbes 2015, 6, 234–242. [Google Scholar] [CrossRef]
- Zeng, Q.; Li, D.; He, Y.; Li, Y.; Yang, Z.; Zhao, X.; Liu, Y.; Wang, Y.; Sun, J.; Feng, X.; et al. Discrepant Gut Microbiota Markers for the Classification of Obesity-Related Metabolic Abnormalities. Sci. Rep. 2019, 9, 13424. [Google Scholar] [CrossRef]
- Borton, M.A.; Sabag-Daigle, A.; Wu, J.; Solden, L.M.; O’Banion, B.S.; Daly, R.A.; Wolfe, R.A.; Gonzalez, J.F.; Wysocki, V.H.; Ahmer, B.M.M.; et al. Chemical and Pathogen-Induced Inflammation Disrupt the Murine Intestinal Microbiome. Microbiome 2017, 5, 47. [Google Scholar] [CrossRef]
- Rogers, M.B.; Firek, B.; Shi, M.; Yeh, A.; Brower-Sinning, R.; Aveson, V.; Kohl, B.L.; Fabio, A.; Carcillo, J.A.; Morowitz, M.J. Disruption of the Microbiota across Multiple Body Sites in Critically Ill Children. Microbiome 2016, 4, 66. [Google Scholar] [CrossRef]
- Stoffel, M.A.; Acevedo-Whitehouse, K.; Morales-Durán, N.; Grosser, S.; Chakarov, N.; Krüger, O.; Nichols, H.J.; Elorriaga-Verplancken, F.R.; Hoffman, J.I. Early Sexual Dimorphism in the Developing Gut Microbiome of Northern Elephant Seals. Mol. Ecol. 2020, 29, 2109–2122. [Google Scholar] [CrossRef]
- Sun, B.; Gu, Z.; Wang, X.; Huffman, M.A.; Garber, P.A.; Sheeran, L.K.; Zhang, D.; Zhu, Y.; Xia, D.-P.; Li, J. Season, Age, and Sex Affect the Fecal Mycobiota of Free-Ranging Tibetan Macaques (Macaca thibetana). Am. J. Primatol. 2018, 80, e22880. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, X.; Ding, Y.; Hu, Y.; Nie, Y.; Wei, W.; Ma, S.; Yan, L.; Zhu, L.; Wei, F. Seasonal Variation in Nutrient Utilization Shapes Gut Microbiome Structure and Function in Wild Giant Pandas. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170955. [Google Scholar] [CrossRef]
- Tang, K.-Y.; Wang, Z.-W.; Wan, Q.-H.; Fang, S.-G. Metagenomics Reveals Seasonal Functional Adaptation of the Gut Microbiome to Host Feeding and Fasting in the Chinese Alligator. Front. Microbiol. 2019, 10, 2409. [Google Scholar] [CrossRef]
- Orkin, J.D.; Campos, F.A.; Myers, M.S.; Cheves Hernandez, S.E.; Guadamuz, A.; Melin, A.D. Seasonality of the Gut Microbiota of Free-Ranging White-Faced Capuchins in a Tropical Dry Forest. ISME J. 2019, 13, 183–196. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Ambudkar, S.V. Overview: ABC Transporters and Human Disease. J. Bioenerg. Biomembr. 2001, 33, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Yao, Y.; Wang, C.; Dong, M.; Wu, Y.; Li, D.; Xie, M.; Ni, Q.; Zhang, M.; Xu, H. Seasonal Dynamics of Gut Microbiota in a Cohort of Wild Tibetan Macaques (Macaca thibetana) in Western China. Glob. Ecol. Conserv. 2021, 25, e01409. [Google Scholar] [CrossRef]
- Lazar, J.T.; Tabor, J.J. Bacterial Two-Component Systems as Sensors for Synthetic Biology Applications. Curr. Opin. Syst. Biol. 2021, 28, 100398. [Google Scholar] [CrossRef] [PubMed]
- Hersbach, H.; Bell, B.; Berrisford, P.; Biavati, G.; Horányi, A.; Muñoz Sabater, J.; Nicolas, J.; Peubey, C.; Radu, R.; Rozum, I.; et al. 2023: ERA5 Hourly Data on Pressure Levels from 1940 to Present. Copernicus Climate Change Service (C3S) Climate Data Store (CDS). Available online: https://mirror-earth.com/ (accessed on 15 August 2025).
- Feh, C.; Munkhtuya, B.; Enkhbold, S.; Sukhbaatar, T. Ecology and Social Structure of the Gobi Khulan Equus Hemionus Subsp. in the Gobi B National Park, Mongolia. Biol. Conserv. 2001, 101, 51–61. [Google Scholar] [CrossRef]
- Liu, W.; Yang, W.; Xu, W. Food habits of the Kulan (Equus hemionus) in autumn. Acta Theriol. Sin. 2008, 28, 33–36. [Google Scholar] [CrossRef]
- Liu, Y.; Si, X.; Bi, J.; Wu, X. Wintering Feeding Habits of Asiatic Wild Ass (Equus hemionus hemionus) in Central Inner Mongolia. J. Northeast For. Univ. 2015, 43, 88–91. [Google Scholar] [CrossRef]
- Bi, S.; Bi, J.; Gao, F. Summer food-habits of Asiatic wild ass (Equus hemionus hemionus) in central area of Inner Mongolia. J. Biol. 2009, 26, 30–33. [Google Scholar]
- Kartzinel, T.R.; Hsing, J.C.; Musili, P.M.; Brown, B.R.P.; Pringle, R.M. Covariation of Diet and Gut Microbiome in African Megafauna. Proc. Natl. Acad. Sci. USA 2019, 116, 23588–23593. [Google Scholar] [CrossRef] [PubMed]
| KEGG Level 2 Pathways | WS (Mean) | WS (SD) | CS (Mean) | CS (SD) | Wilcoxon _adj.p |
|---|---|---|---|---|---|
| Carbohydrate metabolism | 14.2188 | 0.2585 | 14.1086 | 0.0982 | 0.5486 |
| Amino acid metabolism | 7.8327 | 0.3125 | 8.5900 | 0.0774 | 0.0021 |
| Glycan biosynthesis and metabolism | 8.3952 | 0.2417 | 7.1323 | 0.1417 | 0.0021 |
| Metabolism of cofactors and vitamins | 5.9110 | 0.0531 | 5.8319 | 0.0520 | 0.0342 |
| Replication and repair | 5.7053 | 0.2280 | 5.6533 | 0.1044 | 0.9433 |
| Energy metabolism | 4.5317 | 0.0714 | 5.4279 | 0.0960 | 0.0021 |
| Translation | 3.9471 | 0.0972 | 4.8214 | 0.1235 | 0.0021 |
| Nucleotide metabolism | 4.2836 | 0.1373 | 4.7683 | 0.0543 | 0.0021 |
| Cellular community—prokaryotes | 4.7670 | 0.1461 | 3.8899 | 0.0393 | 0.0021 |
| Signal transduction | 5.0111 | 0.2804 | 3.9948 | 0.0834 | 0.0021 |
| Membrane transport | 3.6382 | 0.0994 | 4.3342 | 0.1052 | 0.0021 |
| Lipid metabolism | 3.9895 | 0.1303 | 4.0340 | 0.0645 | 0.5486 |
| Biosynthesis of other secondary metabolites | 3.2261 | 0.0751 | 2.9493 | 0.0421 | 0.0021 |
| Folding, sorting and degradation | 2.8743 | 0.0424 | 2.9547 | 0.0207 | 0.0080 |
| Metabolism of other amino acids | 2.6941 | 0.0472 | 2.8986 | 0.0502 | 0.0021 |
| Cell growth and death | 2.5095 | 0.0350 | 2.3095 | 0.0484 | 0.0021 |
| Drug resistance: antimicrobial | 2.3286 | 0.0686 | 2.5330 | 0.0311 | 0.0021 |
| Transport and catabolism | 1.9091 | 0.1134 | 1.5829 | 0.0594 | 0.0021 |
| Metabolism of terpenoids and polyketides | 1.9260 | 0.1857 | 1.5668 | 0.0179 | 0.0021 |
| Infectious disease: bacterial | 1.4953 | 0.0379 | 1.4124 | 0.0159 | 0.0021 |
| Xenobiotics biodegradation and metabolism | 1.0368 | 0.0239 | 1.2379 | 0.0204 | 0.0021 |
| Endocrine system | 1.0108 | 0.0350 | 1.0598 | 0.0223 | 0.0342 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Gu, H.; Gao, H.; Zhang, T.; Jiang, F.; Song, P.; Liu, Y.; Fan, Q.; Xu, Y.; Zhang, R. Insights into Cold-Season Adaptation of Mongolian Wild Asses Revealed by Gut Microbiome Metagenomics. Microorganisms 2025, 13, 2304. https://doi.org/10.3390/microorganisms13102304
Wang J, Gu H, Gao H, Zhang T, Jiang F, Song P, Liu Y, Fan Q, Xu Y, Zhang R. Insights into Cold-Season Adaptation of Mongolian Wild Asses Revealed by Gut Microbiome Metagenomics. Microorganisms. 2025; 13(10):2304. https://doi.org/10.3390/microorganisms13102304
Chicago/Turabian StyleWang, Jianeng, Haifeng Gu, Hongmei Gao, Tongzuo Zhang, Feng Jiang, Pengfei Song, Yan Liu, Qing Fan, Youjie Xu, and Ruidong Zhang. 2025. "Insights into Cold-Season Adaptation of Mongolian Wild Asses Revealed by Gut Microbiome Metagenomics" Microorganisms 13, no. 10: 2304. https://doi.org/10.3390/microorganisms13102304
APA StyleWang, J., Gu, H., Gao, H., Zhang, T., Jiang, F., Song, P., Liu, Y., Fan, Q., Xu, Y., & Zhang, R. (2025). Insights into Cold-Season Adaptation of Mongolian Wild Asses Revealed by Gut Microbiome Metagenomics. Microorganisms, 13(10), 2304. https://doi.org/10.3390/microorganisms13102304






