Morphological, Molecular, and Alkaloid Gene Profiling of Epichloë Endophytes in Elymus cylindricus and Elymus tangutorum from China
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Preservation of Plant Materials and Endophytes
2.2. Isolation of Endophytes and Morphological Examination
2.3. DNA Extraction, PCR Analyses, and Phylogenetic Analysis
2.4. Alkaloid Gene Detection
3. Results
3.1. Isolation of Endophytes
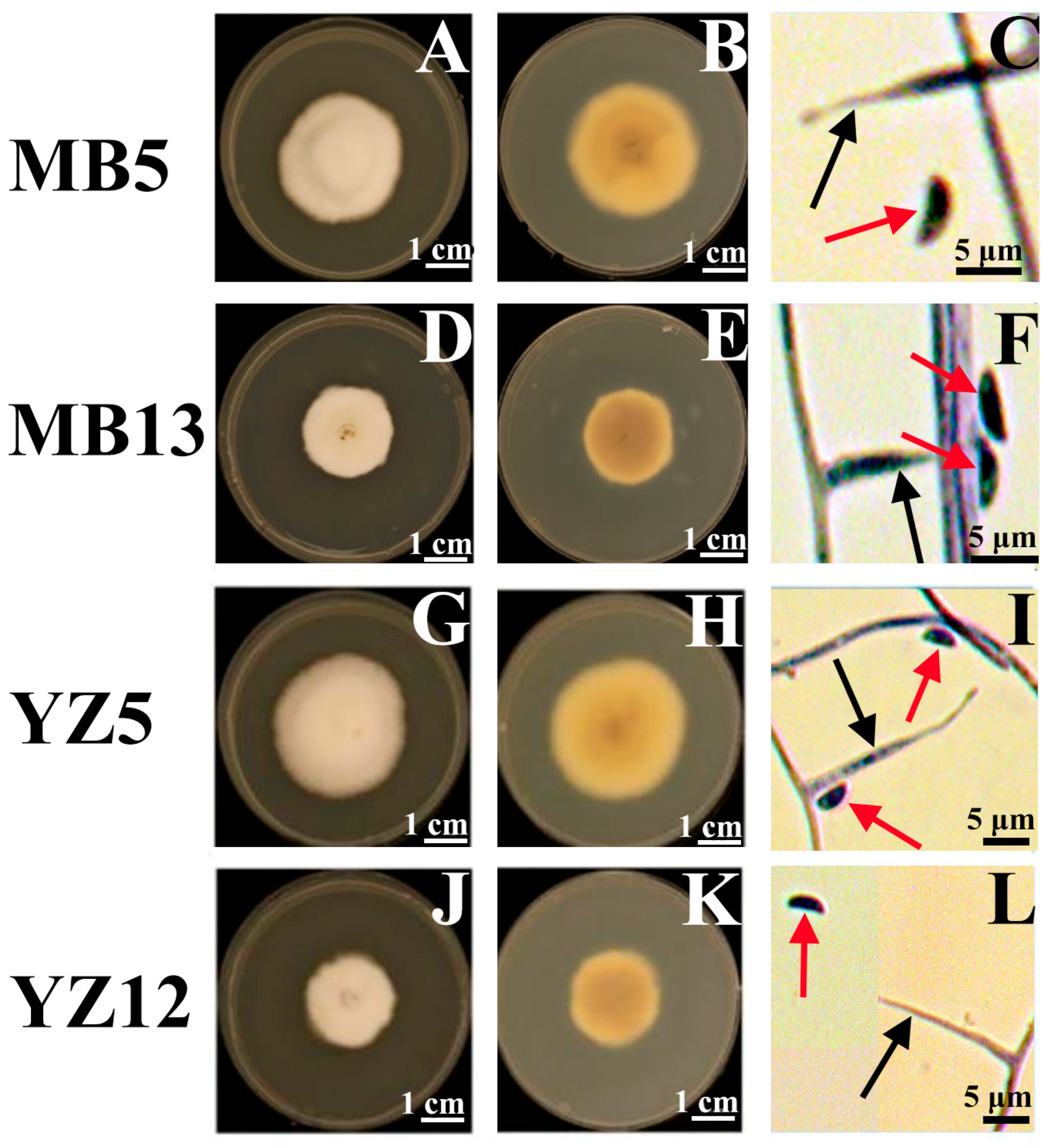
3.2. Morphological Examination
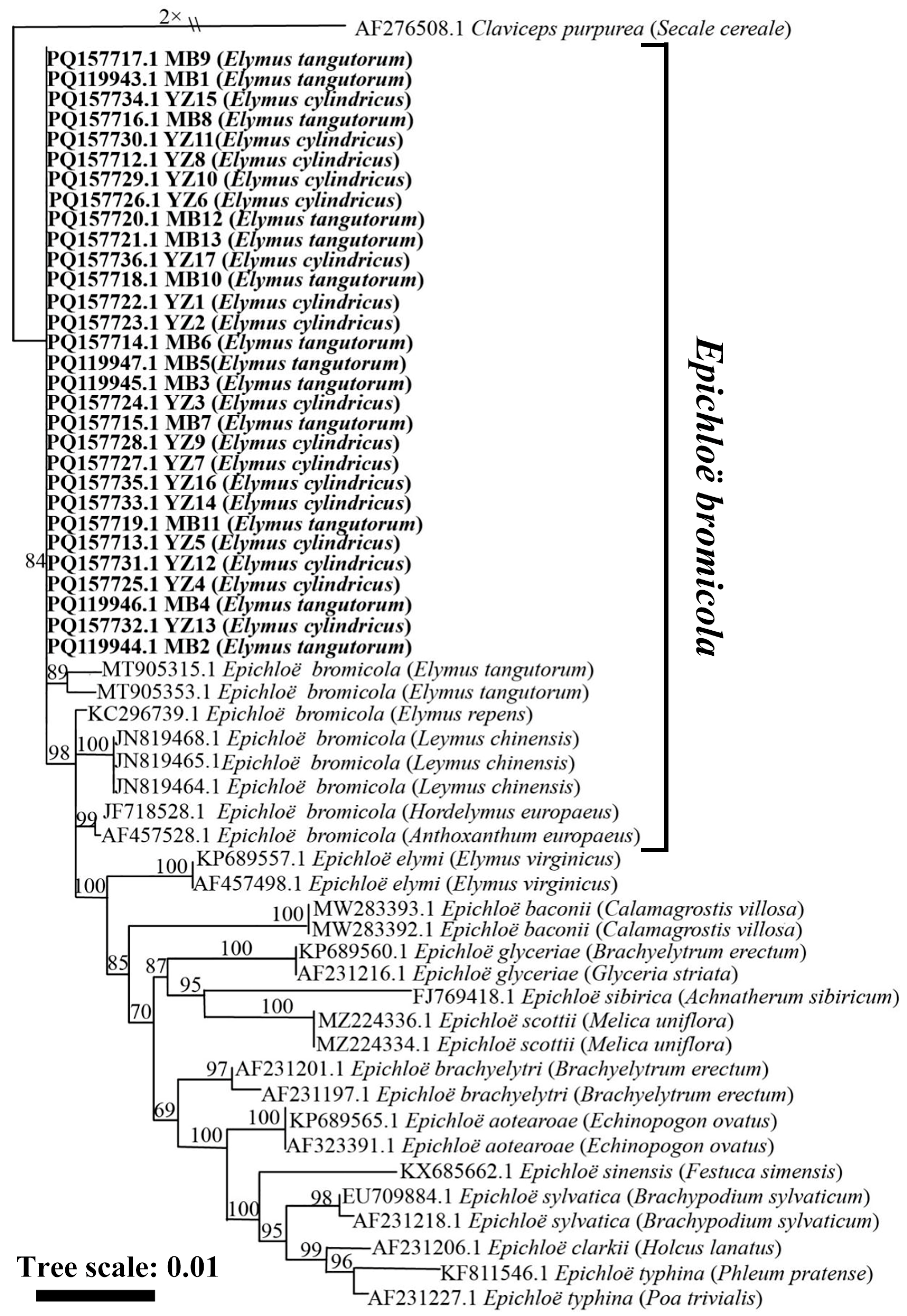
3.3. Phylogenetic Analysis
3.4. Alkaloid Gene Detection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, M.R.; Schardl, C.L.; Johnson, M.C. Fungal Endophytes of Grasses 1987. Annu. Rev. Phytopathol. Previous Issues 1987, 25, 293–315. [Google Scholar] [CrossRef]
- Schardl, C.L. The Epichloë symbionts of the grass subfamily Poöideae. Ann. Mo. Bot. Gard. 2010, 97, 646–665. [Google Scholar] [CrossRef]
- Christensen, M.J.; Bennett, R.J.; Ansari, H.A.; Koga, H.; Johnson, R.D.; Bryan, G.T.; Simpson, W.R.; Koolaard, J.P.; Nickless, E.M.; Voisey, C.R. Epichloë endophytes grow by intercalary hyphal extension in elongating grass leaves. Fungal Genet. Biol. 2008, 45, 84–93. [Google Scholar] [CrossRef]
- Tanaka, A.; Takemoto, D.; Chujo, T.; Scott, B. Fungal endophytes of grasses. Curr. Opin. Plant Biol. 2012, 15, 462–468. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Bacon, C.W.; Schardl, C.L.; White, J.F.; Tadych, M. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 2014, 106, 202–215. [Google Scholar] [CrossRef]
- Campbell, M.A.; Tapper, B.A.; Simpson, W.R.; Johnson, R.D.; Mace, W.; Ram, A.; Lukito, Y.; Dupont, P.Y.; Johnson, L.J.; Scott, D.B.; et al. Epichloë hybrida, sp. nov., an emerging model system for investigating fungal allopolyploidy. Mycologia 2017, 109, 715–729. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Young, C.A.; Stewart, A.V.; Simpson, W.R.; Hume, D.E.; Scott, B. Epichloë novae-zelandiae, a new endophyte from the endemic New Zealand grass Poa matthewsii. New Zealand J. Bot. 2019, 57, 271–288. [Google Scholar] [CrossRef]
- Tintjer, T.; Leuchtmann, A.; Clay, K. Variation in horizontal and vertical transmission of the endophyte Epichloë elymi infecting the grass Elymus hystrix. New Phytol. 2008, 179, 236–246. [Google Scholar] [CrossRef]
- Gundel, P.E.; Rudgers, J.A.; Whitney, K.D. Vertically transmitted symbionts as mechanisms of transgenerational effects. Am. J. Bot. 2017, 104, 787–792. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Li, C.; Swoboda, G.A.; Young, C.A.; Sugawara, K.; Leuchtmann, A.; Schardl, C.L. Two distinct Epichloë species symbiotic with Achnatherum inebrians, drunken horse grass. Mycologia 2015, 107, 863–873. [Google Scholar] [CrossRef]
- Kendall, L.; Ali, M.; Kishan, M.; Holly, P.; Marin, L. Interaction between grasses and Epichloë endophytes and its significance to biotic and abiotic stress tolerance and the rhizosphere. Microorganisms 2021, 9, 2186. [Google Scholar] [CrossRef]
- Krauss, J.; Vikuk, V.; Young, C.A.; Krischke, M.; Mueller, M.J.; Baerenfaller, K. Epichloë endophyte infection rates and alkaloid content in commercially available grass seed mixtures in europe. Microorganisms 2020, 8, 498. [Google Scholar] [CrossRef]
- Bastias, D.A.; Ueno, A.C.; Gundel, P.E. Global change factors influence plant-Epichloë associations. J. Fungi 2023, 9, 446. [Google Scholar] [CrossRef]
- Wang, J.; Hou, W.; Christensen, M.J.; Li, X.; Xia, C.; Li, C.; Nan, Z. Role of Epichloë endophytes in improving host grass resistance ability and soil properties. J. Agric. Food Chem. 2020, 68, 6944–6955. [Google Scholar] [CrossRef]
- Schardl, C.L.; Florea, S.; Pan, J.; Nagabhyru, P.; Bec, S.; Calie, P.J. The epichloae: Alkaloid diversity and roles in symbiosis with grasses. Curr. Opin. Plant Biol. 2013, 16, 480–488. [Google Scholar] [CrossRef]
- Song, H.; Nan, Z.B.; Song, Q.Y.; Xia, C.; Li, X.Z.; Yao, X.; Xu, W.B.; Kuang, Y.; Tian, P.; Zhang, Q.P. Advances in research on Epichloë endophytes in Chinese native grasses. Front. Microbiol. 2016, 7, 1399. [Google Scholar] [CrossRef]
- Bacon, C.W.; Porter, J.K.; Robbins, J.D.; Luttrell, E.S. Epichloë typhina from toxic tall fescue grasses. Appl. Environ. Microbiol. 1977, 34, 576–581. [Google Scholar] [CrossRef]
- Rowan, D.D.; Gaynor, D.L. Isolation of feeding deterrents against argentine stem weevil from ryegrass infected with the endophyte Acremonium loliae. J. Chem. Ecol. 1986, 12, 647–658. [Google Scholar] [CrossRef]
- Schardl, C.L.; Grossman, R.B.; Nagabhyru, P.; Faulkner, J.R.; Mallik, U.P. Loline alkaloids: Currencies of mutualism. Phytochemistry 2007, 68, 980–996. [Google Scholar] [CrossRef]
- Tanaka, A.; Tapper, B.A.; Popay, A.; Parker, E.J.; Scott, B. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 2005, 57, 1036–1050. [Google Scholar] [CrossRef]
- Berry, D.; Mace, W.; Grage, K.; Wesche, F.; Gore, S.; Schardl, C.L.; Young, C.A.; Dijkwel, P.P.; Leuchtmann, A.; Bode, H.B.; et al. Efficient nonenzymatic cyclization and domain shuffling drive pyrrolopyrazine diversity from truncated variants of a fungal NRPS. Proc. Natl. Acad. Sci. USA 2019, 116, 25614–25623. [Google Scholar] [CrossRef]
- Young, C.A.; Bryant, M.K.; Christensen, M.J.; Tapper, B.A.; Bryan, G.T.; Scott, B. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Genet. Genom. 2005, 274, 13–29. [Google Scholar] [CrossRef]
- Young, C.A.; Felitti, S.; Shields, K.; Spangenberg, G.; Johnson, R.D.; Bryan, G.T.; Saikia, S.; Scott, B. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet. Biol. 2006, 43, 679–693. [Google Scholar] [CrossRef]
- Young, C.A.; Tapper, B.A.; May, K.; Moon, C.D.; Schardl, C.L.; Scott, B. Indole-diterpene biosynthetic capability of Epichloë endophytes as predicted by ltm gene analysis. Appl. Environ. Microbiol. 2009, 75, 2200–2211. [Google Scholar] [CrossRef]
- Fleetwood, D.J.; Scott, B.; Lane, G.A.; Tanaka, A.; Johnson, R.D. A complex ergovaline gene cluster in Epichloë endophytes of grasses. Appl. Environ. Microbiol. 2007, 73, 2571–2579. [Google Scholar]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-symbiotic fungi as chemical engineers: Multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef]
- Charlton, N.D.; Craven, K.D.; Afkhami, M.E.; Hall, B.A.; Ghimire, S.R.; Young, C.A. Interspecific hybridization and bioactive alkaloid variation increases diversity in endophytic Epichloë species of Bromus laevipes. FEMS Microbiol. Ecol. 2014, 90, 276–289. [Google Scholar] [CrossRef]
- Takach, J.E.; Young, C.A. Alkaloid genotype diversity of tall fescue endophytes. Crop Sci. 2014, 54, 667–678. [Google Scholar] [CrossRef]
- Easton, H.S. Grasses and Neotyphodium endophytes: Co-adaptation and adaptive breeding. Euphytica 2007, 154, 295–306. [Google Scholar] [CrossRef]
- Bamisile, B.S.; Dash, C.K.; Akutse, K.S.; Keppanan, R.; Afolabi, O.G.; Hussain, M.; Qasim, M.; Wang, L. Prospects of endophytic fungal entomopathogens as biocontrol and plant growth promoting agents: An insight on how artificial inoculation methods affect endophytic colonization of host plants. Microbiol. Res. 2018, 217, 34–50. [Google Scholar] [CrossRef]
- Saikkonen, K.; Phillips, T.D.; Faeth, S.H.; McCulley, R.L.; Saloniemi, I.; Helander, M. Performance of endophyte infected tall fescue in Europe and north America. PLoS ONE 2016, 11, e0157382. [Google Scholar] [CrossRef]
- Cagnano, G.; Vazquez-de-Aldana, B.R.; Asp, T.; Roulund, N.; Jensen, C.S.; Soto-Barajas, M.C. Determination of loline alkaloids and mycelial biomass in endophyte-infected Schedonorus pratensis by Near-Infrared Spectroscopy and Chemometrics. Microorganisms 2020, 8, 776. [Google Scholar] [CrossRef]
- Becker, Y.; Green, K.A.; Scott, B.; Becker, A.M. Artificial inoculation of Epichloë festucae into Lolium perenne, and visualisation of endophytic and epiphyllous fungal growth. Bio-Protoc. 2018, 8, e2990. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, L.; Liu, Y.; Huang, Z.; Shi, J.; Wang, Y.; Ma, Y.; Esteban Lucas-Borja, M.; Lopez-Vicente, M.; Wu, G.L. Restoration of a hillslope grassland with an ecological grass species (Elymus tangutorum) favors rainfall interception and water infiltration and reduces soil loss on the Qinghai-Tibetan Plateau. Catena 2022, 219, 106632. [Google Scholar] [CrossRef]
- Song, H.; Nan, Z. Origin, divergence, and phylogeny of asexual Epichloë endophyte in Elymus Species from Western China. PLoS ONE 2015, 10, e0127096. [Google Scholar] [CrossRef]
- Du, M.X.; Wang, T.; Li, C.J.; Chen, T.X. Discovery and characterization of Epichloë fungal endophytes from Elymus spp. in Northwest China. Microorganisms 2024, 12, 1497. [Google Scholar] [CrossRef]
- Li, C.J.; Nan, Z.B.; Li, F. Biological and physiological characteristics of Neotyphodium gansuense symbiotic with Achnatherum inebrians. Microbiol. Res. 2008, 163, 431–440. [Google Scholar] [CrossRef]
- Latch, G.C.M.; Christensen, M.J. Artificial infection of grasses with endophytes. Ann. Appl. Biol. 1985, 107, 17–24. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Schardl, C.L. Mating compatibility and phylogenetic relationships among two new species of Epichloë and other congeneric European species. Mycol. Res. 1998, 102, 1169–1182. [Google Scholar] [CrossRef]
- White, J.F. Widespread distribution of endophytes in the Poaceae. Plant Dis. 1987, 71, 340–342. [Google Scholar] [CrossRef]
- Craven, K.D.; Blankenship, J.D.; Leuchtmann, A.; Hignight, K.; Schardl, C.L. Hybrid fungal endophytes symbiotic with the grass Lolium pratense. Sydowia 2001, 53, 44–73. [Google Scholar]
- Chen, T.X.; Simpson, W.R.; Song, Q.Y.; Chen, S.H.; Li, C.J.; Ahmad, R.Z. Identification of Epichloë endophytes associated with wild barley (Hordeum brevisubulatum) and characterisation of their alkaloid biosynthesis. New Zealand J. Agric. Res. 2019, 62, 131–149. [Google Scholar] [CrossRef]
- Leuchtmann, A.; Oberhofer, M. The Epichloë endophytes associated with the woodland grass Hordelymus europaeus including four new taxa. Mycologia 2013, 105, 1315–1324. [Google Scholar] [CrossRef]
- Yi, M.; Hendricks, W.Q.; Kaste, J.; Charlton, N.D.; Nagabhyru, P.; Panaccione, D.G.; Young, C.A. Molecular identification and characterization of endophytes from uncultivated barley. Mycologia 2018, 110, 453–472. [Google Scholar] [CrossRef]
- Zhu, M.J.; Ren, A.Z.; Wen, W.; Gao, Y.B. Diversity and taxonomy of endophytes from Leymus chinensis in the Inner Mongolia steppe of China. FEMS Microbiol. Lett. 2013, 340, 135–145. [Google Scholar] [CrossRef]
- Chen, T.X.; Wang, T.; Du, M.X.; Malik, K.; Li, C.J.; Bao, G.S. Discovery of Epichloë as novel endophytes of Psathyrostachys lanuginosa in China and their alkaloid profiling. Front. Microbiol. 2024, 15, 1383923. [Google Scholar] [CrossRef]
- Li, W.; Ji, Y.L.; Yu, H.S.; Wang, Z.W. A new species of Epichloë symbiotic with Chinese grasses. Mycologia 2006, 98, 560–570. [Google Scholar] [CrossRef]
- Kang, Y.; Ji, Y.; Sun, X.; Zhan, L.; Li, W.; Yu, H.; Wang, Z. Taxonomy of Neotyphodium endophytes of Chinese native Roegneria plants. Mycologia 2009, 101, 211–219. [Google Scholar] [CrossRef]
- Shi, C.; An, S.; Yao, Z.; Young, C.A.; Panaccione, D.G.; Lee, S.T.; Schardl, C.L.; Li, C. Toxin-producing Epichloë bromicola strains symbiotic with the forage grass Elymus dahuricus in China. Mycologia 2017, 109, 847–859. [Google Scholar] [CrossRef]
- Schardl, C.L.; Leuchtmann, A. Three new species of Epichloë symbiotic with North American grasses. Mycologia 1999, 91, 95–107. [Google Scholar] [CrossRef]
- Charlton, N.D.; Craven, K.D.; Mittal, S.; Hopkins, A.A.; Young, C.A. Epichloë canadensis, a new interspecific epichloid hybrid symbiotic with Canada wildrye (Elymus canadensis). Mycologia 2012, 104, 1187–1199. [Google Scholar] [CrossRef]
- Moon, C.D.; Miles, C.O.; Järlfors, U.; Schardl, C.L. The evolutionary origins of three new Neotyphodium endophyte species from grasses indigenous to the Southern Hemisphere. Mycologia 2002, 94, 694–711. [Google Scholar] [CrossRef] [PubMed]
- Moon, C.D.; Guillaumin, J.J.; Ravel, C.; Li, C.; Craven, K.D.; Schardl, C.L. New Neotyphodium endophyte species from the grass tribes Stipeae and Meliceae. Mycologia 2007, 99, 895–905. [Google Scholar] [CrossRef]
- Gentile, A.; Rossi, M.S.; Cabral, D.; Craven, K.D.; Schardl, C.L. Origin, divergence, and phylogeny of Epichloë endophytes of native Argentine grasses. Mol. Phylogenetics Evol. 2005, 35, 196–208. [Google Scholar] [CrossRef]
- Zhang, D.X.; Nagabhyru, P.; Schardl, C.L. Regulation of a chemical defense against herbivory produced by symbiotic fungi in grass plants. Plant Physiol. 2009, 150, 1072–1082. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Tippmann, H.F. Analysis for free: Comparing programs for sequence analysis. Brief. Bioinform. 2004, 5, 82–87. [Google Scholar] [CrossRef]
- Bui Quang, M.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 2461. [Google Scholar] [CrossRef]
- Zhou, T.; Xu, K.; Zhao, F.; Liu, W.; Li, L.; Hua, Z.; Zhou, X. Itol. toolkit accelerates working with iTOL (Interactive Tree of Life) by an automated generation of annotation files. Bioinformatics 2023, 39, btad339. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Takach, J.E.; Schardl, C.L.; Charlton, N.D.; Scott, B.; Young, C.A. Disparate independent genetic events disrupt the secondary metabolism gene perA in certain symbiotic Epichloë species. Appl. Environ. Microbiol. 2015, 81, 2797–2807. [Google Scholar] [CrossRef] [PubMed]
- Young, C.A.; Hume, D.E.; Mcculley, R.L. Forages and pastures symposium: Fungal endophytes of tall fescue and perennial ryegrass: Pasture friend or foe? J. Anim. Sci. 2013, 91, 2379–2394. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y. Characteristics of Epichloë Endophyte–Festuca sinensis Symbiote. Master’s Thesis, Lanzhou University, Lanzhou, China, 2016. [Google Scholar]
- Wei, Y.K.; Gao, Y.B.; Xu, H.; Su, D.; Zhang, X.; Wang, Y.H.; Lin, F.; Chen, L.; Nie, L.Y.; Ren, A.Z. Occurrence of endophytes in grasses native to northern China. Grass Forage Sci. 2006, 61, 422–429. [Google Scholar] [CrossRef]
- Schulthess, F.M.; Faeth, S.H. Distribution, abundances, and associations of the endophytic fungal community of Arizona fescue (Festuca arizonica). Mycologia 1998, 90, 569–578. [Google Scholar] [CrossRef]
- Wang, T.; Chen, T.X.; White, J.F.; Li, C.J. Identification of three Epichloë endophytes from Hordeum bogdanii Wilensky in China. J. Fungi 2022, 8, 928. [Google Scholar] [CrossRef]
- Li, C.J.; Nan, Z.B.; Paul, V.H.; Dapprich, P.D.; Liu, Y. A new Neotyphodium species symbiotic with drunken horse grass (Achnatherum inebrians) in China. Mycotaxon 2004, 90, 141–147. [Google Scholar]
- Saikkonen, K.; Gundel, P.E.; Helander, M. Chemical ecology mediated by fungal endophytes in grasses. J. Chem. Ecol. 2013, 39, 962–968. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Faulkner, J.R.; Florea, S.; Pan, J. Chemotypic diversity of epichloae, fungal symbionts of grasses. Fungal Ecol. 2012, 5, 331–344. [Google Scholar] [CrossRef]
- Christensen, M.J. Variation in the ability of Acremonium endophytes of Lolium perenne, Festuca arundinacea and F. pratensis to form compatible associations in the 3 grasses. Mycol. Res. 1995, 99, 466–470. [Google Scholar] [CrossRef]
- Hopkins, A.A.; Young, C.A.; Panaccione, D.G.; Simpson, W.R.; Mittal, S.; Bouton, J.H. Agronomic performance and lamb health among several tall fescue novel endophyte combinations in the South-Central USA. Crop Sci. 2010, 50, 1552–1561. [Google Scholar] [CrossRef]
- Klotz, J.L. Activities and effects of ergot alkaloids on livestock physiology and production. Toxins 2015, 7, 2801–2821. [Google Scholar] [CrossRef]
- Shi, Q.; Matthew, C.; Liu, W.H.; Nan, Z.B. Alkaloid contents in Epichloë endophyte-infected Elymus tangutorum sampled along an elevation gradient on the Qinghai-Tibetan Plateau. Agronomy 2020, 10, 1812. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Zhang, Q.W.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K. Review: Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019, 75, 2543–2548. [Google Scholar] [CrossRef]
- Simpson, W.R.; Faville, M.J.; Moraga, R.A.; Williams, W.M.; McManus, M.T.; Johnson, R.D. Epichloë fungal endophytes and the formation of synthetic symbioses in Hordeeae (=Triticeae) grasses. J. Syst. Evol. 2014, 52, 794–806. [Google Scholar] [CrossRef]
- Caradus, J.R.; Card, S.D.; Hewitt, K.G.; Hume, D.E.; Johnson, L.J. Asexual Epichloë fungi—Obligate mutualists. Encyclopedia 2021, 1, 1084–1100. [Google Scholar] [CrossRef]
- Li, C.J.; Wang, Z.F.; Chen, T.X.; and Nan, Z.B. Creation of novel barley germplasm using an Epichloë endophyte. Chin. Sci. Bull. 2021, 66, 2608–2617. [Google Scholar] [CrossRef]

| Host | Location | Longitude | Latitude | Altitude (m) | No. of Samples | No. of Infected Samples | Infection Frequency (%) | No. of Isolated Strains |
|---|---|---|---|---|---|---|---|---|
| Elymus cylindricus | Qinghai, Haixi | 98.5042° E | 36.9185° N | 2924.2 | 71 | 14 | 19.72 | 3 |
| 98.5582° E | 37.0036° N | 3074.2 | 27 | 4 | 14.81 | 2 | ||
| 97.3324° E | 37.3628° N | 2932.5 | 25 | 2 | 8.00 | 1 | ||
| 97.2155° E | 37.3197° N | 2841.8 | 20 | 12 | 60.00 | 2 | ||
| Qinghai, Hainan | 101.3243° E | 35.8219° N | 2906.3 | 60 | 10 | 16.67 | 2 | |
| 101.0984° E | 35.7741° N | 3268.1 | 30 | 6 | 20.00 | 2 | ||
| 100.8079° E | 34.7338° N | 3344.6 | 30 | 2 | 6.67 | 1 | ||
| Qinghai, Guoluo | 100.6322° E | 34.6656° N | 3212.1 | 30 | 4 | 13.33 | 1 | |
| 100.6789° E | 33.0516° N | 3574.8 | 32 | 4 | 12.50 | 1 | ||
| Qinghai, Yushu | 97.0268° E | 32.9638° N | 3694.9 | 17 | 1 | 5.88 | 1 | |
| 96.4477° E | 32.3355° N | 3625.2 | 30 | 2 | 6.67 | 1 | ||
| Elymus tangutorum | Qinghai, Haixi | 97.2155° E | 37.3197° N | 2841.8 | 33 | 3 | 9.09 | 2 |
| 97.7083° E | 36.0147° N | 3004.9 | 10 | 4 | 40.00 | 2 | ||
| 97.8082° E | 36.0430° N | 2981.3 | 28 | 9 | 32.14 | 3 | ||
| 98.1301° E | 36.4181° N | 3071.5 | 18 | 1 | 5.56 | 2 | ||
| Qinghai, Hainan | 100.6789° E | 33.0515° N | 3329.7 | 24 | 3 | 12.50 | 2 | |
| 96.5569° E | 32.6282° N | 3574.8 | 6 | 1 | 16.67 | 1 | ||
| Qinghai, Yushu | 97.0268° E | 32.9638° N | 3694.9 | 23 | 1 | 4.34 | 1 |
| Host | Endophyte | Conidia Size (μm) | Length of Conidiogenous Cell (μm) | Growth Rate (mm/d, 25 °C) | Reference or Source | |
|---|---|---|---|---|---|---|
| Length | Width | |||||
| Elymus cylindricus | YZ1 (ns) 1 | 3.71 ± 0.12 (ef) 1 | 1.68 ± 0.17 (kl) | 9.34 ± 0.38 (kl) | 1.05 ± 0.02 (abcd) | This study |
| YZ2 (ns) | 4.64 ± 0.16 (b) | 2.49 ± 0.16 (fgh) | 10.47 ± 0.18 (j) | 0.88 ± 0.08 (cdef) | ||
| YZ3 (ns) | 4.58 ± 0.11 (bc) | 2.56 ± 0.20 (efgh) | 13.40 ± 0.24 (e) | 1.01 ± 0.08 (abcd) | ||
| YZ4 (ns) | 4.52 ± 0.19 (bc) | 2.56 ± 0.19 (efgh) | 14.37 ± 0.22 (d) | 1.00 ± 0.09 (abcde) | ||
| YZ5 (ns) | 4.60 ± 0.18 (bc) | 2.53 ± 0.21 (efgh) | 15.16 ± 0.22 (c) | 1.03 ± 0.06 (abcd) | ||
| YZ6 (ns) | 3.45 ± 0.13 (g) | 1.51 ± 0.18 (m) | 11.48 ± 0.25 (h) | 1.18 ± 0.06 (a) | ||
| YZ7 (ns) | 3.60 ± 0.13 (fg) | 1.76 ± 0.14 (jk) | 12.38 ± 0.43 (fg) | 0.87 ± 0.23 (def) | ||
| YZ8 (ns) | 3.49 ± 0.23 (g) | 1.27 ± 0.04 (n) | 9.09 ± 0.63 (l) | 0.84 ± 0.05 (defg) | ||
| YZ9 (ns) | 3.53 ± 0.16 (fg) | 1.53 ± 0.04 (lm) | 15.48 ± 0.26 (bc) | 0.66 ± 0.11 (fgh) | ||
| YZ10 (ns) | 3.53 ± 0.15 (fg) | 1.86 ± 0.03 (j) | 10.50 ± 0.22 (i) | 0.67 ± 0.07 (fgh) | ||
| YZ11 (ns) | 4.55 ± 0.17 (bc) | 2.54 ± 0.09 (efgh) | 9.38 ± 0.30 (kl) | 0.64 ± 0.03 (gh) | ||
| YZ12 (ns) | 4.39 ± 0.57 (c) | 2.40 ± 0.13 (hi) | 12.55 ± 0.29 (f) | 0.68 ± 0.03 (fgh) | ||
| YZ13 (ns) | 4.52 ± 0.20 (bc) | 2.45 ± 0.07 (h) | 12.69 ± 0.83 (f) | 0.67 ± 0.03 (fgh) | ||
| YZ14 (ns) | 4.63 ± 0.22 (b) | 2.79 ± 0.07 (b) | 13.37 ± 0.17 (e) | 0.61 ± 0.04 (h) | ||
| YZ15 (ns) | 4.51 ± 0.25 (bc) | 2.51 ± 0.14 (efgh) | 13.75 ± 0.28 (e) | 0.90 ± 0.19 (bcde) | ||
| YZ16 (ns) | 4.39 ± 0.17 (c) | 2.60 ± 0.17 (defg) | 12.59 ± 0.24 (f) | 1.07 ± 0.07 (abcd) | ||
| YZ17 (ns) | 3.59 ± 0.24 (fg) | 2.29 ± 0.15 (i) | 12.67 ± 0.23 (f) | 0.63 ± 0.34 (gh) | ||
| Elymus tangutorm | MB1 (ns) | 5.42 ± 0.21 (a) | 3.28 ± 0.15 (a) | 16.55 ± 0.23 (a) | 0.99 ± 0.35 (abcde) | This study |
| MB2 (ns) | 3.63 ± 0.23 (fg) | 1.66 ± 0.24 (kl) | 15.83 ± 0.60 (b) | 0.91 ± 0.13 (bcde) | ||
| MB3 (ns) | 3.50 ± 0.22 (fg) | 1.79 ± 0.17 (jk) | 15.47 ± 0.23 (bc) | 1.06 ± 0.17 (abcd) | ||
| MB4 (ns) | 5.47 ± 0.17 (a) | 3.37 ± 0.25 (a) | 10.91 ± 0.57 (h) | 1.03 ± 0.09 (abcd) | ||
| MB5 (ns) | 4.50 ± 0.15 (bc) | 2.56 ± 0.19 (efgh) | 12.07 ± 0.63 (g) | 0.93 ± 0.12 (bcde) | ||
| MB6 (ns) | 4.52 ± 0.20 (bc) | 2.51 ± 0.10 (efgh) | 11.61 ± 0.34 (h) | 1.13 ± 0.04 (ab) | ||
| MB7 (ns) | 4.61 ± 0.14 (bc) | 2.43 ± 0.32 (hi) | 9.56 ± 0.29 (k) | 1.02 ± 0.14 (abcd) | ||
| MB8 (ns) | 4.50 ± 0.09 (bc) | 2.63 ± 0.15 (cdef) | 12.47 ± 0.15 (f) | 1.01 ± 0.13 (abcd) | ||
| MB9 (ns) | 4.18 ± 0.35 (d) | 2.66 ± 0.13 (bcde) | 12.51 ± 0.21 (f) | 1.01 ± 0.04 (abcd) | ||
| MB10 (ns) | 5.34 ± 0.21 (a) | 2.55 ± 0.16 (efgh) | 10.49 ± 0.29 (j) | 0.90 ± 0.06 (bcde) | ||
| MB11 (ns) | 4.59 ± 0.16 (bc) | 2.60 ± 0.17 (defg) | 11.64 ± 0.74 (h) | 1.02 ± 0.04 (abcd) | ||
| MB12 (ns) | 4.58 ± 0.14 (bc) | 2.76 ± 0.08 (bcd) | 14.37 ± 0.72 (d) | 1.11 ± 0.05 (abc) | ||
| MB13 (ns) | 3.86 ± 0.14 (e) | 2.77 ± 0.07 (bc) | 13.64 ± 0.52 (e) | 0.78 ± 0.21 (efg) | ||
| Bromus ramosus (Bromus benekenii) | Epichloë bromicola (ns) | 4.2 ± 0.5 | 2.0 ± 0.3 | nt | 0.90 | [39] |
| Bromus erectum | Epichloë bromicola (s) 1 | 3.8 ± 0.4 | 2.0 ± 0.3 | 8–23 | 2.29–2.48 (PDA, 24 °C) | [39] |
| Hordeum brevisubulatum | Epichloë bromicola WBE1 (ns) | 5.17 ± 0.06 | 2.87 ± 0.17 | 19.50 ± 1.06 | 0.88 ± 0.01 (PDA, 25 °C) | [42] |
| Hordelymus europaeus | Epichloë bromicola (ns) | 4.2 ± 0.4 | 2.1 ± 0.2 | 20.2 ± 4.7 | 1.43–1.67 (PDA, 24 °C) | [43] |
| Hordeum bogdanii | Epichloë bromicola (ns) | 4.6 ± 0.4 | 2.7 ± 0.3 | 14.0 ± 3.5 | 1.21 ± 0.1 (PDA, 25 °C) | [44] |
| Leymus chinensis | Epichloë bromicola (s) | 5.3 ± 0.1 | 3.5 ± 0.1 | 29.0–31.0 | 1.7 ± 0.07 (PDA, 25 °C) | [45] |
| Psathyrostachys lanuginosa | Epichloë bromicola PF9 (ns) | 3.6 ± 0.07 | 1.8 ± 0.04 | 12.3 ± 0.07 | 2.23 ± 0.05 (PDA, 25 °C) | [46] |
| Roegneria kamoji | Epichloë bromicola (s) | 4.7–5.2 | 2.0–2.9 | 16.5–25.8 | 0.83–2.58 (PDA, 25 °C) | [47,48] |
| Elymus cylindricus | Epichloë bromicola AD3 (ns) | 4.16 ± 0.07 | nt | 10.75 ± 0.76 | 1.01 ± 0.04 (PDA, 25 °C) | [36] |
| Elymus dahuricus | Epichloë bromicola LE1 (ns) | 3.33 ± 0.09 | nt | 11.85 ± 0.70 | 2.32 ± 0.06 (PDA, 25 °C) | [36] |
| Elymus dahuricus | Epichloë bromicola (s) | 4.5–6.5 | 2.5–4 | 9–27 | 0.78–1.05 (PDA, 25 °C) | [49] |
| Elymus nutans | Epichloë bromicola GA2 (ns) | 3.75 ± 0.10 | nt | 10.76 ± 0.47 | 0.83 ± 0.03 (PDA, 25 °C) | [36] |
| Elymus sibiricus | Epichloë bromicola FC1 (ns) | 3.78 ± 0.09 | nt | 13.59 ± 0.91 | 1.13 ± 0.03 (PDA, 25 °C) | [36] |
| Elymus tangutorum | Epichloë bromicola KE1 (ns) | 5.09 ± 0.13 | nt | 13.33 ± 0.62 | 1.29 ± 0.09 (PDA, 25 °C) | [36] |
| Elymus virginicus | Epichloë elymi (s) | 4.0 ± 0.4 | 2.2 ± 0.2 | 17.0 ± 3.0 | 1.95–2.85, (PDA, 24 °C) | [50] |
| Elymus canadensis | Epichloë canadensis (ns) | 5.8–8 | 2.5–4.0 | 12.5–41.5 | 0.96–1.71, (PDA, 23 °C) | [51] |
| Elymus nutans | Epichloë glyceriae (ns) | 3.8–6.2 | 2.2–2.8 | 31.0 ± 5.0 | 2.47–3, (PDA, 24 °C) | [50] |
| Endophyte Isolate | Mating Type Genes | Segments of ppzA Gene | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mtAC | mtBA | A1 | T1 | C | A2 | M | T2 | R | ΔR | |
| YZ8, MB8, MB11 | + | - | - | - | - | - | - | - | - | - |
| YZ1, MB1 | + | - | - | - | - | - | - | - | - | + |
| YZ11, YZ13, MB13 | + | - | - | - | + | - | - | - | - | - |
| YZ9, YZ14 | + | - | - | - | - | - | - | + | - | + |
| YZ12, MB12 | + | - | - | + | - | - | - | - | - | + |
| YZ7, MB7, MB9 | + | - | - | - | + | - | - | + | - | + |
| YZ2, YZ4, MB2, MB4 | + | - | - | - | + | + | - | + | - | + |
| YZ10 | + | - | + | - | - | - | - | + | + | + |
| YZ5, MB5 | + | - | + | - | + | + | - | + | - | + |
| YZ15 | + | - | - | - | + | + | - | + | + | + |
| MB10 | + | - | + | - | + | - | - | + | + | + |
| MB6 | + | - | + | + | + | + | + | + | - | + |
| YZ3, YZ6, YZ16, YZ17, MB3 | + | - | + | + | + | + | + | + | + | + |
| Endophye | Ergot Alkaloid (EAS) Genes | Predicted Ergot-Alkaloid-Producing Type | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dmaW | easF | easC | easE | easD | easA | easG | cloA | lpsB | lpsA | easH | lpsC | easO | easP | ||
| YZ5 | + | - | + | - | - | - | - | - | - | - | + | - | - | - | --- |
| MB5 | + | + | + | + | - | + | - | - | - | - | - | - | - | - | CC |
| YZ9 | + | - | + | + | + | + | + | + | - | - | - | - | - | - | --- |
| YZ13 | + | + | + | + | + | - | - | + | - | + | + | - | - | - | CC |
| MB6 | + | + | + | + | + | + | + | + | - | - | - | - | - | - | CC, D-LA |
| MB12 | + | + | + | + | + | + | - | + | + | - | - | - | - | - | CC |
| MB13 | + | + | + | + | + | + | - | + | - | + | - | - | - | - | CC |
| YZ8 | + | - | + | + | + | + | + | + | + | - | + | - | - | - | --- |
| YZ12 | + | + | + | + | + | + | - | + | - | + | + | - | - | - | CC |
| YZ14 | + | - | + | + | + | + | + | + | - | + | + | - | - | - | --- |
| MB2 | + | + | + | + | + | + | + | + | + | - | - | - | - | - | CC, D-LA |
| YZ7 | + | - | + | + | + | + | + | + | + | + | + | - | - | - | --- |
| YZ11 | + | + | + | + | + | + | + | + | + | - | + | - | - | - | CC, D-LA |
| MB3, MB7, MB11 | + | + | + | + | + | + | + | + | - | + | + | - | - | - | CC, D-LA |
| MB4 | + | + | + | + | + | + | - | + | + | + | + | - | - | - | CC |
| MB8 | + | + | + | + | + | + | + | + | + | + | - | - | - | - | CC, D-LA |
| YZ1, YZ2, YZ3, YZ4, YZ6, YZ10, YZ15, YZ16, YZ17, MB1, MB9, MB10 | + | + | + | + | + | + | + | + | + | + | + | - | - | - | CC, D-LA, ERV |
| Endophyte | Indole-Diterpene (IDT/LTM) Genes | Predicted Indole-Diterpene-Producing Type | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| idtG | idtB | idtM | idtC | idtS | idtP | idtQ | idtF | idtK | idtE | idtJ | ||
| MB5, MB6, MB7 | + | - | - | - | - | - | - | - | - | - | - | --- |
| YZ9 | - | + | - | - | + | - | - | - | - | - | - | --- |
| YZ14 | + | + | - | - | - | - | - | - | - | - | - | --- |
| YZ1, YZ2 | - | + | - | + | + | - | - | - | - | - | - | --- |
| YZ8 | + | + | - | - | - | - | - | + | - | - | - | --- |
| MB1 | - | + | - | - | + | - | - | + | - | - | - | --- |
| MB9 | + | + | - | - | - | - | - | + | - | - | - | --- |
| MB12 | - | + | - | - | + | - | - | + | - | - | - | --- |
| MB13 | + | - | - | + | - | - | - | + | - | - | - | --- |
| YZ4 | - | + | - | + | + | - | - | - | + | - | - | --- |
| YZ7 | + | + | - | + | + | - | - | - | - | - | - | --- |
| YZ15 | + | + | - | - | + | - | - | + | - | - | - | --- |
| MB4 | + | + | - | - | - | - | - | + | + | - | - | --- |
| YZ3, YZ5 | - | + | + | + | + | - | - | - | + | - | - | --- |
| YZ13 | + | + | - | + | - | - | + | + | - | - | - | --- |
| MB2 | + | - | - | + | + | - | - | + | + | - | - | --- |
| YZ10, YZ11, YZ12 | + | + | - | + | + | - | - | + | + | - | - | --- |
| YZ6 | + | + | + | + | - | - | + | + | + | - | - | --- |
| MB3 | - | + | + | + | + | - | + | + | + | - | - | --- |
| MB8, MB11 | + | + | - | + | + | - | + | + | + | - | - | --- |
| YZ16, YZ17, MB10 | + | + | + | + | + | - | + | + | + | - | - | paspaline |
| Type | Total No. of Strains | Mating Type Genes | Endophyte | Epichloë Species | Predicted Alkaloid-Producing Type |
|---|---|---|---|---|---|
| I | 3 | mtAC | YZ16, YZ17, MB10 | E. bromicola | PAS + CC + D-LA + ERV |
| II | 9 | mtAC | YZ1, YZ2, YZ3, YZ4, YZ6, YZ10, YZ15, MB1, MB9 | E. bromicola | CC + D-LA + ERV |
| III | 7 | mtAC | YZ11, MB2, MB3, MB6, MB7, MB8, MB11 | E. bromicola | CC + D-LC |
| IV | 6 | mtAC | YZ12, YZ13, MB4, MB5, MB12, MB13 | E. bromicola | CC |
| V | 5 | mtAC | YZ5, YZ7, YZ8, YZ9, YZ14 | E. bromicola | --- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Liu, W.; Huang, K.; Bao, G.; Li, C. Morphological, Molecular, and Alkaloid Gene Profiling of Epichloë Endophytes in Elymus cylindricus and Elymus tangutorum from China. Microorganisms 2025, 13, 2275. https://doi.org/10.3390/microorganisms13102275
Chen T, Liu W, Huang K, Bao G, Li C. Morphological, Molecular, and Alkaloid Gene Profiling of Epichloë Endophytes in Elymus cylindricus and Elymus tangutorum from China. Microorganisms. 2025; 13(10):2275. https://doi.org/10.3390/microorganisms13102275
Chicago/Turabian StyleChen, Taixiang, Wencong Liu, Kai Huang, Gensheng Bao, and Chunjie Li. 2025. "Morphological, Molecular, and Alkaloid Gene Profiling of Epichloë Endophytes in Elymus cylindricus and Elymus tangutorum from China" Microorganisms 13, no. 10: 2275. https://doi.org/10.3390/microorganisms13102275
APA StyleChen, T., Liu, W., Huang, K., Bao, G., & Li, C. (2025). Morphological, Molecular, and Alkaloid Gene Profiling of Epichloë Endophytes in Elymus cylindricus and Elymus tangutorum from China. Microorganisms, 13(10), 2275. https://doi.org/10.3390/microorganisms13102275






