In Vitro Evaluation of Halotolerant Bacillus velezensis 24.5 as a Promising Probiotic with Broad-Spectrum Antimicrobial Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Bacterial Strains and Growth Conditions
2.2. Molecular Characterization of 24.5
2.2.1. Molecular Identification of Strain 24.5
2.2.2. Detection of Bioactive Metabolite-Encoding Genes
2.3. Functional Properties of 24.5 Strains
2.3.1. Antimicrobial Activity
2.3.2. Gastrointestinal Tolerance Assay
2.3.3. Antioxidant Effect
2.3.4. Enzymatic Activity Assays
2.3.5. Assessment of Cell Surface Properties of Bacillus velezensis 24.5
2.4. Safety Assessment
2.4.1. Haemolytic Activity Assay
2.4.2. Antibiotic Susceptibility Testing
2.5. Statistical Analysis
3. Results
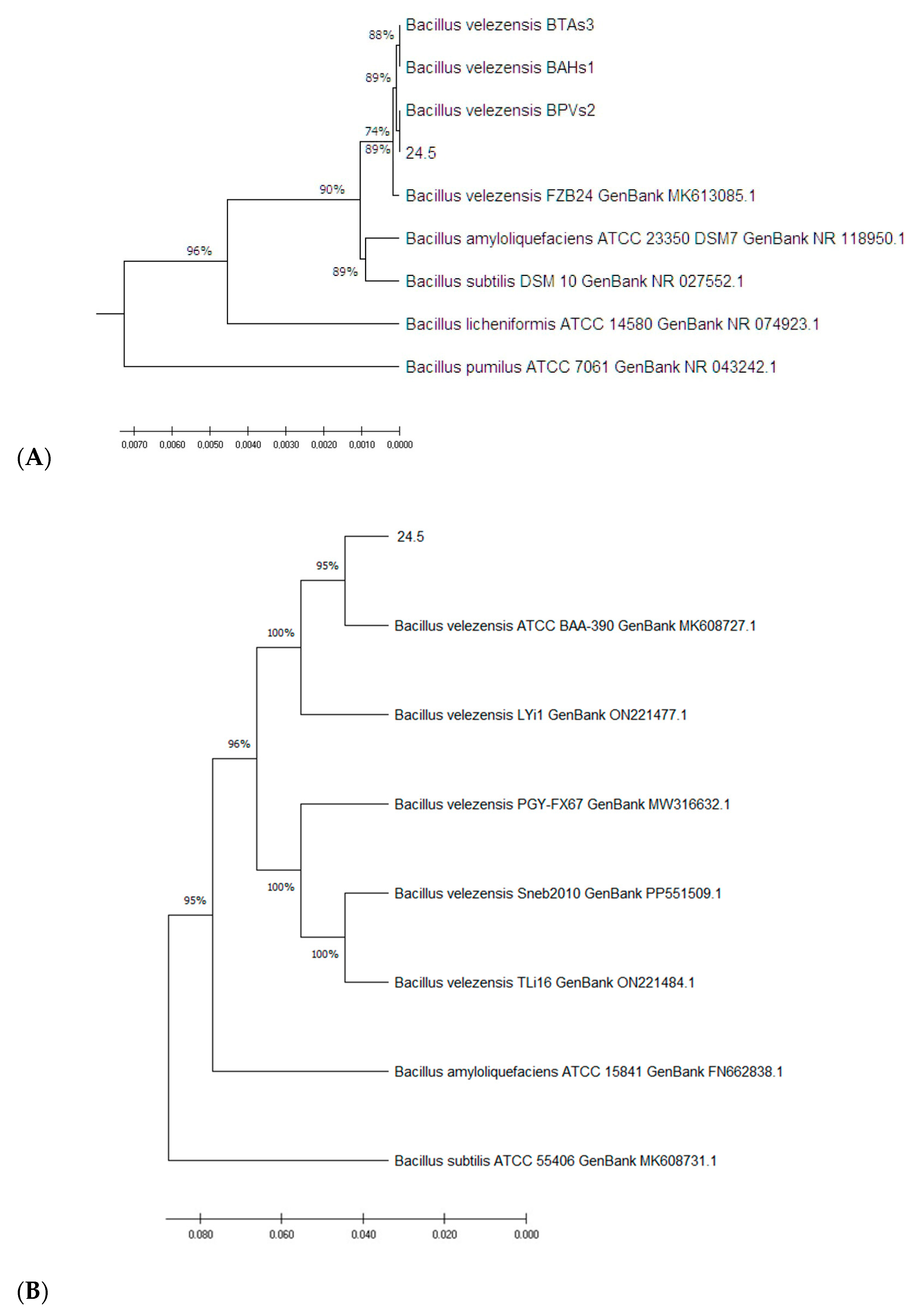
3.1. Molecular Characterization of the 24.5 Strain
3.1.1. Molecular Identification
3.1.2. PCR Detection of Biosynthetic Genes for Secondary Metabolites
3.2. Probiotic Patterns of Strain 24.5
3.2.1. Antimicrobial Pattern
3.2.2. Resistance to Simulated Gastrointestinal Conditions
3.2.3. Antioxidant Activity
3.2.4. Enzymatic Profile of Strain 24.5
3.2.5. Cell Surface Properties of Halotolerant Strain 24.5
3.3. Safety Traits of Strain 24.5
3.3.1. Haemolytic Activity
3.3.2. Antibiotic Susceptibility Profile
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO/WHO. Guidelines for the Evaluation of Probiotics in Food. In Proceedings of the Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food, London, ON, Canada, 30 April–1 May 2002. [Google Scholar]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Das, T.K.; Pradhan, S.; Chakrabarti, S.; Mondal, K.C.; Ghosh, K. Current status of probiotic and related health benefits. Appl. Food Res. 2022, 2, 100185. [Google Scholar] [CrossRef]
- Dobreva, L.; Atanasova, N.; Donchev, P.; Krumova, E.; Abrashev, R.; Karakirova, Y.; Mladenova, R.; Tolchkov, V.; Ralchev, N.; Dishliyska, V.; et al. Candidate-Probiotic Lactobacilli and Their Postbiotics as Health-Benefit Promoters. Microorganisms 2024, 12, 1910. [Google Scholar] [CrossRef]
- Maftei, N.-M.; Raileanu, C.R.; Balta, A.A.; Ambrose, L.; Boev, M.; Marin, D.B.; Lisa, E.L. The Potential Impact of Probiotics on Human Health: An Update on Their Health-Promoting Properties. Microorganisms 2024, 12, 234. [Google Scholar] [CrossRef]
- Baliyan, N.; Kumari, M.; Kumari, P.; Dindhoria, K.; Mukhia, S.; Kumar, S.; Kumar, R. Probiotics in fermented products and supplements. In Current Developments in Bioengineering; Elsevier: Amsterdam, The Netherlands, 2022; pp. 73–107. [Google Scholar]
- Pompa, L.; Montanari, A.; Tomassini, A.; Bianchi, M.M.; Aureli, W.; Miccheli, A.; Uccelletti, D.; Schifano, E. In Vitro Probiotic Properties and In Vivo Anti-Ageing Effects of Lactoplantibacillus plantarum PFA2018AU Strain Isolated from Carrots on Caenorhabditis elegans. Microorganisms 2023, 11, 1087. [Google Scholar] [CrossRef] [PubMed]
- Mariarosaria, M. Bifidobacteria, Lactobacilli… when, how and why to use them. Glob. Pediatr. 2024, 8, 100139. [Google Scholar] [CrossRef]
- Pristavu, M.-C.; Diguță, F.C.; Aldea, A.C.; Badea, F.; Dragoi Cudalbeanu, M.; Ortan, A.; Matei, F. Functional Profiling of Enterococcus and Pediococcus Strains: An In Vitro Study on Probiotic and Postbiotic Properties. Microorganisms 2025, 13, 1348. [Google Scholar] [CrossRef] [PubMed]
- Pais, P.; Almeida, V.; Yılmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What makes it tick as successful probiotic? J. Fungi 2020, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pacheco, P.; Pintado, C.; Briones Pérez, A.; Arévalo-Villena, M. Potential Probiotic Strains of Saccharomyces and Non-Saccharomyces: Functional and Biotechnological Characteristics. J. Fungi 2021, 7, 177. [Google Scholar] [CrossRef]
- Fernández-Pacheco, P.; Ramos Monge, I.M.; Fernández-González, M.; Poveda Colado, J.M.; Arévalo-Villena, M. Safety Evaluation of Yeasts With Probiotic Potential. Front. Nutr. 2021, 8, 659328. [Google Scholar] [CrossRef]
- Vrînceanu, C.-R.; Diguță, F.C.; Cudalbeanu, M.D.; Ortan, A.; Mihai, C.; Bărbulescu, I.D.; Frîncu, M.; Begea, M.; Matei, F.; Teodorescu, R.I. Exploring the Potential of Torulaspora delbrueckii, Starmerella bacillaris, and Saccharomyces cerevisiae as a Probiotic Starter for Craft Beer Production. Foods 2025, 14, 1608. [Google Scholar] [CrossRef]
- Tegegne, B.A.; Kebede, B. Probiotics, their prophylactic and therapeutic applications in human health development: A review of the literature. Heliyon 2022, 8, e09725. [Google Scholar] [CrossRef] [PubMed]
- Evivie, S.E.; Huo, G.C.; Igene, J.O.; Bian, X. Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr. Res. 2017, 61, 1318034. [Google Scholar] [CrossRef]
- Liang, D.; Wu, F.; Zhou, D.; Tan, B.; Chen, T. Commercial probiotic products in public health: Current status and potential limitations. Crit. Rev. Food Sci. Nutr. 2024, 64, 6455–6476. [Google Scholar] [CrossRef]
- Su, T.; Shen, B.; Hu, X.; Teng, Y.; Weng, P.; Wu, Z.; Liu, L. Research advance of Bacillus velezensis: Bioinformatics, characteristics, and applications. Food Sci. Hum. Wellness 2024, 13, 1756–1766. [Google Scholar] [CrossRef]
- Fan, B.; Blom, J.; Klenk, H.-P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an “Operational Group B. amyloliquefaciens” within the B. subtilis Species Complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef]
- Payne, J.; Bellmer, D.; Jadeja, R.; Muriana, P. The Potential of Bacillus Species as Probiotics in the Food Industry: A Review. Foods 2024, 13, 2444. [Google Scholar] [CrossRef] [PubMed]
- Khalid, F.; Khalid, A.; Fu, Y.; Hu, Q.; Zheng, Y.; Khan, S.; Wang, Z. Potential of Bacillus velezensis as a Probiotic in Animal Feed: A Review. J. Microbiol. 2021, 59, 627–633. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Gebrechristos, S.; Garvey, S.M.; Spears, J.L. Genetic and Phenotypic Characterization of Bacillus velezensis Strain BV379 for Human Probiotic Applications. Microorganisms 2024, 12, 436. [Google Scholar] [CrossRef]
- Lu, S.; Na, K.; Li, Y.; Zhang, L.; Fang, Y.; Guo, X. Bacillus-derived probiotics: Metabolites and mechanisms involved in bacteria–host interactions. Crit. Rev. Food Sci. Nutr. 2024, 64, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Guimarães Sousa, E.; Munis Campos, G.; Quaresma, L.S.; Mendonça Mota, T.F.; de Castilhos Ghisi, N.; Camargos Gomes, G.; Viegas Santos, R.C.V.; de Souza, B.G.R.; Guédon, É.; de Castro Soares, S.; et al. Exploring Bacillus velezensis in biomedical context: A systematic review. Acad. Mol. Biol. Genom. 2025, 2. [Google Scholar] [CrossRef]
- FDA GRN. GRAS Notification 1131. Available online: https://www.fda.gov/media/175167/download (accessed on 7 July 2025).
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 11: Suitability of taxonomic units notified to EFSA until September 2019. EFSA J. 2020, 18, 5965. [Google Scholar]
- Vörös, M.; Manczinger, L.; Kredics, L.; Szekeres, A.; Shine, K.; Alharbi, N.S.; Khaled, J.M.; Vágvölgyi, C. Influence of agro-environmental pollutants on a biocontrol strain of Bacillus velezensis. Microbiol. Open 2019, 8, e00660. [Google Scholar] [CrossRef]
- Boiu-Sicuia, O.A.; Cornea, C.P. Bacterial strains involved in soilborne phytopathogens inhibition. Sci. Pap. Ser. A Agron. 2021, 64, 641–646. [Google Scholar]
- Boiu-Sicuia, O.-A.; Toma, R.C.; Diguță, C.F.; Matei, F.; Cornea, C.P. In Vitro Evaluation of Some Endophytic Bacillus to Potentially Inhibit Grape and Grapevine Fungal Pathogens. Plants 2023, 12, 2553. [Google Scholar] [CrossRef]
- Proca, I.; Diguta, F.C.; Cornea, C.P.; Jurcoane, S.; Matei, F. Halotolerant Bacillus amyloliquefaciens 24.5 is helpful as a biological agent to control phyto-pathogenic fungi. Rom. Biotechnol. Lett. 2020, 25, 1744–1753. [Google Scholar]
- Chen, B.; Zeng, Y.; Wang, J.; Lei, M.; Gan, B.; Wan, Z.; Wu, L.; Luo, G.; Cao, S.; An, T.; et al. Targeted Screening of Fiber Degrading Bacteria with Probiotic Function in Herbivore Feces. Probiotics Antimicrob. Proteins 2025, 17, 1473–1497. [Google Scholar] [CrossRef]
- Sam-On, M.F.S.; Mustafa, S.; Hashim, A.M.; Malek, A.Z.A. Probiogenomic insights into Bacillus velezensis MFSS1 for controlling aquaculture pathogens. Microb. Pathog. 2025, 205, 107645. [Google Scholar] [CrossRef]
- da Rosa, C.E.; Pinilla, C.M.B.; Toss, L.D.; Brandelli, A. In Silico and In Vitro Characterization of Bacillus velezensis P45: Screening for a Novel Probiotic Candidate. Foods 2025, 14, 2334. [Google Scholar] [CrossRef]
- Harwood, C.R.; Mouillon, J.-M.; Pohl, S.; Arnau, J. Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 2018, 42, 721–738. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.-h. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed]
- Ngalimat, M.S.; Yahaya, R.S.R.; Baharudin, M.M.A.-a.; Yaminudin, S.M.; Karim, M.; Ahmad, S.A.; Sabri, S. A Review on the Biotechnological Applications of the Operational Group Bacillus amyloliquefaciens. Microorganisms 2021, 9, 614. [Google Scholar] [CrossRef]
- Xiong, Q.; Liu, D.; Zhang, H.; Dong, X.; Zhang, G.; Liu, Y.; Zhang, R. Quorum sensing signal autoinducer-2 promotes root colonization of Bacillus velezensis SQR9 by affecting biofilm formation and motility. Appl. Microbiol. Biotechnol. 2020, 104, 7177–7185. [Google Scholar] [CrossRef] [PubMed]
- Alenezi, F.N.; Slama, H.B.; Bouket, A.C.; Cherif-Silini, H.; Silini, A.; Luptakova, L.; Nowakowska, J.A.; Oszako, T.; Belbahri, L. Bacillus velezensis: A Treasure House of Bioactive Compounds of Medicinal, Biocontrol and Environmental Importance. Forests 2021, 12, 1714. [Google Scholar] [CrossRef]
- Nicolas, G.M. Secondary Metabolites from Bacillus spp. probiotics as potential treatments for multidrug-resistant pathogens: A comprehensive review. Curr. Res. Microb. Sci. 2025, 8, 100392. [Google Scholar] [CrossRef] [PubMed]
- Danilova, I.; Sharipova, M. The practical potential of bacilli and their enzymes for industrial production. Front. Microbiol. 2020, 11, 1782. [Google Scholar] [CrossRef]
- Li, C.; Li, S.; Dang, G.; Jia, R.; Chen, S.; Deng, X.; Liu, G.; Beckers, Y.; Cai, H. Screening and characterization of Bacillus velezensis LB-Y-1 toward selection as a potential probiotic for poultry with multi-enzyme production property. Front. Microbiol. 2023, 14, 1143265. [Google Scholar] [CrossRef]
- Prazdnova, E.V.; Mazanko, M.S.; Shevchenko, V.N.; Skripnichenko, R.V.; Kulikov, M.P.; Golovko, L.S.; Grigoriev, V.A.; Maltseva, T.A.; Kulikova, D.B.; Rudoy, D.V. Genomic Characterization of Four Novel Probiotic Strains with Enzymatic Activity and Their Effects on Carp (Cyprinus carpio). Animals 2025, 15, 1998. [Google Scholar] [CrossRef]
- Hrabar, J.; Babić, I.; Jozić, S.; Trumbić, Ž.; Pioppi, A.; Nielsen, L.J.D.; Maravić, A.; Tomašević, T.; Kovacs, Á.T. Mladineo, I. Prospecting microbiota of Adriatic fish: Bacillus velezensis as a potential probiotic candidate. Anim. Microbiome 2025, 7, 64. [Google Scholar] [CrossRef]
- Chun, J.; Bae, K.S. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 2000, 78, 123–127. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tapi, A.; Chollet-Imbert, M.; Scherens, B.; Jacques, P. New approach for the detection of non-ribosomal peptide synthetase genes in Bacillus strains by polymerase chain reaction. Appl. Microbiol. Biotechnol. 2010, 85, 1521–1531. [Google Scholar] [CrossRef]
- Chung, S.; Kong, H.; Buyer, J.S.; Lakshman, D.K.; Lydon, J.; Kim, S.-D.; Roberts, D.P. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soilborne pathogens of cucumber and pepper. Appl. Microbiol. Biotechnol. 2008, 80, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Athukorala, S.N.P.; Dilantha, F.W.G.; Khalid, Y.R. Identification of antifungal antibiotics of Bacillus species isolated from different microhabitats using polymerase chain reaction and MALDI-TOF mass spectrometry. Can. J. Microbiol. 2009, 55, 1021–1032. [Google Scholar] [CrossRef]
- Compaoré, C.S.; Nielsen, D.S.; Sawadogo-Lingani, H.; Berner, T.S.; Nielsen, K.F.; Adimpong, D.B.; Diawara, B.; Ouédraogo, G.A.; Jakobsen, M.; Thorsen, L. Bacillus amyloliquefaciens ssp. plantarum strains as potential protective starter cultures for the production of Bikalga, an alkaline fermented food. J. Appl. Microbiol. 2013, 115, 133–146. [Google Scholar]
- Toma, C.T.; Boiu-Sicuia, O.-A.; Diguță, C.F.; Ciucă, M.; Matei, F.; Cornea, C.P. Selected plant protection Bacillus strains increase food safeness by inhibiting human pathogenic bacteria. Rom. Agric. Res. 2023, 40, 609–619. [Google Scholar] [CrossRef]
- Borah, T.; Gogoi, B.; Khataniar, A.; Gogoi, M.; Das, A.; Borah, D. Probiotic characterization of indigenous Bacillus velezensis strain DU14 isolated from Apong, a traditionally fermented rice beer of Assam. Biocatal. Agric. Biotechnol. 2019, 18, 101008. [Google Scholar] [CrossRef]
- Coulibaly, W.H.; Kouadio, N.R.; Camara, F.; Diguță, C.; Matei, F. Functional properties of lactic acid bacteria isolated from Tilapia (Oreochromis niloticus) in Ivory Coast. BMC Microbiol. 2023, 23, 152. [Google Scholar] [CrossRef] [PubMed]
- Proca, I.G.; Diguta, C.F.; Jurcoane, S.; Matei, F. Screening of halotolerant bacteria producing hydrolytic enzymes with biotechnological applications. Sci. Bull. Ser. F Biotechnol. 2020, XXIV, 197–204. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI document M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Amerikova, M.; Pencheva El-Tibi, I.; Maslarska, V.; Bozhanov, S.; Tachkov, K. Antimicrobial activity, mechanism of action, and methods for stabilisation of defensins as new therapeutic agents. Biotechnol. Biotechnol. Equip. 2019, 33, 671–682. [Google Scholar] [CrossRef]
- Elsadek, M.M.; Wang, S.; Wu, Z.; Wang, J.; Wang, X.; Zhang, Y.; Yu, M.; Guo, Z.; Wang, Q.; Wang, G.; et al. Characterization of Bacillus spp. isolated from the intestines of Rhynchocypris lagowskii as a potential probiotic and their effects on fish pathogens. Microb. Pathog. 2023, 180, 106163. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh Behbahani, B.; Noshad, M.; Vasiee, A.; Brück, W.M. Probiotic Bacillus Strains Inhibit Growth, Biofilm Formation, and Virulence Gene Expression of Listeria monocytogenes. LWT 2024, 191, 115596. [Google Scholar] [CrossRef]
- Nwagu, T.N.; Ugwuodo, C.J.; Onwosi, C.O.; Inyima, O.; Uchendu, O.C.; Akpuru, C. Evaluation of the probiotic attributes of Bacillus strains isolated from traditional fermented African locust bean seeds (Parkia biglobosa), “daddawa”. Ann. Microbiol. 2020, 70, 20. [Google Scholar] [CrossRef]
- Arslan, N.P.; Azad, F.; Orak, T.; Budak-Savas, A.; Ortucu, S.; Dawar, P.; Baltaci, M.O.; Ozkan, H.; Esim, N.; Taskin, M. A review on bacteria-derived antioxidant metabolites: Their production, purification, characterization, potential applications, and limitations. Arch. Pharm. Res. 2025, 48, 253–292. [Google Scholar] [CrossRef]
- Khan, M.M.; Kim, Y.K.; Bilkis, T.; Suh, J.-W.; Lee, D.Y.; Yoo, J.C. Reduction of Oxidative Stress through Activating the Nrf2 mediated HO-1 Antioxidant Efficacy Signaling Pathway by MS15, an Antimicrobial Peptide from Bacillus velezensis. Antioxidants 2020, 9, 934. [Google Scholar] [CrossRef]
- Shivangi, S.; Devi, P.B.; Ragul, K.; Shetty, P.H. Probiotic Potential of Bacillus Strains Isolated from an Acidic Fermented Food Idli. Probiotics Antimicrob. Proteins 2020, 12, 1502–1513. [Google Scholar] [CrossRef]
- Kang, M.; Su, X.; Yun, L.; Shen, Y.; Feng, J.; Yang, G.; Meng, X.; Zhang, J.; Chang, X. Evaluation of probiotic characteristics and whole genome analysis of Bacillus velezensis R-71003 isolated from the intestine of common carp (Cyprinus carpio L.) for its use as a probiotic in aquaculture. Aquac. Rep. 2022, 25, 101254. [Google Scholar] [CrossRef]
- Pełka, K.; Bin Hafeez, A.; Worobo, R.W.; Szweda, P. Probiotic Potential of Bacillus Isolates from Polish Bee Pollen and Bee Bread. Probiotics Antimicrob. Proteins 2023, 17, 364–377. [Google Scholar] [CrossRef]

| Secondary Metabolite Groups | Bioactive Metabolites | Genes | Primers | Primer Sequence 5′-3′ | Alignment Temperature | References |
|---|---|---|---|---|---|---|
| Lipopeptides | Fengycin | fen | Af2 F | GAATAYMTCGGMCGTMTKGA | 45 °C | [46] |
| Tf1 R | GCTTTWADKGAATSBCCGCC | |||||
| Iturin | ituD | ITUD F1 | TTGAAYGTCAGYGCSCCTTT | 55 °C | [47] | |
| ITUD R1 | TGCGMAAATAATGGSGTCGT | |||||
| Iturin | ituC | ITUC F1 | CCCCCTCGGTCAAGTGAATA | 55 °C | [47] | |
| ITUC R1 | TTGGTTAAGCCCTGATGCTC | |||||
| Iturin A | ituA | ITUD1 F ITUD1 R | GATGCGATCTCCTTGGATGT | 55 °C | [48] | |
| ATCGTCATGTGCTGCTTGAG | ||||||
| Surfactin | srf/lch | As1 F | CGCGGMTACCGVATYGAGC | 43 °C | [46] | |
| Ts2 R | ATBCCTTTBTWDGAATGTCCGCC | |||||
| Surfactin | srfA | SrfA F | AGAGCACATTGAGCGTTACAAA | 55 °C | [47] | |
| SrfA R | CAGCATCTCGTTCAACTTTCAC | |||||
| Bacillomycin | bmyA | bmyA F | CTCATTGCTGCCGCTCAATC | 55 °C | [49] | |
| bmyA R | CCGAATCTACGAGGGGAACG | |||||
| Mycosubtilin | myc | Am1 F | CAKCARGTSAAAATYCGMGG | 45 °C | [47] | |
| Tm1 R | CCDASATCAAARAADTTATC | |||||
| Polyketides | Difficidin | dfnA | dfnA F | GGATTCAGGAGGGCATACCG | 55 °C | [49] |
| dfnA R | ATTGATTAAACGCGCCGAGC | |||||
| Macrolactin | mnlA | mlnA F | CCGTGATCGGACTGGATGAG | 55 °C | [49] | |
| mlnA R | CATCGCACCTGCCAAATACG | |||||
| Bacillaene | baeA | BaeR F | ATGTCAGCTCAGTTTCCGCA | 55 °C | [49] | |
| BaeR R | GATCGCCGTCTTCAATTGCC | |||||
| Dipeptide | Bacilysin | bacA/B bac B | baeA/B F | TGCTCTGTTATAGCGCGGAG | 55 °C | [49] |
| baeA/B R | GTCATCGTATCCCACCCGTC |
| Secondary Metabolite Groups | Bioactive Metabolites | Genes | Expected Product Size (bp) | Detected PCR Product |
|---|---|---|---|---|
| Lipopeptides | Fengycin | fen | 443, 452 | yes * |
| Iturin | ituD | 482 | yes * | |
| Iturin | ituC | 594 | no | |
| Iturin A | ituA | 647 | yes * | |
| Surfactin | srf/lch | 419–431 | yes * | |
| Surfactin | srfA | 626 | yes | |
| Bacillomycin | bmyA | 853 | yes * | |
| Mycosubtilin | myc | 416, 419 | yes | |
| Polyketides | Difficidin | dfnA | 653 | yes |
| Macrolactin | mnlA | 668 | yes | |
| Bacillaene | baeA | 688 | yes | |
| Dipeptides | Bacilysin | bacA/B bac B | 910 | yes |
| Bacteria Gram-Positive | Bacteria Gram-Negative | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| Pathogens | Aspect | Inhibition Halo (mm) | Pathogens | Aspect | Inhibition Halo (mm) | Candida | Aspect | Inhibition Halo (mm) |
| B. cereus ATCC 11778 |  | 2.00 ± 0.00 f | E. coli ATCC 11229 |  | 0.00 ± 0.00 g | C. albicans ATCC 10231 |  | 15.33 ± 0.94 b |
| Ent. faecium ATCC 6057 |  | 9.00 ± 0.82 cd | E. coli ATCC 8739 |  | 0.00 ± 0.00 g | C. glabrata ATCC 2001 |  | 3.00 ± 0.82 f |
| Ent. faecalis ATCC 29212 |  | 2.33 ± 0.47 f | Ps. aeruginosa ATCC 15442 |  | 1.00 ± 0.00 f | C. parapsilopsis ATCC 20019 |  | 8.33 ± 0.47 cd |
| Ent. hirae ATCC 10541 |  | 5.33 ± 0.47 e | Ps. aeruginosa ATCC 27853 |  | 2.00 ± 0.00 f | C. tropicalis ATCC 44508 |  | 0.00 ± 0.00 g |
| L. ivanovii ATCC 19119 |  | 4.00 ± 0.00 ef | Pr. vulgaris ATCC 13315 |  | 5.67 ± 0.47 de | |||
| L. monocytogens ATCC 7644 |  | 9.00 ± 0.00 cd | S. enterica Typhimurium ATCC 14028 |  | 1.67 ± 0.47 f | |||
| L. innocua ATCC 33090 |  | 1.00 ± 0.00 f | S. enterica Enteritidis ATCC 13076 |  | 2.67 ± 0.47 f | |||
| Staph. aureus ATCC 25923 |  | 10.67 ± 0.94 bc | S. marcescens ATCC 14756 |  | 2.33 ± 0.47 f | |||
| Staph. aureus ATCC 33592 MRSA |  | 10.67 ± 0.94 bc | ||||||
| Staph. epidermidis ATCC 51625 |  | 12.33 ± 0.47 b | ||||||
| Staph. epidermidis ATCC 12228 |  | 13.00 ± 0.82 b | ||||||
| Strep. pyogenes ATCC 19615 |  | 4.67 ± 0.47 ef | ||||||
| R. equi ATCC 8939 |  | 16.67 ± 0.94 a | ||||||
| Bile Salt Concentrations | Initial Time | Simulated Intestinal Conditions | |||
|---|---|---|---|---|---|
| 2 h | 4 h | ||||
| Log10 CFU/mL | % Viability | Log10 CFU/mL | % Viability | ||
| Control | 7.50 ± 0.09 | 8.64 ± 0.08 | 115.11 ± 0.93 | 10.56 ± 0.10 | 140.70 ± 0.28 |
| 0.3% | 7.37 ± 0.08 | 8.15 ± 0.15 | 110.59 ± 0.94 | 9.88 ± 0.07 | 134.02 ± 1.67 |
| 0.5% | 7.32 ± 0.09 | 7.38 ± 0.15 | 100.77 ± 0.92 | 8.52 ± 0.11 | 116.39 ± 1.50 |
| 1% | 7.39 ± 0.02 | 7.75 ± 0.14 | 104.97 ± 1.70 | 8.21 ± 0.02 | 111.10 ± 0.88 |
| 2% | 7.29 0.09 | 7.34 ± 0.07 | 100.60 ± 0.75 | 7.61 ± 0.05 | 104.40 ± 1.60 |
| Strain | Sample Type | Antioxidant Activity (% ± SD) |
|---|---|---|
| 24.5 | Intact cells | 34.68 ± 1.33 a |
| Cell-free extract | 18.47 ± 1.69 b |
| Strain | Plate Screening | Api Zym Kit | ||||||
|---|---|---|---|---|---|---|---|---|
| Amylase | Cellulase | Lipase | Protease | Alkaline Phosphatase 2 | Esterase (C4) 3 | Esterase Lipase (C8) 4 | Naphthol-AS-BI-Phosphohydrolase 11 | |
| 24.5 |  |  | 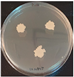 | 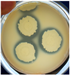 |  | |||
| + | + | - | + | ± | + | + | + | |
| Strain | Auto-Aggregation (%) | Hydrophobicity (%) | Co-Aggregation (%) | |||||
|---|---|---|---|---|---|---|---|---|
| 2 h | 5 h | 24 h | Hexane | Xylene | Ethyl Acetate | S. aureus | E. coli | |
| 24.5 | 18.60 ± 0.31 c | 27.97 ± 0.12 b | 84.42 ± 0.27 a | 71.62 ±0.32 a | 34.34 ± 0.13 c | 53.89 ± 0.15 b | 39.42 ± 0.45 a | 28.25 ± 0.29 b |
| Drug Class | Antibiotic (Code) | Interpretation * |
|---|---|---|
| β-lactams | Amoxicillin/clavulanic acid (AMC20/10) | I |
| Ampicillin (AM10) | I | |
| Cefalexin (CL30) | S | |
| Ceftriaxone (CRO30) | S | |
| Cefuroxime sodium (CXM30) | I | |
| Penicillin (P2) | I | |
| Aminoglycosides | Amikacin (AK10) | S |
| Gentamicin (CN30) | S | |
| Kanamycin (K30) | S | |
| Streptomycin (S10) | I | |
| Polypeptides | Bacitracin (B10) | R |
| Colistin (CT10) | I | |
| Fluoroquinolones/Quinolones | Ciprofloxacin (CIP1) | S |
| Nalidixic acid (NA30) | I | |
| Norfloxacin (NOR30) | S | |
| Phenicols | Chloramphenicol (C30) | S |
| Macrolides | Erythromycin (E10) | I |
| Lincosamides | Lincomycin (L10) | I |
| Nitrofurans | Nitrofurantoin (F300) | I |
| Tetracyclines | Oxytetracycline (T30) | I |
| Tetracycline (TE30) | I | |
| Folate pathway inhibitors | Trimethoprim/sulphamethoxazole (SXT25) | S |
| Glycopeptides | Vancomycin (VA10) | I |
| Antifungals | Fluconazole (FLU10) | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diguță, F.C.; Toma, R.C.; Matei, F. In Vitro Evaluation of Halotolerant Bacillus velezensis 24.5 as a Promising Probiotic with Broad-Spectrum Antimicrobial Activity. Microorganisms 2025, 13, 2240. https://doi.org/10.3390/microorganisms13102240
Diguță FC, Toma RC, Matei F. In Vitro Evaluation of Halotolerant Bacillus velezensis 24.5 as a Promising Probiotic with Broad-Spectrum Antimicrobial Activity. Microorganisms. 2025; 13(10):2240. https://doi.org/10.3390/microorganisms13102240
Chicago/Turabian StyleDiguță, Filofteia Camelia, Radu Cristian Toma, and Florentina Matei. 2025. "In Vitro Evaluation of Halotolerant Bacillus velezensis 24.5 as a Promising Probiotic with Broad-Spectrum Antimicrobial Activity" Microorganisms 13, no. 10: 2240. https://doi.org/10.3390/microorganisms13102240
APA StyleDiguță, F. C., Toma, R. C., & Matei, F. (2025). In Vitro Evaluation of Halotolerant Bacillus velezensis 24.5 as a Promising Probiotic with Broad-Spectrum Antimicrobial Activity. Microorganisms, 13(10), 2240. https://doi.org/10.3390/microorganisms13102240







