Isolation of Endophytic Phosphate-Solubilizing Bacteria from Chinese Cymbidium (Cymbidium spp.) Orchid Roots
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Treatment
2.2. DNA Extraction and PCR Amplification
2.3. Selection of PSB Strains in Solid and Liquid Media
2.4. Measurement of Plant Growth Parameters
2.5. Measurements of Soluble Phosphorus in Liquid Medium and Total Available Phosphorus in Rhizosphere Soils
2.6. Measurements of Exopolysaccharides
2.7. Analysis of Organic Acid Production in Liquid Cultures by HPLC
2.8. Measurements of Alkaline Phosphatase Activities
2.9. Colonization Dynamics of PSB in Orchid Roots and Rhizosphere Soil
2.10. Quantitative RT-PCR Analysis and Statistical Analysis
2.11. Statistical Analysis
3. Results
3.1. Bacterial Identification
3.2. Evaluation of Phosphate-Solubilizing Ability
3.2.1. Ability to Dissolve Inorganic Phosphorus
3.2.2. Ability to Mineralize Organic Phosphorus
3.3. Effects of Endophytic PSB Inoculation on P Uptake and Orchid Growth
3.4. Effects of Endophytic PSB Inoculation on the Properties and Phosphorus Availability in the Rhizosphere Soil of Orchids
3.5. Gene Transcript Levels Involved in Phosphorus Transport in Chinese Cymbidium Inoculated with Different PSB Strains
3.6. Colonization of PSB in Orchid Roots and Rhizosphere Soil
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, F.; Gao, J.; Wei, Y.; Ren, R.; Zhang, G.; Lu, C.; Jin, J.; Ai, Y.; Wang, Y.; Chen, L.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Shenoy, V.; Kalagudi, G.M. Enhancing plant phosphorus use efficiency for sustainable cropping. Biotechnol. Adv. 2005, 23, 501–513. [Google Scholar] [CrossRef]
- Herrera-Estrella, L.; López-Arredondo, D. Phosphorus: The underrated element for feeding the world. Trends Plant Sci. 2016, 21, 461–463. [Google Scholar] [CrossRef]
- Zhang, A.; Zhang, M.; Zhu, J.; Mei, Y.; Xu, F.; Bai, H.; Sun, K.; Zhang, W.; Dai, C.; Jia, Y. Endofungal bacterial microbiota promotes the absorption of chelated inorganic phosphorus by host pine through the ectomycorrhizal system. Microbiol. Spectr. 2023, 11, e00162-23. [Google Scholar] [CrossRef]
- Mei, Y.; Zhang, M.; Cao, G.; Zhu, J.; Zhang, A.; Bai, H.; Dai, C.; Jia, Y. Endofungal bacteria and ectomycorrhizal fungi synergistically promote the absorption of organic phosphorus in Pinus massoniana. Plant Cell Environ. 2024, 47, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Ahemad, M.; Zaidi, A.; Khan, M.S.; Oves, M. Biological importance of phosphorus and phosphate solubilizing microorganisms-an overview. In Phosphate Solubilizing Microbes for Crop Improvement; Khan, M.S., Zaidi, A., Eds.; Nova Science Publishers Inc.: New York, NY, USA, 2009; pp. 1–4. [Google Scholar]
- Zaidi, A.; Khan, M.S.; Ahmad, E.; Saif, S.; Rizvi, A.; Shahid, M. Growth stimulation and management of diseases of ornamental plants using phosphate solubilizing microorganisms: Current perspective. Acta Physiol. Plant 2016, 38, 117. [Google Scholar] [CrossRef]
- Singh, N.; Pandey, P.; Dubey, R.C.; Maheshwari, D.K. Biological control of root rot fungus Macrophomina phaseolina and growth enhancement of Pinus roxburghii (Sarg.) by rhizosphere competent Bacillus subtilis BN1. World J. Microb. Biot. 2008, 24, 1669–1679. [Google Scholar] [CrossRef]
- Bargaz, A.; Elhaissoufi, W.; Khourchi, S.; Benmrid, B.; Borden, K.A.; Rchiad, Z. Benefits of phosphate solubilizing bacteria on belowground crop performance for improved crop acquisition of phosphorus. Microbiol. Res. 2021, 252, 126842. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef]
- Cheng, Y.; Narayanan, M.; Shi, X.; Chen, X.; Li, Z.; Ma, Y. Phosphate-solubilizing bacteria: Their agroecological function and optimistic application for enhancing agro-productivity. Sci. Total Environ. 2023, 901, 166468. [Google Scholar] [CrossRef]
- Nico, M.; Ribaudo, C.M.; Gori, J.I.; Cantore, M.; Curá, J.A. Uptake of phosphate and promotion of vegetative growth in glucose-exuding rice plants (Oryza sativa) inoculated with plant growth-promoting bacteria. Appl. Soil Ecol. 2012, 61, 190–195. [Google Scholar] [CrossRef]
- Costa, E.M.; Lima, W.; Oliveira-Longatti, S.M.; Souza, F.M. Phosphate-solubilising bacteria enhance Oryza sativa growth and nutrient accumulation in an oxisol fertilized with rock phosphate. Ecol. Eng. 2015, 83, 380–385. [Google Scholar] [CrossRef]
- Pradhan, M.; Sahoo, R.K.; Pradhan, C.; Tuteja, N.; Mohanty, S. Contribution of native phosphorous-solubilizing bacteria of acid soils on phosphorous acquisition in peanut (Arachis hypogaea L.). Protoplasma 2017, 254, 2225–2236. [Google Scholar] [CrossRef]
- Li, H.; Ding, X.; Chen, C.; Zheng, X.; Han, H.; Li, C.; Gong, J.; Xu, T.; Li, Q.; Ding, G.; et al. Enrichment of phosphate solubilizing bacteria during late developmental stages of eggplant (Solanum melongena L.). FEMS Microbiol. Ecol. 2019, 95, fiz023. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, J.; Jia, P.; Yang, T.; Zeng, Q.; Zhang, S.; Liao, B.; Shu, W.; Li, J. Novel phosphate-solubilizing bacteria enhance soil phosphorus cycling following ecological restoration of land degraded by mining. ISME J. 2020, 14, 1600–1613. [Google Scholar] [CrossRef]
- Khan, I.; Zada, S.; Rafiq, M.; Sajjad, W.; Zaman, S.; Hasan, F. Phosphate solubilizing epilithic and endolithic bacteria isolated from clastic sedimentary rocks, Murree lower Himalaya, Pakistan. Arch. Microbiol. 2022, 204, 332. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Z.; Ahmad, M.; Raza, M.A.; Hilger, T.; Rasche, F. Phosphate-solubilizing bacillus sp. modulate soil exoenzyme activities and improve wheat growth. Microb. Ecol. 2024, 87, 31. [Google Scholar] [CrossRef]
- Kochian, L.V. Molecular physiology of mineral nutrient acquisition, transport, and utilization. In Biochemistry and Molecular Biology of Plants; Buchanan, B., Gruissem, W., Jones, R., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; pp. 1222–1230. [Google Scholar]
- Cameron, D.D.; Johnson, I.; Leake, J.R.; Read, D.J.; Ren, Y.F. Mycorrhizal acquisition of inorganic phosphorus by the green-leaved terrestrial orchid Goodyera repens. Ann. Bot. 2007, 99, 831–834. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Puga, M.I.; Rojas-Triana, M.; Martinez-Hevia, I.; Diaz, S.; Poza-Carrión, C.; Miñambres, M.; Leyva, A. Plant adaptation to low phosphorus availability: Core signaling, crosstalks, and applied implications. Mol. Plant 2022, 15, 104–124. [Google Scholar] [CrossRef]
- Othman, R.; Panhwar, Q.A. Phosphate-solubilizing bacteria improves nutrient uptake in Aerobic Rice. In Phosphate Solubilizing Microorganisms; Khan, M., Zaidi, A., Musarrat, J., Eds.; Springer: Cham, Switzerland, 2014. [Google Scholar] [CrossRef]
- Pande, A.; Pandey, P.; Mehra, S.; Singh, M.; Kaushik, S. Phenotypic and genotypic characterization of phosphate solubilizing bacteria and their efficiency on the growth of maize. J. Genet. Eng. Biotechnol. 2017, 15, 379–391. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, X.; He, X.; Cao, Y.; Guo, T.; Li, T.; Ni, H.; Tang, X. Phosphate-solubilizing Pseudomonas sp. strain P34-L promotes wheat growth by colonizing the wheat rhizosphere and improving the wheat root system and soil phosphorus nutritional status. J. Plant Growth Regul. 2019, 38, 1314–1324. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Liu, L.; Li, S.; Xie, J.; Xue, X.; Jiang, Y. Screening of phosphate-solubilizing bacteria and their abilities of phosphorus solubilization and wheat growth promotion. BMC Microbiol. 2022, 22, 296. [Google Scholar] [CrossRef]
- McCormick, M.K.; Jacquemyn, H. What constrains the distribution of orchid populations? New Phytol. 2015, 202, 392–400. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Fochi, V.; Chitarra, W.; Kohler, A.; Voyron, S.; Singan, V.R.; Lindquist, E.A.; Barry, K.W.; Girlanda, M.; Grigoriev, I.V.; Martin, F.; et al. Fungal and plant gene expression in the Tulasnella calospora-Serapias vomeracea symbiosis provides clues about nitrogen pathways in orchid mycorrhizas. New Phytol. 2017, 213, 365–379. [Google Scholar] [CrossRef]
- Selosse, M.A.; Petrolli, R.; Mujica, M.I.; Laurent, L.; Perez-Lamarque, B.; Figura, T.; Bourceret, A.; Jacquemyn, H.; Li, T.; Gao, J.; et al. The waiting room hypothesis revisited by orchids: Were orchid mycorrhizal fungi recruited among root endophytes? Ann. Bot. 2022, 129, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, P.; Qin, J.; Guo, J.; Deng, J. High-throughput sequencing-based analysis of the composition and diversity of endophytic bacteria community in tubers of Gastrodia elata f.glauca. Front. Microbiol. 2023, 13, 1092552. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, A.; Diez-Galán, A.; Cobos, R.; Calvo-Peña, C.; Barreiro, C.; Medina-Turienzo, J.; Sánchez-García, M.; Coque, J.J.R. Using rhizosphere phosphate solubilizing bacteria to improve barley (Hordeum vulgare) plant Productivity. Microorganisms 2021, 9, 1619. [Google Scholar] [CrossRef]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef]
- Altschul, S.; Modden, T.; Schafer, A.; Zhang, J.; Miller, W.; Lipman, D. Gapped BLAST and PSIBLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3404. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Li, Z.; Li, J.; Liu, G.; Li, Y.; Wu, X.; Liang, J.; Wang, Z.; Chen, Q.; Peng, F. Isolation, Characterization and Growth-Promoting Properties of Phosphate-Solubilizing Bacteria (PSBs) Derived from Peach Tree Rhizosphere. Microorganisms 2025, 13, 718. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Dong, X.; Yang, H.; Chang, Y.S.; Xu, Z.; Che, F.; Wang, S.; Huang, W. Characterization of phosphate solubilizing bacteria in the sediments of eutrophic lakes and their potential for cyanobacterial recruitment. Chemosphere 2024, 352, 141276. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Mehlich, A. Mehlich 3 soil test extractant: A modification of Mehlich 2 extractant. Commun. Soil Sci. Plan. 1984, 15, 1409–1416. [Google Scholar] [CrossRef]
- Gauri, S.S.; Mandal, S.S.; Gauri, S.M.; Mondal, K.C.; Dey, S.; Pati, B.R. Enhanced production and partial characterization of an extracellular polysaccharide from newly isolated Azotobacter sp. SSB81. Bioresour. Technol. 2009, 100, 4240–4243. [Google Scholar] [CrossRef]
- Fiori, J.; Amadesi, E.; Fanelli, F.; Tropeano, C.V.; Rugolo, M.; Gotti, R. Cellular and mitochondrial determination of low molecular mass organic acids by LC-MS/MS. J. Pharm. Biom. 2018, 150, 33–38. [Google Scholar] [CrossRef]
- Qvirist, L.; Carlsson, N.G.; Andlid, T. Assessing phytase activity-methods, definitions and pitfalls. J. Biol. Methods 2015, 2, e16. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Coy, R.M.; Held, D.W.; Kloepper, J.W. Rhizobacterial colonization of bermudagrass by Bacillus spp. in a Marvyn loamy sand soil. Appl. Soil Ecol. 2019, 141, 10–17. [Google Scholar] [CrossRef]
- Lin, Z.; Zhu, G.; Lu, C.Q.; Gao, J.; Li, J.; Xie, Q.; Wei, Y.; Jin, J.; Wang, F.; Yang, F. Functional conservation and divergence of SEPALLATA-like genes in floral development in Cymbidium sinense. Front. Plant Sci. 2023, 14, 1209834. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D.L. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lin, W.; Hsiao, Y.; Chiou, T. Milestones in understanding transport, sensing, and signaling of the plant nutrient phosphorus. Plant Cell 2024, 36, 1504–1523. [Google Scholar] [CrossRef]
- Raymond, N.S.; Gómez-Muñoz, B.; van der Bom, F.J.T.; Nybroe, O.; Jensen, L.S.; Müller-Stöver, D.S.; Oberson, A.; Richardson, A.E. Phosphate-solubilising microorganisms for improved crop productivity: A critical assessment. New Phytol. 2021, 229, 1268–1277. [Google Scholar] [CrossRef]
- Lambers, H. Phosphorus acquisition and utilization in plants. Annu. Rev. Plant Biol. 2022, 73, 17–42. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Cai, B. Phosphate-solubilizing bacteria: Advances in their physiology, molecular mechanisms and microbial community effects. Microorganisms 2023, 11, 2904. [Google Scholar] [CrossRef]
- Fan, Y.; Lin, F.; Yang, L.; Zhong, X.; Wang, M.H.; Zhou, J.; Chen, Y.M.; Yang, Y. Decreased soil organic P fraction associated with ectomycorrhizal fungal activity to meet increased P demand under N application in a subtropical forest ecosystem. Biol. Fertil. Soils 2018, 54, 149–161. [Google Scholar] [CrossRef]
- Marra, L.M.; Sousa Soares, C.R.F.; de Oliveira, S.M.; Ferreira, P.A.A.; Soares, B.L.; de Carvalho, R.F.; de Moreira, F.M.S. Biological nitrogen fixation and phosphate solubilization by bacteria isolated from tropical soils. Plant Soil 2012, 357, 289–307. [Google Scholar] [CrossRef]
- Nautiyal, C.S.; Bhadauria, S.; Kumar, P.; Lal, H.; Mondal, R.; Verma, D. Stress induced phosphate solubilization in bacteria isolated from alkaline soils. FEMS Microbiol. Lett. 2000, 182, 291–296. [Google Scholar] [CrossRef]
- Chen, Y.P.; Rekha, P.D.; Arun, A.B.; Shen, F.T.; La, W.-A.; Young, C.C. Phosphate solubilizing bacteria from subtropical soil and their tricalcium phosphate solubilizing abilities. Appl. Soil Ecol. 2006, 34, 33–41. [Google Scholar] [CrossRef]
- Yi, Y.; Huang, W.; Ge, Y. Exopolysaccharide: A novel important factor in the microbial dissolution of tricalcium phosphate. World J. Microb. Biot. 2008, 24, 1059–1065. [Google Scholar] [CrossRef]
- Fatima, T.; Mishra, I.; Verma, R.; Arora, N.K. Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. Biotech 2020, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Han, Q.; Liu, Q.; Gan, Y.; Rensing, C.; Rivera, W.; Zhao, Q.; Zhang, J. Roles of phosphate-solubilizing bacteria in mediating soil legacy phosphorus availability. Microbiol. Res. 2023, 272, 127375. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, G.; Wei, Y.; Dong, Y.; Hou, L.; Jiao, R. Isolation and screening of multifunctional phosphate solubilizing bacteria and its growth-promoting effect on Chinese fir seedling. Sci. Rep. 2021, 11, 9081. [Google Scholar] [CrossRef]
- González, E.; Solano, R.; Rubio, V.; Leyva, A.; Paz-Ares, J. Phosphate transporter traffic facilitator is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 2005, 17, 3500–3512. [Google Scholar] [CrossRef]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Theresa, F.; Law, T.F.; de Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Anawar, H.M.; Rengel, Z.; Damon, P.; Tibbett, M. Arsenic-phosphorus interactions in the soil-plant-microbe system: Dynamics of uptake, suppression and toxicity to plants. Environ. Pollut. 2018, 233, 1003–1012. [Google Scholar] [CrossRef]
- Dissanayaka, D.M.S.B.; Ghahremani, M.; Siebers, M.; Wasaki, J.; Plaxton, W.C. Recent insights into the metabolic adaptations of phosphorus-deprived plants. J. Exp. Bot. 2020, 72, 199–223. [Google Scholar] [CrossRef] [PubMed]


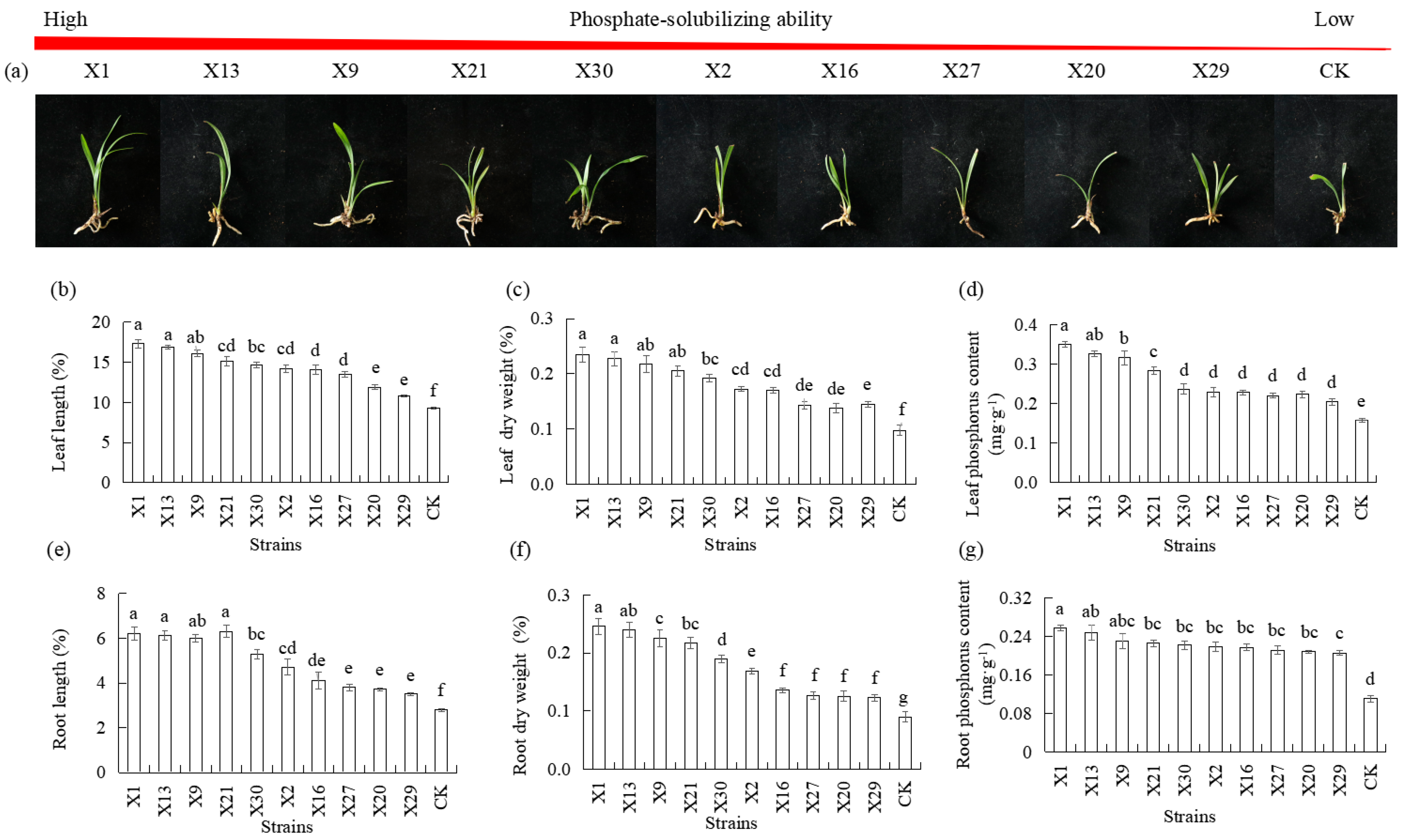
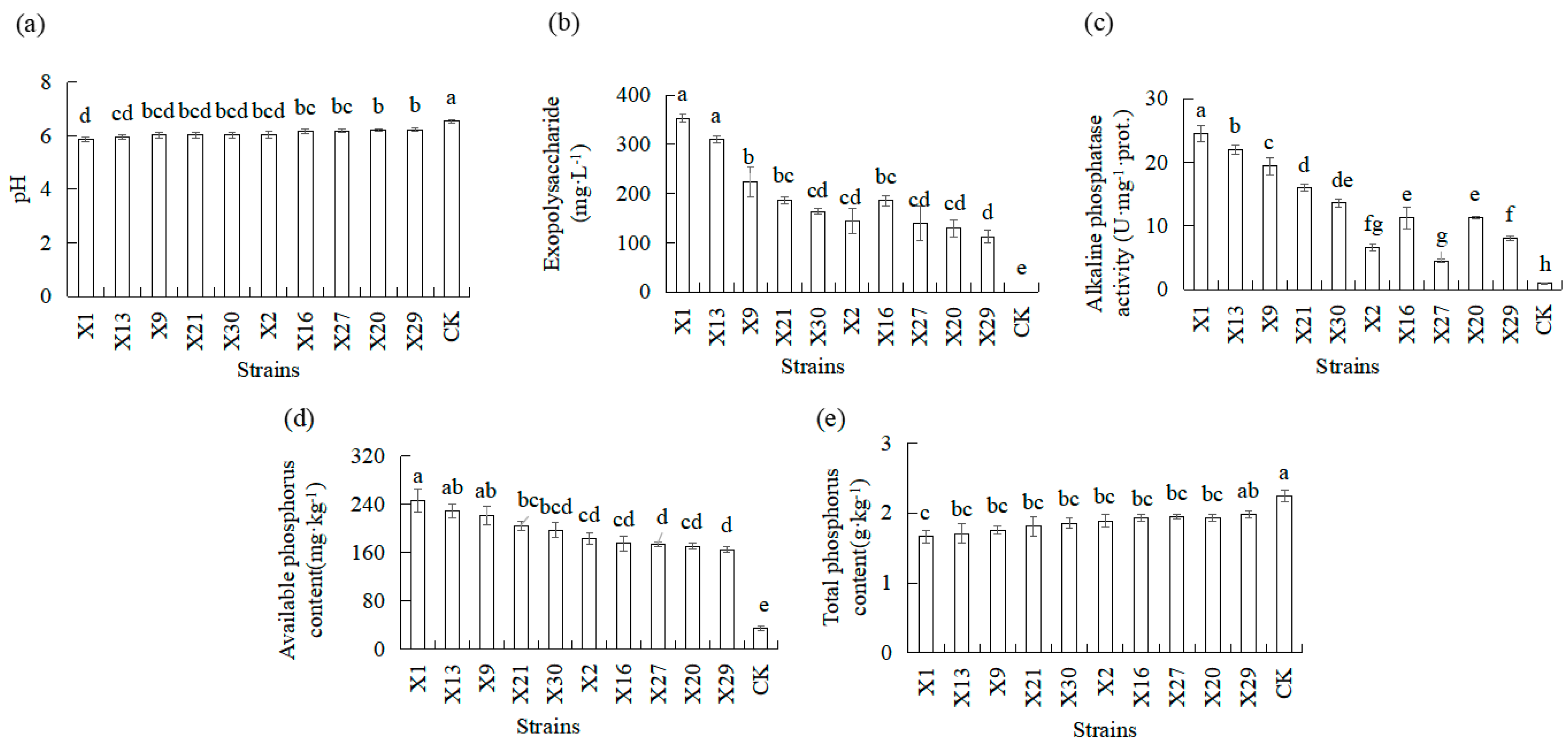


| Organic Acid | X1 | X13 | X9 | X21 | X30 | X2 | X16 | X27 | X20 | X29 |
|---|---|---|---|---|---|---|---|---|---|---|
| Lactic acid | 9.00±1.80cd | 17.25 ± 1.95 a | 14.14 ± 2.86 abc | 16.76 ± 0.31 ab | 10.22 ± 2.42 bcd | 7.50 ± 1.84 cd | 13.39 ± 3.05 abcd | 13.14 ± 2.72 abcd | 6.97 ± 1.38 d | 11.09 ± 0.44 abcd |
| Maleic acid | 0.02±0.00c | 0.03 ± 0.00 c | 0.04 ± 0.00 c | 0.27 ± 0.06 b | 1.95 ± 0.08 a | 0.01 ± 0.00 c | 0.00 ± 0.00 c | 0.02 ± 0.00 c | 0.03 ± 0.01 c | 0.02 ± 0.00 c |
| Fumaric acid | 0.05±0.01b | 0.09 ± 0.02 a | 0.03 ± 0.00 bc | 0.03 ± 0.01 bcd | 0.02 ± 0.00 bcd | 0.03 ± 0.01 bcd | 0.01 ± 0.00 cd | 0.02 ±0.01 cd | 0.00 ± 0.00 d | 0.00 ± 0.00 d |
| Succinic acid | 2.02 ± 0.23 a | 1.42 ± 0.01 b | 0.81 ± 0.02 c | 0.73 ± 0.09 cd | 0.53 ± 0.04 def | 0.29 ± 0.01 fg | 0.38 ± 0.01 ef | 0.53 ±0.02 def | 0.62 ± 0.02 cde | 0.07 ± 0.01 gh |
| Nicotinic acid | 0.12 ± 0.05 ab | 0.05 ± 0.02 bcd | 0.05 ± 0.01 bcd | 0.01 ± 0.01 cd | 0.00 ± 0.00 cd | 0.11 ± 0.05 ab | 0.16 ± 0.05 a | 0.04 ± 0.02 bcd | 0.04 ± 0.00 bcd | 0.10 ± 0.00 abc |
| Pyroglutamic acid | 0.24 ± 0.04 b | 0.12 ± 0.02 c | 0.19 ± 0.01 b | 0.00 ± 0.00 f | 0.01 ± 0.00 ef | 0.06 ± 0.01 cdef | 0.45 ± 0.06 a | 0.08 ± 0.00 cde | 0.03 ± 0.00 def | 0.10 ± 0.00 cd |
| Ethylmalonic acid | 0.02 ± 0.00 a | 0.01 ± 0.01 b | 0.01 ± 0.00 b | 0.01 ± 0.00 b | 0.00 ± 0.00 b | 0.01 ± 0.01 b | 0.01 ± 0.01 b | 0.01 ± 0.00 b | 0.01 ± 0.00 b | 0.01 ± 0.00 ab |
| Glutaric acid | 0.08 ± 0.04 a | 0.03 ± 0.01 bc | 0.06 ± 0.00 ab | 0.01 ± 0.00 c | 0.00 ± 0.00 c | 0.03 ± 0.01 bc | 0.03 ± 0.00 bc | 0.03 ± 0.00 bc | 0.03 ± 0.00 bc | 0.03 ± 0.00 bc |
| Malic acid | 0.45 ± 0.075 a | 0.25 ± 0.04 b | 0.13 ± 0.02 c | 0.11 ± 0.03 c | 0.12 ± 0.02 c | 0.02 ± 0.01 d | 0.00 ± 0.00 d | 0.08 ± 0.01 cd | 0.16 ± 0.01 c | 0.01 ± 0.00 d |
| 5-HydroXymethyl-2-furoic acid | 0.38 ± 0.04 a | 0.28 ± 0.00 ab | 0.25 ± 0.02 abc | 0.19 ± 0.00 bc | 0.13 ± 0.08 c | 0.16 ± 0.05 bc | 0.26 ± 0.01 abc | 0.24 ± 0.01 abc | 0.17 ± 0.06 bc | 0.18 ± 0.04 bc |
| Tartaric acid | 14.52 ± 2.80 ab | 7.60 ± 0.19 bcde | 9.18 ± 0.11 cd | 12.00 ± 1.66 bc | 13.83 ± 0.22 b | 19.04 ± 2.73 a | 4.36 ± 0.04 ef | 5.44 ± 0.11 def | 3.29 ± 0.25 fg | 2.03 ± 0.05 fg |
| Phenylpyruvic acid | 0.24 ± 0.02 c | 2.70 ± 0.13 a | 0.24 ± 0.08c | 0.08 ± 0.03 c | 0.04 ± 0.02 c | 0.05 ± 0.01 c | 2.14 ± 0.18 b | 0.15 ± 0.00 c | 0.10 ± 0.01 c | 0.16 ± 0.03 c |
| Phenyllactic acid | 0.06 ± 0.00 b | 2.75 ± 1.09 a | 0.09 ± 0.00 b | 0.16 ± 0.00 b | 0.08 ± 0.04 b | 0.04 ± 0.03 b | 0.33 ± 0.02 b | 0.14 ± 0.05 b | 0.07 ± 0.03 b | 0.17 ± 0.00 b |
| Vanillic acid | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.90 ± 0.18 a | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00b |
| PyridoXine | 0.02 ± 0.01 a | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b |
| Suberic acid | 0.01 ± 0.00 cd | 0.02 ± 0.00 cd | 0.00 ± 0.00 d | 0.00 ± 0.00 d | 0.00 ± 0.00 d | 0.04 ± 0.00 bc | 0.07 ± 0.01 a | 0.06 ± 0.03 ab | 0.04 ± 0.01 bc | 0.00 ± 0.00 d |
| Hippuric acid | 0.00 ± 0.00 d | 0.01 ± 0.00 d | 0.00 ± 0.00 d | 0.00 ± 0.00 d | 0.01 ± 0.01 d | 0.00 ±0.00 d | 0.02 ± 0.00 cd | 0.06 ± 0.03 ab | 0.07 ± 0.01 a | 0.04 ± 0.00 bc |
| Citric acid | 19.37 ± 1.14 a | 3.52 ± 0.25 bc | 4.21 ± 0.92 b | 2.18 ± 0.74 cd | 3.94 ± 0.72 bc | 1.33 ± 0.07 de | 1.32 ± 0.50 de | 0.31 ± 0.01 e | 3.76 ± 0.09 bc | 0.69 ± 0.01 de |
| D-Glucuronic acid | 27.65 ± 3.82 a | 23.21 ± 0.33 b | 19.21 ± 1.48 c | 14.74 ± 0.68 d | 14.99 ± 1.60 d | 11.60 ± 1.80de | 12.61 ± 0.94 de | 12.01 ± 0.92 de | 9.01 ± 1.84 ef | 5.83 ± 0.70 f |
| Pantothenic acid | 1.23 ± 0.35 c | 3.75 ± 0.29 a | 1.22 ± 0.18 c | 0.16 ± 0.04 d | 0.62 ± 0.51 cd | 0.53 ± 0.08 cd | 3.75 ± 0.26 a | 1.24 ± 0.63 c | 1.22 ± 0.14 c | 2.39 ± 0.07 b |
| Total organic acids | 75.40 ± 10.41 a | 63.08 ± 4.35 ab | 49.83 ± 5.71 bc | 48.33 ± 3.85 bc | 46.69 ± 5.75 bc | 40.84 ± 6.71 cd | 39.30 ± 5.15 cd | 33.60 ± 4.57 cd | 25.63 ± 3.85 d | 22.92 ± 1.36 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Jin, J.; Wang, X.; Zhu, W.; Gao, J.; Li, J.; Xie, Q.; Wei, Y.; Lu, C.; Zhu, G.; et al. Isolation of Endophytic Phosphate-Solubilizing Bacteria from Chinese Cymbidium (Cymbidium spp.) Orchid Roots. Microorganisms 2025, 13, 2229. https://doi.org/10.3390/microorganisms13102229
Sun Y, Jin J, Wang X, Zhu W, Gao J, Li J, Xie Q, Wei Y, Lu C, Zhu G, et al. Isolation of Endophytic Phosphate-Solubilizing Bacteria from Chinese Cymbidium (Cymbidium spp.) Orchid Roots. Microorganisms. 2025; 13(10):2229. https://doi.org/10.3390/microorganisms13102229
Chicago/Turabian StyleSun, Yanmei, Jianpeng Jin, Xiting Wang, Wei Zhu, Jie Gao, Jie Li, Qi Xie, Yonglu Wei, Chuqiao Lu, Genfa Zhu, and et al. 2025. "Isolation of Endophytic Phosphate-Solubilizing Bacteria from Chinese Cymbidium (Cymbidium spp.) Orchid Roots" Microorganisms 13, no. 10: 2229. https://doi.org/10.3390/microorganisms13102229
APA StyleSun, Y., Jin, J., Wang, X., Zhu, W., Gao, J., Li, J., Xie, Q., Wei, Y., Lu, C., Zhu, G., & Yang, F. (2025). Isolation of Endophytic Phosphate-Solubilizing Bacteria from Chinese Cymbidium (Cymbidium spp.) Orchid Roots. Microorganisms, 13(10), 2229. https://doi.org/10.3390/microorganisms13102229








