Abstract
Archaea have been identified as early colonizers of the human intestine, appearing from the first days of life. It is hypothesized that the origin of many of these archaea is through vertical transmission during breastfeeding. In this study, we aimed to characterize the archaeal composition in samples of mother-neonate pairs to observe the potential vertical transmission. We performed a cross-sectional study characterizing the archaeal diversity of 40 human colostrum-neonatal stool samples by next-generation sequencing of V5–V6 16S rDNA libraries. Intra- and inter-sample analyses were carried out to describe the Archaeal diversity in each sample type. Human colostrum and neonatal stools presented similar core microbiota, mainly composed of the methanogens Methanoculleus and Methanosarcina. Beta diversity and metabolic prediction results suggest homogeneity between sample types. Further, the co-occurrence network analysis showed associations between Archaea and Bacteria, which might be relevant for these organisms’ presence in the human milk and neonatal stool ecosystems. According to relative abundance proportions, beta diversity, and co-occurrence analyses, the similarities found imply that there is vertical transmission of archaea through breastfeeding. Nonetheless, differential abundances between the sample types suggest other relevant sources for colonizing archaea to the neonatal gut.
Keywords:
breastfeeding; human milk; microbiota; Archaea; neonatal gut; 16S rDNA; vertical transmission 1. Introduction
Human milk is composed of essential nutrients and bioactive constituents, including proteins, carbohydrates, fatty acids [1], cytokines [2], oligosaccharides, immune factors [3], and microbiota [4] that cater to the evolving needs of the growing infant [5]. Human milk can be classified into colostrum, transitional, and mature milk according to the composition and lactation stage [6]. For instance, colostrum is the first milk produced during the first days after birth [7]. It is characterized by a high content of growth factors and immunoglobulins, which provide passive immunity to newborns, protecting them against infections [8].
Human milk’s microbiota includes bacteria, eukaryotes, fungi, and archaea [9]. Most studies have focused on the former [10], thus revealing the central bacteriome of human milk and its impact on newborn health [11]. The origin of bacteria in human milk is not fully understood, and two non-exclusive sources have been proposed: the entero-mammary pathway and the retrograde flow [12,13,14]. The first consists of immune cell-mediated bacterial translocation from the maternal gastrointestinal tract to the mammary gland [4,12,13,14]. In more detail, dendritic cells penetrate the gut epithelium and select bacteria, which are then transported to the mammary gland through lymphatic circulation [4,15,16]. The second refers to external contamination or the transfer from the mother’s skin or infant’s mouth to the mammary gland [14,16].
In contrast to bacteria, archaea diversity in human milk has long been neglected [9]. The first evidence of archaea presence in human milk came from metagenomics studies [9,17]. Nevertheless, a recent study proved archaea viability by cultivating Methanobrevibacter smithii, a methanogenic archaeon, from colostrum and mature milk [18]. Interestingly, the presence of archaea is reported in the gastrointestinal tract from the first days of life [19,20,21,22]. Accordingly, M. smithii has been identified as an early colonizer, establishing in the gastric mucosa just after birth [20].
Methanogenic archaea (MA) are prevalent archaea in the digestive tract of adults, particularly M. smithii and Methanosphaera stadmanae [23,24,25,26]. MA plays a fundamental role in the human gut, as they are capable of producing methane (CH4) through the assimilation of H2 and CO2, which are products of polysaccharide fermentation by bacteria [27,28,29,30]. They use hydrogen as an electron donor, reducing CO2, acetate, and multiple methyl-containing compounds into CH4 [30,31]. This metabolic activity facilitates the growth of fermentative bacteria in the gut, thus conforming to a syntropic relationship [31,32]. MA has been associated with various diseases such as diverticulosis [33,34], inflammatory bowel disease [35,36], atherosclerosis [37,38,39], malnutrition [40], and obesity [32,41,42,43]. However, the relationship between MA and illness is not entirely understood and can be contradictory. For example, MA has been associated with obese and normal-weight individuals [18,41,42,43,44,45,46]. Although the role played by archaea in disease is not clear, they appear to be key microbiota components of the human gastrointestinal tract [47,48].
Given its significance for human health, its presence in human milk, and its role in the gastrointestinal tract from the earliest days of life, this study aimed to characterize the diversity of the archaeal community in human colostrum and neonatal stool samples. This was achieved through Ion Torrent semiconductor DNA sequencing of V5–V6 regions of 16S rRNA gene libraries. Our findings suggest that human milk serves as a primary source of archaea for the neonatal gut, supporting the concept of vertical transmission of archaea during breastfeeding.
2. Materials and Methods
2.1. Study Design and Selection of Subjects
The cross-sectional descriptive study consisted of 40 mother-neonatal pairs of patients from the “Dr. José María Rodríguez” General Hospital, located in Ecatepec-de-Morelos, State of Mexico (19°36′35″ N, 99°3′36″ W). The samples were obtained from healthy lactating women and exclusively breastfed newborns. Colostrum and neonatal stool samples were collected from 0 to 3 days after birth, up to 2 h after the newborn was breastfed, from November 2017 to January 2018. The inclusion criteria were as follows: (1) Mexican origin with at least two generations of ancestry, (2) gestational age between 37 and 41 weeks, (3) birth weight between 2500–4500 g, (4) Apgar score greater than 7 at 5 min after birth. Exclusion criteria: (1) Maternal probiotic and alcohol consumption, (2) smoking, (3) diabetes before or during pregnancy, (4) antibiotic use during the last trimester of pregnancy and before sampling. The participants were given a survey recording sociodemographic and clinical data (maternal age, gestational age at delivery, type of delivery, newborn sex, and age). Written informed consent was obtained from all participants before the study, following the 2013 Declaration of Helsinki. The protocol was approved by the Ethics Committee of the General Hospital “Dr José María Rodríguez” (Project identification code: 217B560002018006).
2.2. Sample Collection
The colostrum-neonatal stool sample pairs were collected on the same day up to 2 h after the newborn was breastfed. Human colostrum (HC) was collected manually into a sterile polypropylene tube (~5 to 10 mL). Breast sanitation was not conducted in order to obtain the microorganisms transferred during breastfeeding. The neonatal stool (NS) was recovered from diapers and transferred to sterile containers using sterile tongue depressors. The samples were sent to the laboratory in a cold environment, distributed in aliquots of 1 mL (HC) or 200 mg (NS), and stored at −20 °C until DNA was extracted within 24 h of arrival.
2.3. DNA Extraction
First, 1 mL of HC was centrifuged at 4 °C, 10,000× g for 15 min in a refrigerated centrifuge (Eppendorf 5415R) and the fat was removed with a roll of sterile dental cotton. The aqueous supernatant was removed by decantation, the pellet was resuspended in 1 mL of sterile PBS pH 7.4, and then centrifuged again at 10,000× g for 15 min. The obtained pellet was resuspended in 300 μL of PBS pH 7.4 and processed for DNA extraction using FavorPrep Milk bacterial DNA extraction kit (Cat: FAMBD001, Favorgen, Biotech Corp, Taiwan) following the manufacturer’s instructions. Fecal DNA was extracted from 200 mg NS samples using a QIAamp DNA Stool Mini Kit (Cat.: 12830-50, Qiagen group, Venlo, Netherlands), following the manufacturer’s instructions. In two cases, 300 μL of PBS pH 7.4 was used as a negative control for DNA extraction. The DNA concentration in samples was measured at 260/280 absorbance using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA); no absorbance was detected for negative controls. DNA integrity was assessed by electrophoretic fractionation of 5 μL of DNA sample in 0.5% agarose gel stained with 0.80 μL of Midori Green advanced dye (1:15) using TBE buffer. DNA was visualized using the MolecularImager® Gel DocTM XR System program (Bio-Rad Laboratories, Chicago, IL, USA). Extracted DNA was stored at −20 °C.
2.4. Preparation of the 16S rRNA Gene Library and Next-Generation Sequencing
The forward Arc787F (5′-ATT-AgA-TAC-CCG-BgT-AgT-CC-3′) and reverse primer Arc1059R (5′-gCC-ATg-CAC-CWC-CTC-T-3′) were used for the polymerase chain reactions (PCR) [49]. The PCR reactions were performed using the Phusion High-Fidelity PCR Kit (Cat F-530S), ThermoFisher Scientific, Waltham, MA, USA). The reaction mixture consisted of 4.0 µL of 1× HF buffer, 0.4 µL of dNTPs (200 µM), 0.2 µL of Phusion polymerase (0.02 U/µL), 1.0 µL of each Forward and Reverse primer (10 µM), and 0.2 µL of MgCl2 (0.5 mM). The DNA template volume was adjusted to 13.2 µL with nuclease-free water for a final concentration of 8.0 ng in 20.0 µL. The reactions were programmed in 2720 Thermal Cycler (Applied Biosystems™, ThermoFisher Scientific, Waltham, MA, USA) with a 5 min 95 °C hot start, followed by 5 min initial denaturation at 95° C, 25× (94 °C, 15 s denaturation, 56 °C, 15 s annealing, 72 °C for 15 s extension), and a final 7 min extension at 72 °C. Archaeal DNA from a bioreactor [50] was used as positive control. Blank reactions (PCR products with no DNA template from the DNA extraction pipeline) were used as negative controls. The 358 bp amplicons were fractionated in 1.5% agarose gel dyed with Midori Green (Nippon Genetics®, Dueren, Germany) in 0.5× TBE using GeneRulerTM100 bp DNA Ladder (Cat. 15628019, ThermoFisher Scientific, Waltham, MA, USA). Electrophoresis lasted 45 min at 80 V. The DNA was visualized using the Molecular Imager® Gel DocTM XR System program (Bio-Rad Laboratories, Chicago, IL, USA). The library was purified using 2% E-Gel™ EX stained with SYBR GOLD DNA (Cat. G401002, Thermo Scientific, Waltham, MA, USA). The library size and concentration were assessed with the 2100 Bioanalyzer equipment and High Sensitivity DNA kit (Agilent Technologies, Santa Clara, CA, USA). PCR emulsion was carried out with Ion One Touch™ (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. Amplicon enrichment with ionic spheres was carried out using Ion OneTouch ES (Life Technologies, Carlsbad, CA, USA). Sequencing was performed using the Ion 318 Kit V2 Chip (Cat. 4488146, Life Technologies, Carlsbad, CA, USA). Ion Torrent PGM software v4.0.2 was used to demultiplex the sequence data based on their barcodes, reads were filtered to exclude low-quality (quality score ≤ 20), polyclonal sequences (homopolymers > 6), and the adapters were trimmed. The datasets generated for this study can be found in the NCBI BioProject ID PRJNA1018680.
2.5. ASV Determination and Taxonomic Annotation
The FASTQ files were further processed and analyzed with QIIME 2022.2 [51]. The ASVs (Amplicon Sequence Variants) were determined with the QIIME dada2 denoise-single plugin, with sequence truncation at 238 nt. The taxonomic assignation was done with the feature-classifier classify-consensus-blast plugin, with a 97% percentage identity. The Greengenes 2 database was used for analysis with BLAST [52].
2.6. Bioinformatic Analyses
R 4.2.0 [53] in Rstudio [54] was used for the relative abundance, diversity, differential, discriminatory, and co-occurrence analyses. For that purpose, the following packages were used: to import the qiime artifacts, qiime2R [55], for alpha and beta diversity analyses, phyloseq 1.4.0 [56], for differential analyses, DESEq2 1.3.6 [57] and ALDEx2 1.36.0 [58], for discriminatory analysis, lefser 1.15.9 [59], for the heatmap elaboration, ComplexHeatmap 2.12.0 [60], for the ANOSIM, vegan 2.6-2 [61], and for graphical images, tidyverse 1.3.1 [62], dplyr 1.09 (data frame manipulation), ggplot2 3.3.6, scales 1.2.0, ggpubr 0.4.4, and gridExtra 2.3 [63]. PICRUSt2 was executed with the MetaCyc metabolic pathway database option following a published pipeline tutorial [64]. Co-occurrence network analysis was conducted with microeco [65] and meconetcomp [66] packages using 0.6 Spearman’s rank correlation coefficient with a 99% confidence level. All analyses were filtered to the Archaeal Domain except for the co-occurrence analysis. In addition, the reads were filtered using a stringent pipeline to ensure data quality. First, potential contaminants identified in negative PCR sequencing controls were removed using decontam 1.24.0 [67]. Next, low-abundance ASVs (defined as those with a prevalence below 0.1% based on a Phred quality score < 30) were excluded. Finally, sparse ASVs (those > 90% zero values across samples) were filtered out.
2.7. Statistical Methods
Archaeal diversity within samples was estimated with alpha diversity, determining Observed ASVs, Shannon, Simpson, and Fisher indexes. The Shapiro–Wilk preliminary test was applied to test if the data followed a normal distribution; then, the Kruskal–Walli’s test was applied. The differences in beta diversity between samples were evaluated by ANOSIM (Analyses of similarities). A differential abundance analysis (DESeq2) for dependent samples was performed to identify relevant taxa in the distinct sample types and pairs and was evaluated with a Wald test; p-values were adjusted with Holm–Bonferroni method statistics. p- or q-values < 0.05 were considered significant.
3. Results
3.1. Most Participant Mother-Neonate Pairs Were from Urban Areas
Forty colostrum and neonatal stool samples were collected within four days immediately after birth. Most participants came from the State of Mexico (19.4969° N, 99.7233° W) or Mexico City (19.4326° N, 99.1332° W) low-income areas (Table 1). Almost all mothers work at home (95%), and close to 50% of them have a high school education. Most of them were normal weight and more than half of the deliveries were vaginal (Table 1). Regarding the neonate’s anthropometric data, 60% were females and 35% were males; information was not available for two of the samples.

Table 1.
Sociodemographic and clinical characteristics of the study population.
3.2. Methanoculleus and Methanosarcina Lead the Core Archaeal Community in Mother-Neonate Pairs
The archaeal composition and diversity in colostrum and neonatal stool samples from mother-neonate pairs were analyzed using high-throughput sequencing of the V5–V6 regions of the 16S rRNA gene. A total of 4,531,448 raw sequences were generated, including 2,269,715 from colostrum and 2,261,733 from neonatal stool, with a median Phred quality score of 27 (Table S1, Figure S1).
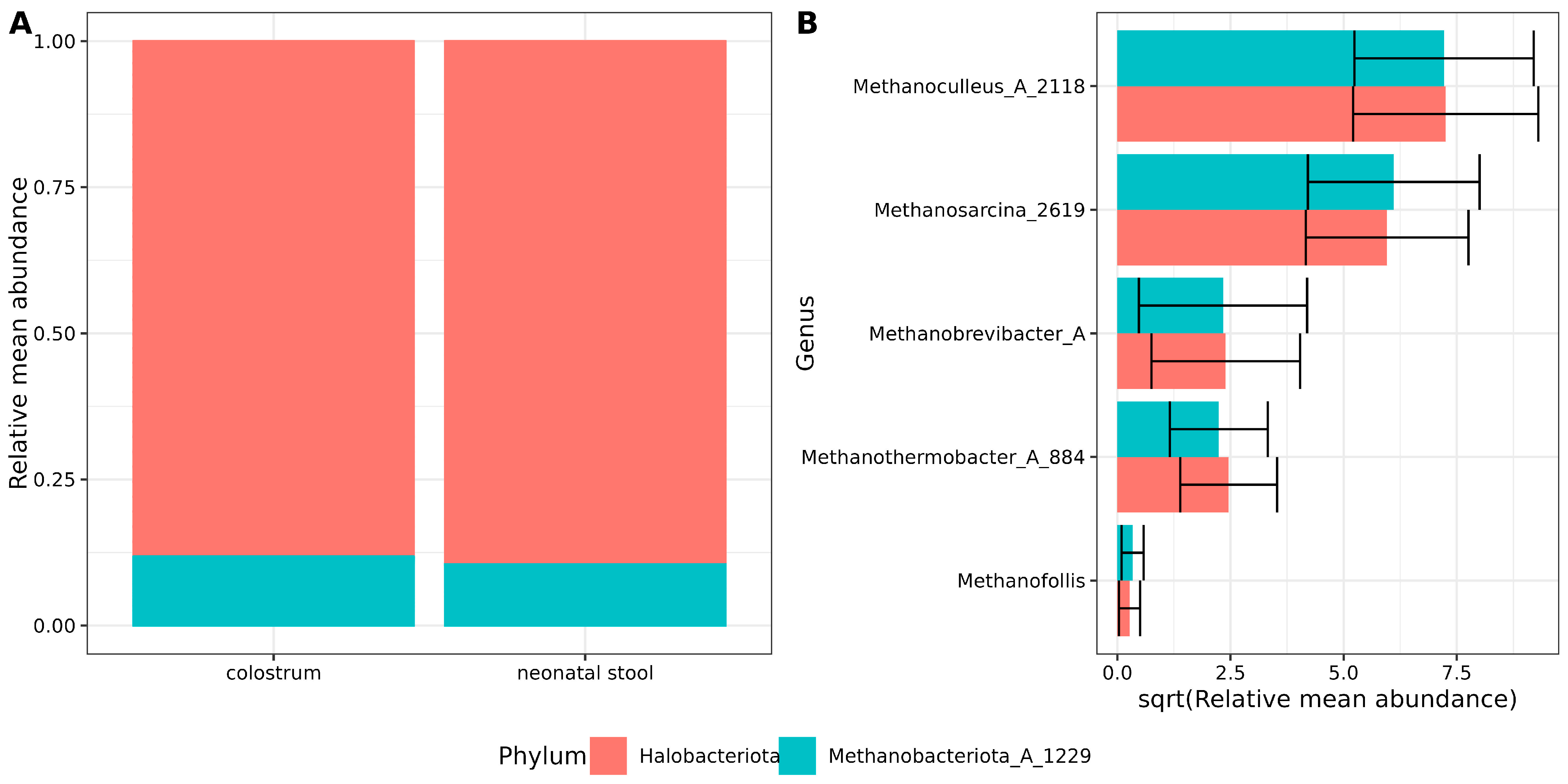
Archaeal sequence reads were filtered and decontaminated using a stringent pipeline as described in the Section 2.6 (Decontam results are shown in Table S2). After filtering, two archaeal phyla were identified in both sample types. Halobacteriota was the predominant phylum, accounting for nearly 89% of the archaeal abundance, followed by Methanobacteriota_A_1229 (~11%) (Figure 1A, Table S3).
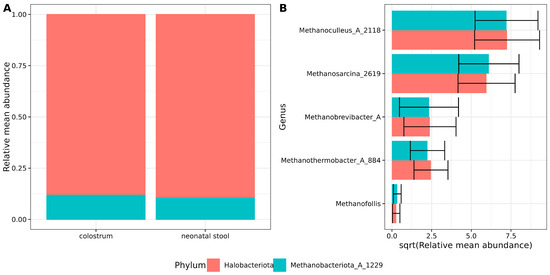
Figure 1.
Archaeal composition in samples. (A) Stacked bar plots showing the archaeal relative mean abundance at the phylum level. Red and green colors indicate sample type and relative abundances are shown on the Y-axis. (B) Bar plots showing the square root relative mean abundance of archaea genera. The Y-axis labels indicate genus, while the X-axis shows the square root-relative and mean abundance. Error bars indicate the standard error of the mean.
At the genus level, the dominance of Methanoculleus_A_2118 and Methanosarcina_2619 was observed in decreasing order of abundance, followed by Methanobrevibacter_A and Methanothermobacter_A_884. The least abundant archaeal genus detected was Methanofollis (Figure 1B, Table S4). No differences or trends with statistical significance were observed in archaeal abundance between the two sample types. These findings suggest a consistent core archaeal community composition between colostrum and neonatal stool.
3.3. Colostrum Is the Primary Source of Neonatal Archaeal Communities
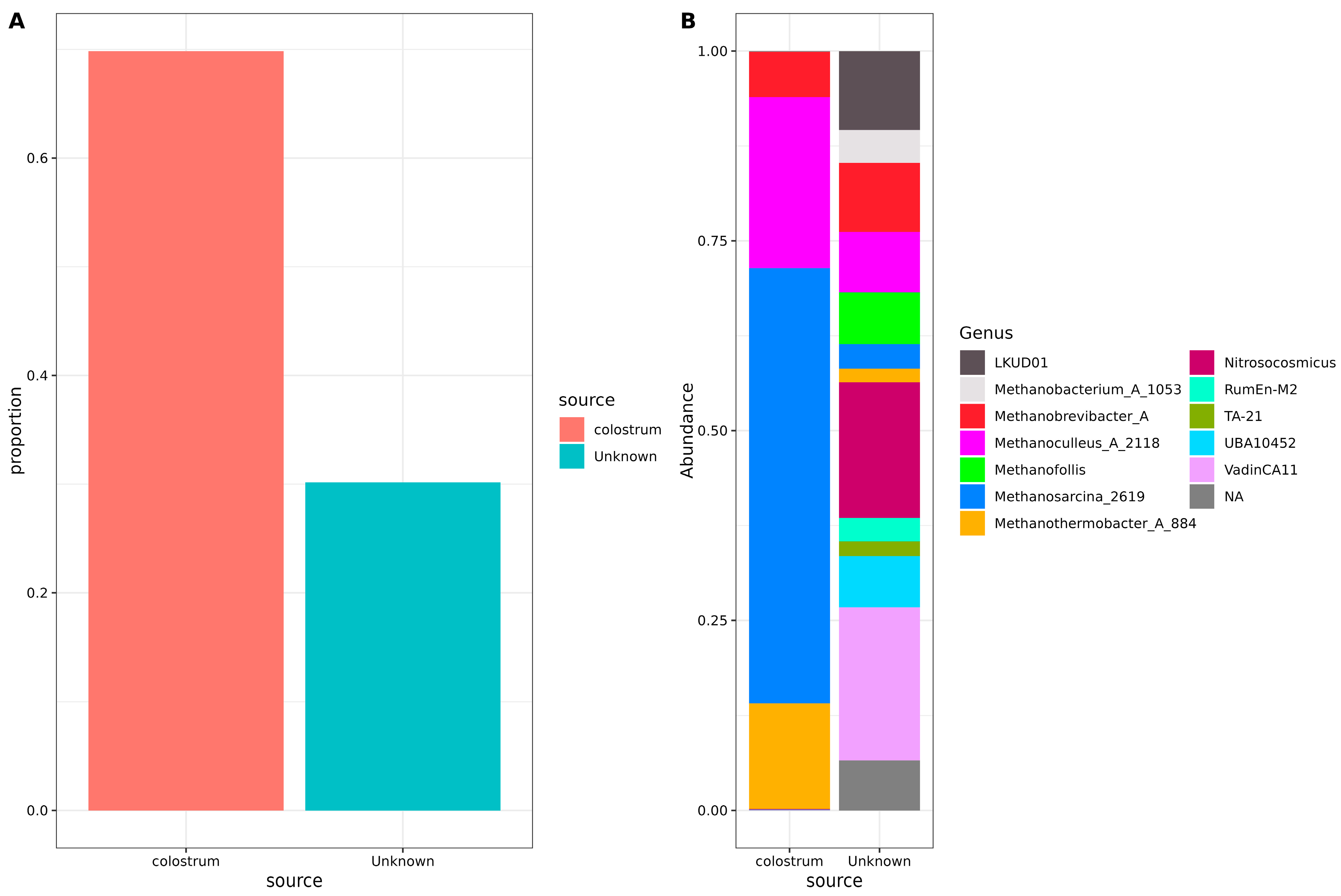
To understand the origin of archaea in neonatal stool samples, a SourceTracker analysis was performed using the unfiltered, non-decontaminated reads (Figure 2). This analysis estimates the proportion of archaea originating from a specific source—in this case, human colostrum. Sequencing results of negative controls from PCR were also included in the study. Our results indicated that nearly three-quarters of the archaeal communities present in the neonatal stool were derived from colostrum. At the same time, the remainder originated from an unidentified source; no trace of contamination from negative controls was observed (Figure 2A).
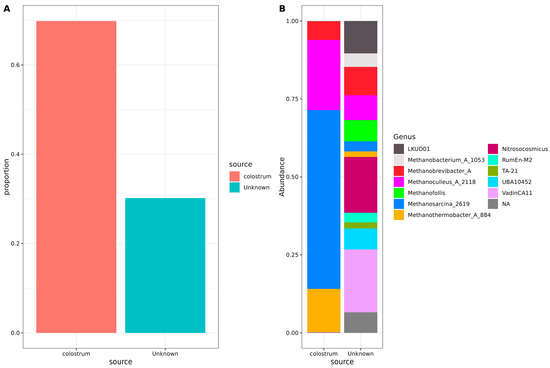
Figure 2.
Source tracker analysis of possible archaeal origin in neonatal stool samples. (A) Bar plots indicate the total source proportion. The Y-axis shows the proportion of the source for the neonatal stool samples, and the X-axis shows the source (colostrum or unknown). (B) Stacked bar plots showing genera composition for each source. The Y-axis shows the genera abundance, and the X-axis shows the source (colostrum or unknown). Color sectors indicate the source or the archaeal genus, according to the labels beside the graphics.
Concerning specific genera, Methanosarcina_2619, Methanoculleus_A_2118, Methanothermobacter_A_884, and Methanobrevibacter_A were found to originate predominantly from colostrum. However, portions of Methanobrevibacter_A, Methanoculleus_A_2118, Methanosarcina_2619, and Methanothermobacter_A_884 ASVs could not be traced back to colostrum or negative controls. In addition, some archaea, such as Methanobacterium_A_1053 and different Methanosarcina and Methanofollis, had unknown origins. Also, several less-known archaeal genera, including Nitrosocosmicus, LDKUD01, RumEn-M2, TA-21, UBA10452, VadinCA11, and various unassigned taxa, had origins that could not be explained by colostrum or any other analyzed source (Figure 2B).
3.4. Colostrum and Neonatal Stool Exhibited Comparable Diversity Metrics
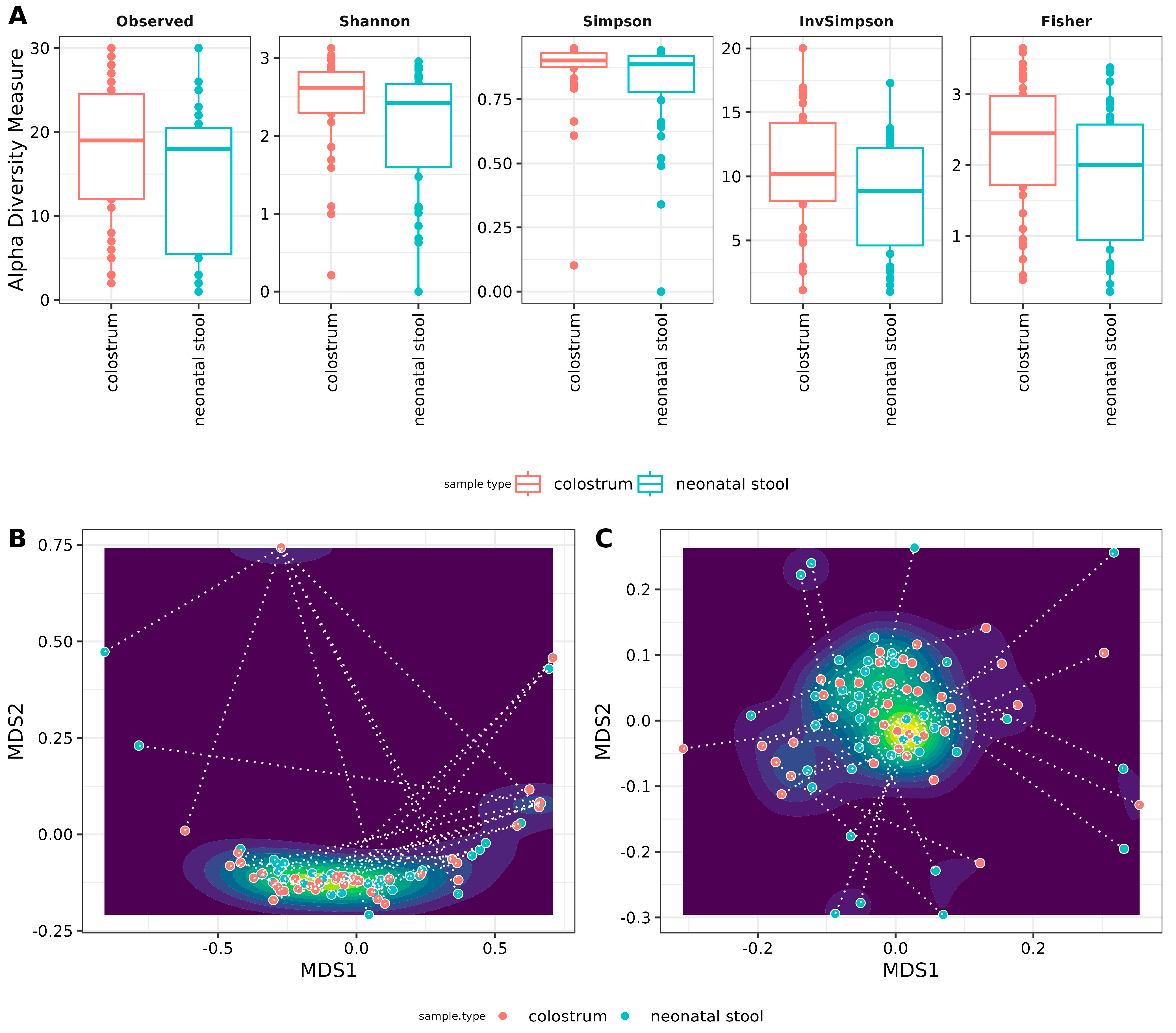
Alpha and beta diversity analyses were conducted to evaluate the homogeneity within and among samples. For the alpha diversity, an interesting trend emerged: colostrum appeared more diverse than neonatal stool across all indices, albeit with a small effect size (Figure 3A). The exception was the Inverse Simpson index, which showed a medium effect size. Statistical tests, including Kruskal–Wallis and ANOVA, revealed significant differences for all indices except the observed number of ASVs. However, after applying Holm correction to adjust the p-values, none of the differences remained statistically significant (Figure 3A, Table S5).
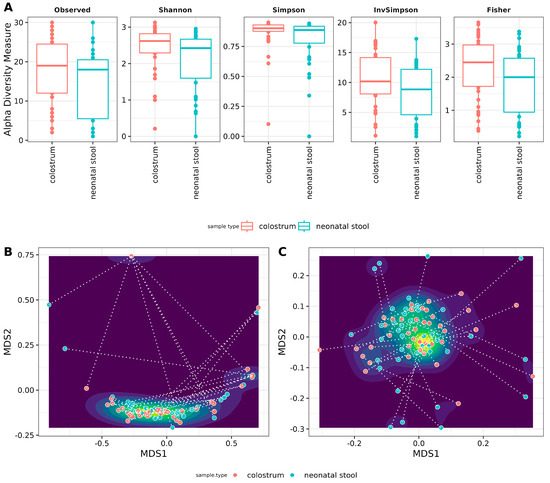
Figure 3.
Archaeal diversity in samples. (A) Alpha diversity box plots. The Y-axis indicates values for the species richness (Observed), diversity indexes (Shannon, Simpson, and Inverse Simpson), and evenness (Fisher). The sample type is shown at the X-axis (see Table S4, for numerical data of indexes) Beta diversity Non-Metric Multidimensional Scaling (NMDS) scatter plots. The graphics show archaeal beta diversity calculated by (B) NMDS ordination based on weighted UniFrac and (C) Jensen–Shannon diverge (JSD) distances. Sample types (colostrum and neonatal stool) are similar according to ANOSIM (weighted UniFrac R = −0.02, p = 0.933 and JSD: R = −0.001 p = 0.494).
Beta diversity was analyzed using weighted UniFrac distance and Jensen–Shannon divergence. It is important to note that filtering the reads resulted in some samples having zero features. To avoid bias in hypothesis testing, complete mother-neonate pairs were retained, including those with zero-feature samples. Our results indicate no significant differences between colostrum and neonatal stool communities, as evidenced by the single cluster formed in Figure 3B,C and the lack of statistical significance.
However, considerable distances were observed between paired samples (mother-neonate binomials), which none of the measured variables explain. These distances are represented by the dotted lines connecting each pair reflected with the low R-values from the ANOSIM analysis (−0.001 for Jensen–Shannon divergence and −0.02 for weighted UniFrac). This variation is likely attributed to the inclusion of zero-feature samples in the analysis (Figure 3B,C).
3.5. There Was a Moderate Positive Correlation in Methanoculleus_A_2118 Abundance in the Binomial
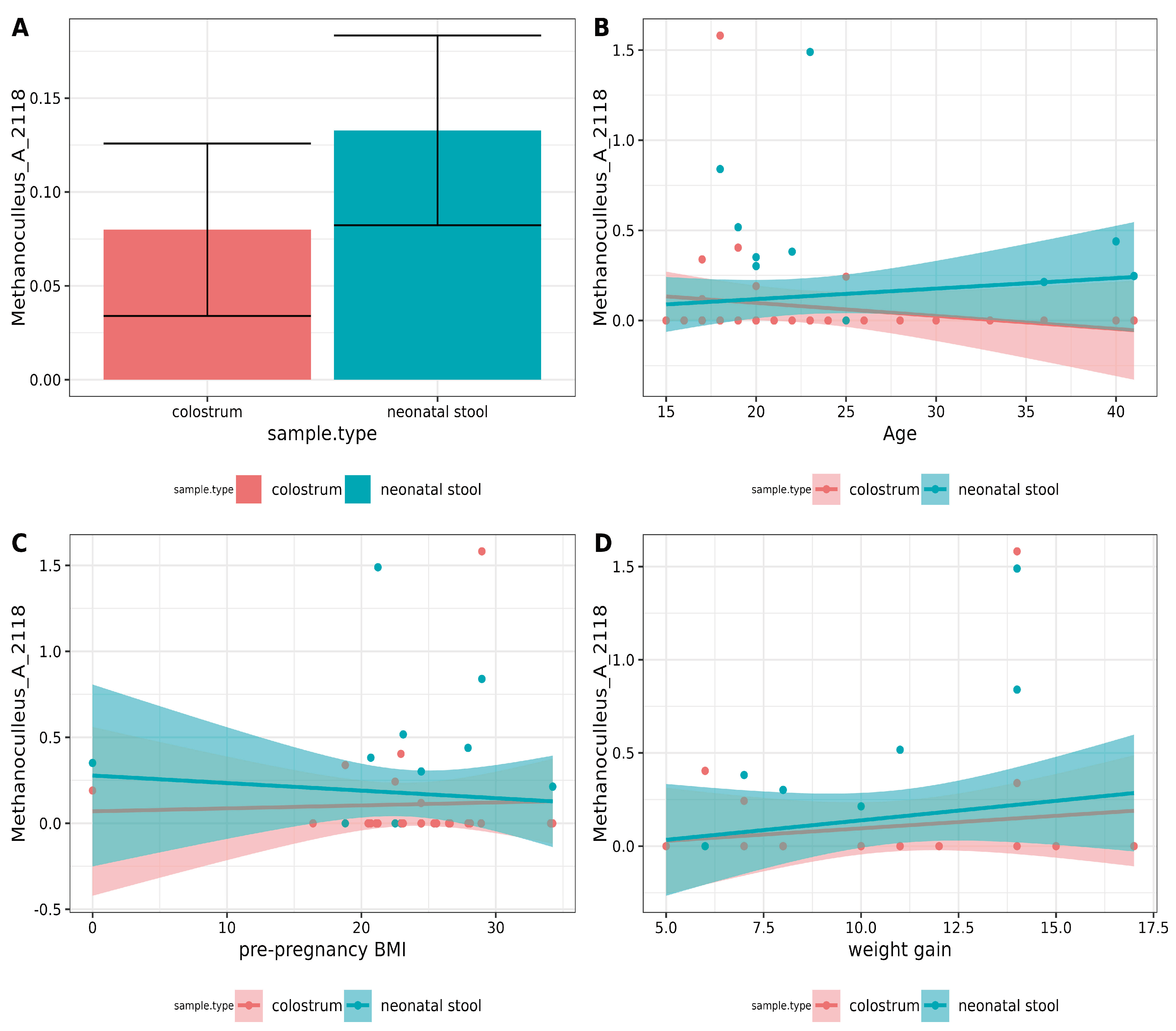
Differential abundance analyses using DESeq2, ALDEx2, and LEFSE did not reveal any significant differences. The two-group Pearson correlation test was used to detect potential similarities. A moderate positive correlation (ρ = 0.35, p = 0.038) for Methanoculleus_A_2118 was found for each pair in the binomials (Figure 4A). However, after applying the Holm correction, this correlation was no longer statistically significant. Correlations between ASVs and variables such as mothers’ age, pre-pregnancy BMI, and weight gain using the ALDEx2 correlation module were also examined, but no associations were found. The results for Methanoculleus_A_2118 are shown in Figure 4B–D.
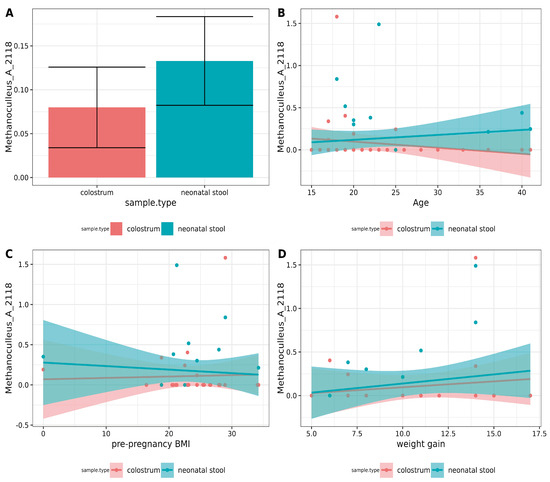
Figure 4.
Correlation analysis of archaea between pairs and metadata. (A) Paired Pearson correlation test between colostrum and neonatal stool. Only p-values < 0.05 were included. The Y-axis shows the square root of mean relative abundance, and the X-axis, sample types. (B–D). Correlation analysis between archaea and metadata using aldex.cor module. No significant correlation was found with the mother’s (A) age, (B) pre-pregnancy BMI, or (D) weight gain during pregnancy.
3.6. Co-Occurrence Networks Show Distinct Microbial Relationships in Colostrum and Neonatal Stool
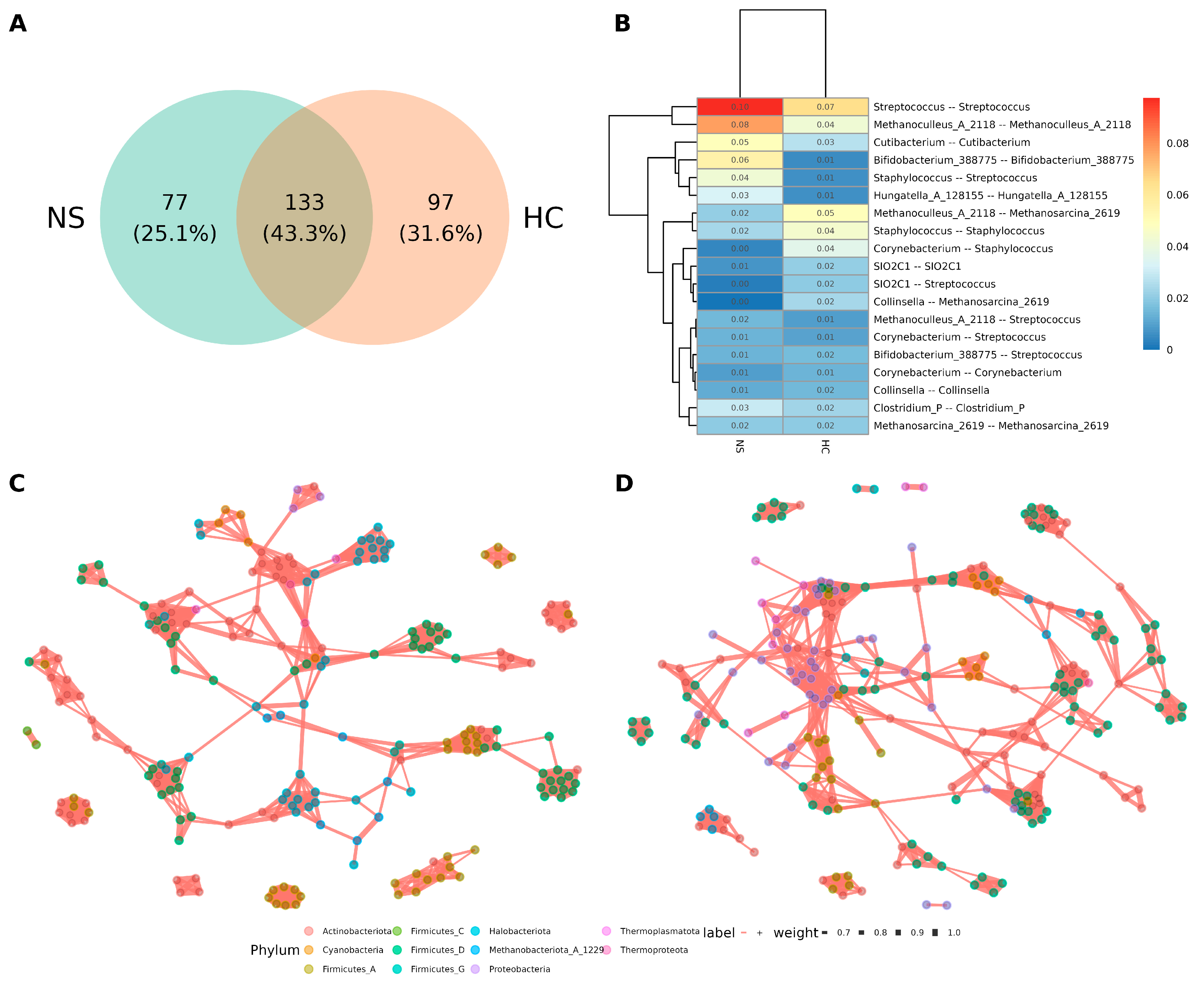
The co-occurrence networks of microbial interactions in each sample type were analyzed. Since the sequencing data included bacterial reads, bacteria were incorporated into the analysis to explore their relationships with archaea. Separate networks were constructed for colostrum and neonatal stool using Pearson correlation with p < 0.01 and ρ > 0.6 thresholds. The colostrum network contained 97 unique edges, while the neonatal stool network had 77, with 133 edges (43.3%) shared between the two (Figure 5A).
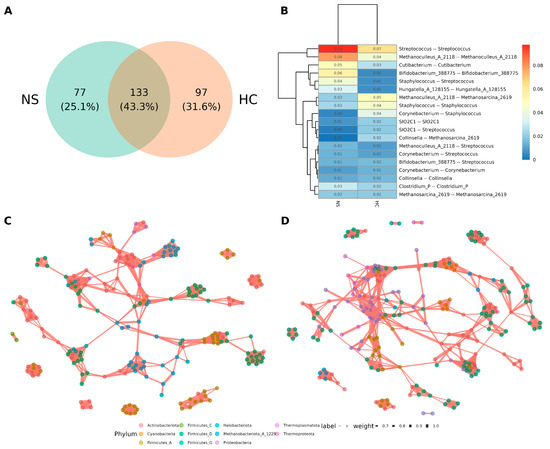
Figure 5.
Microbial co-occurrence network comparison between human milk and neonatal stools. (A) Venn diagram of edges between the networks of neonatal stool (NS) and human colostrum (HC). (B) Number distribution of taxa associated with the linked nodes of positive edges in networks of NS and HC. The number in the plot indicates the ratio of edges against all the positive edges in the network. (C) Microbial co-occurrence network of neonatal stool. A connection between nodes stands for a strong (Spearman’s ρ > 0.6) and significant (p > 0.01) correlation. (D) Microbial co-occurrence network of human colostrum. A connection stands for a strong (Spearman’s ρ > 0.6) and significant (p > 0.01).
The most notable relationships are highlighted in Figure 5B. Many involved ASVs from the same genus (e.g., Streptococcus-Streptococcus, Methanoculleus-Methanoculleus), which may suggest a phylogenetic similarity. However, relationships between different genera in the same phylogenetics domain, such as Staphylococcus-Streptococcus, Methanoculleus-Methanosarcina, Corynebacterium-Staphylococcus, Corynebacterium-Streptococcus, and Bifidobacterium-Streptococcus were observed. Remarkably, a direct inter-domain relationship of bacteria with archaea was limited only to Collinsella-Methanosarcina and Methanoculleus-Streptococcus (Figure 5B).
Examining the overall topology of the individual networks for neonatal stool (Figure 5C, Figure S2) and colostrum (Figure 5D, Figure S3), most edges were between members of the same genus or domain, with only a few hubs showing archaea-bacteria relationships. Notably, bacterial hubs frequently served as bridges for distant connections. Additionally, the hubs in neonatal stool networks were more dispersed, while those in colostrum networks were more closely clustered.
3.7. Predicted Functional Metagenome Highlights Methanogen-Associated Pathways
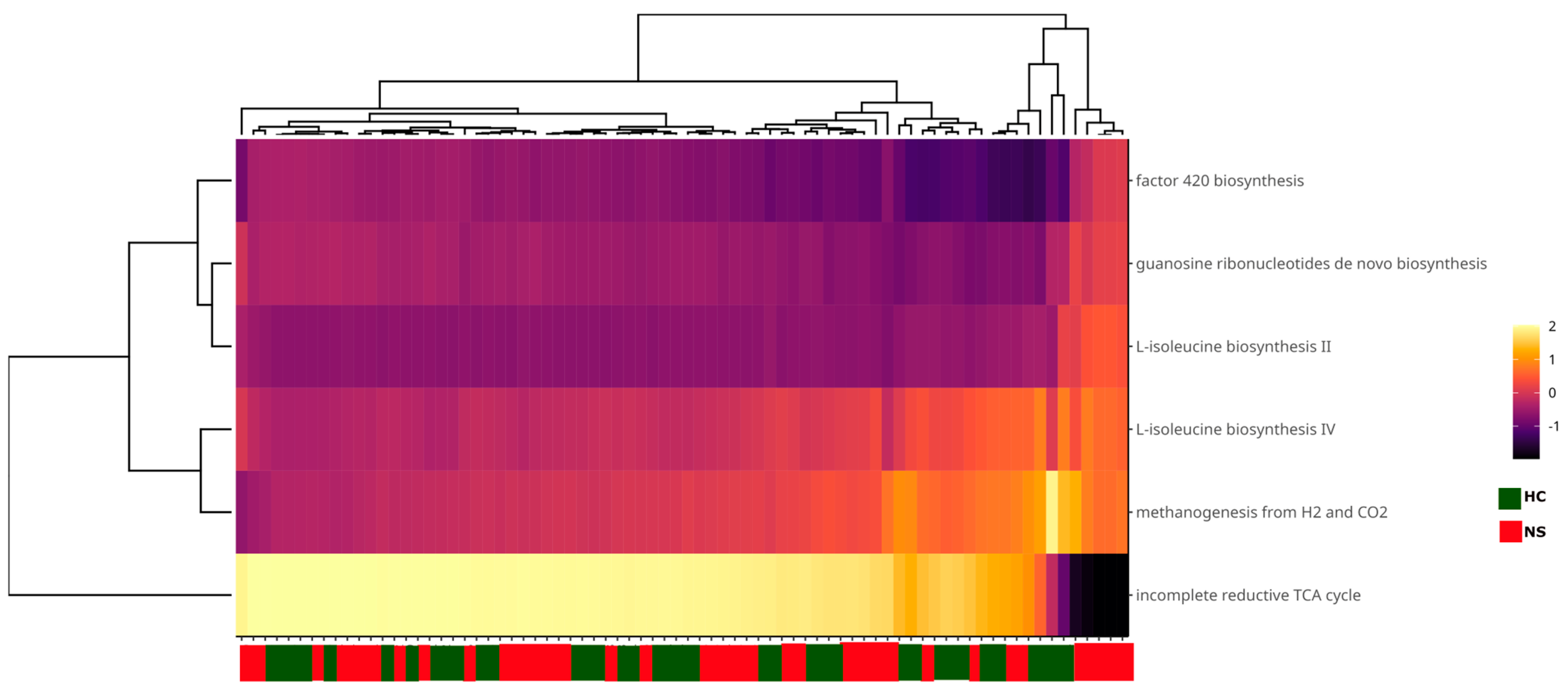
Finally, functional metabolic pathways in the colostrum and neonatal stool microbiota were identified using PICRUSt2 analysis based on the ASV table. No differentially abundant pathways were detected between the two sample types. The primary pathways, present in at least 10% of the samples, were associated with methanogens. These included the incomplete reductive TCA cycle pathway and the L-isoleucine biosynthesis IV and II pathways. Additionally, some pathways were directly related to methanogenesis, such as the factor 420 biosynthesis pathway and the methanogenesis from H₂ to CO₂ pathway (Figure 6).
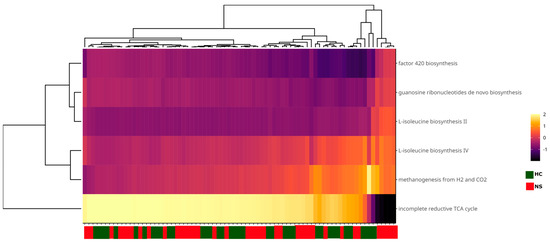
Figure 6.
Heatmap of functional microbial metabolic pathways using PICRUSt2 analysis with MetaCyc database. Columns show the abundance of main metabolic pathways with a prevalence of at least 10% in the samples and an abundance > 1%. Sample names are shown on the X-axis. The color scale from black (−2) to white (2) indicates the relative abundance of the predicted metabolic pathways; the green and red color tags indicate the sample type.
4. Discussion
This work reports the archaeal composition found in colostrum and neonatal stool collected from Mexican mother-neonate pairs. To our knowledge, this is the first report in Mexican binomials. In general, we found a high abundance of phylum Halobacteriota (~89%) and phylum Methanobacteriota_A_1229 (~11%) in colostrum and neonatal stool samples. The main genera were members of this phylum (Methanoculleus and Methanosarcina). A member of the Methanoculleus genus had already been found in the human intestinal mucosa by sequence analysis of the archaeal methyl coenzyme-M reductase (mcrA) gene in colonic biopsies [68]. However, apart from this study, little is known about its presence in the human gut. Similarly, in the case of the Methanosarcina genus, there are no direct reports [39,69]. Although recent studies have not found this genus [25,26], there is one report on human gut methanogens where a mcrA gene phylotype, named Mx-01, has been reported, which was later associated with the Methanosarcinales order [70,71,72,73]. Nevertheless, this phylotype was also suggested to belong to a new order of methanogens [71]. However, it is important to mention that Methanosarcina has been found in the gastrointestinal tract of different animals like goats, termites, and pigs [74,75,76]. Alternatively, Methanosarcina spp. is part of humans’ oral and skin microbiota [77,78,79,80,81,82,83]. Particularly, the species Methanosarcina mazeii and Methanosarcina vacuolata have been observed in subjects affected by periodontitis and healthy subjects [78,82]. It could be the case that the presence of this genus in the neonate’s gut was due to oral or skin microbiota. Another hypothesis would be that it has not been well characterized in the human intestine, its abundance being underestimated in infant populations.
Methanobrevibacter smithii and Methanosphaera stadmanae are considered the most prevalent archaea in the adult human gut [23,25,84]. In addition, in neonates’ gastrointestinal tract, Methanobrevibacter and Methanosphaera, as well as one uncultured phylotype, have been reported [19,20,21,22]. These studies consisted of 16S rRNA gene sequencing [22], multispacer sequence typing [20], clone library sequencing [19], and qPCR targeting [21]. In our work, the genus Methanobrevibacter_A was present in the two sample types but at low abundance (~6.6%), the genus Methanothermobacter_A_884 at ~5.5%, and the genus Methanofollis in less than 0.1% abundance. The disparities in the archaeal genera proportions of this work against previously reported research could be attributed to lifestyle differences, since no previous reports of archaeal composition in Mexican women exist. It is known that hormonal changes during pregnancy affect the microbiota [85,86,87]. Therefore, the archaeal population of a pregnant woman might be distinct. Moreover, the microbiota is also influenced by diet [88,89,90,91], geographical location, and urban or rural lifestyle [92,93]. Considering this, we hypothesized that the differences found in the proportions of archaeal genera might be because we studied human milk and neonatal stool samples of less than four days of age.
The alpha diversity analysis showed that human colostrum had a tendency for higher archaeal diversity than neonatal stools. Accordingly, human milk bacteria in a similar Mexican cohort also showed higher diversity when compared to neonatal stool [94]. We believe this is due to the neonate’s age (<4 days), which indicates that colonization of their gastrointestinal tract was just beginning, explaining the lower diversity [19,22,70]. Apart from this, the significant difference between colostrum’s and neonate stool’s diversity can also be explained by their ecological niches, which might favor the presence of specific archaea [95]. Interestingly, some authors have concluded that alpha diversity measures might underestimate microbiota, and more robust statistical methods might be necessary to assess the differences [96,97]. Colostrum and neonatal stool were found to have a highly similar core archaeal microbiota, consisting of members of Halobacteriota and Methanobacteriota_a_1229 phyla, which included the genera Methanoculleus_A_2118, Methanosarcina_2619, Methanobrevibacter_A, Methanothermobacter_A_884, and Methanofollis. The beta diversity analysis, both by NMDS ordination and Jensen–Shannon diverge (JSD) distances, further explained the similarity between colostrum and neonatal stool samples, with the NMDS ordination showing that the sample types overlapped. The similarity between the two sample types suggests that shared taxa are possibly transmitted via breastfeeding. Moreover, the metagenomic prediction analysis suggested no differentially abundant pathways between the sample types, strengthening our previous results.
Regarding the predicted metabolic routes detected, the low abundance could be due to the combination of the natural low abundance of archaea in the sample types and the lack of archaeal metabolic information in the MetaCyc database. Observing the most prevalent pathways among the samples, we found that all of them were associated with methanogens. The incomplete reductive TCA pathway was present in almost 90% of the samples. This route is characteristic of methanogens and allows for the synthesis of intermediates needed for amino acid production [98,99]. The methanogenesis from H2 and CO2 was also prevalent although less abundant, thus revealing the predominance of hydrogenotrophic archaea, i.e., methanogens that utilize H2, formate, or simple alcohols as electron donors and CO2 as an electron acceptor [100]. This pathway starts with CO2 activation and is followed by numerous transformations, including one aided by the factor 420, which was also prevalent in our study samples, and another methanogenesis indicator [101]. Finally, the L-isoleucine biosynthesis pathways II and IV have been associated with methanogens such as Methanocaldococcus jannaschii [102], Methanothermobacter thermautotrophicus, and Methanobrevibacter arboriphilus [103]. Together, these results suggest that methanogens are vertically transmitted through lactation.
Lastly, the co-occurrence network revealed there were 133 edges or connections (accounting for 43.3%) in common between the human colostrum and neonatal stool networks, supporting the idea of microbial similarity among the sample types. Methanogens and bacteria are known to form syntrophic interactions [24,46,83,104], therefore we sought to see possible Archaea-Bacteria associations. We observed a direct inter-domain relationship of bacteria with archaea only for Collinsella-Methanosarcina and Methanoculleus-Streptococcus. Thus, an interaction between these bacteria and the archaea is plausible. In the human colostrum network, we observed Streptococcus associated with methanogens. Streptococcus co-occurrence with archaea had only been seen in febrile patients’ blood [46,105]. These results showed that some associations found in pathogenesis might also be common in healthy individuals. In the neonatal stool network, we found co-occurring Methanoculleus-Streptococcus.
The presence of archaea in biological samples like human milk could be questioned since these microorganisms are reported to inhabit extremophile environments. It could be argued that the detection of these species is due to contamination during sample handling or other sources like DNA extraction kits [106]. This last possibility was ruled out in this work by using appropriate handling of samples, negative controls for PCR, and massive DNA sequencing followed by downstream bioinformatic strategies used for removing contaminants and low prevalence features, as described previously in the text. A more interesting explanation is that methanogenic archaea are vertically transmitted by the mother to the neonate, favoring the remotion of hydrogen [107]. It is known that under some conditions, the accumulation of hydrogen in the gut has been reported to be associated with human disease [104,108]. Finally, live archaea have been detected and isolated from human colostrum and milk [18,109].
5. Conclusions
In summary, in this study, we characterized the archaeal composition of human colostrum-neonatal stool paired samples, finding evidence of the presence of Methanoculleus_A_2118, Methanosarcina_2619, Methanobrevibacter_A, Methanothermobacter_A_884, and Methanofollis to be the main genera in both sample types. Moreover, the similarities between the sample types suggest there is vertical transmission of archaea during breastfeeding. Differential abundance yielded no significant differences. Finally, the co-occurrence network analysis showed associations between Archaea and Bacteria that might be relevant for these organisms’ presence in the human milk and neonatal stool ecosystems. Future studies should aim to characterize other potential sources of archaea in the neonatal stool as well as their associations with Bacteria. All in all, this study represents a first step in understanding the origin of archaea in the gut from the beginning of life and remarks on the importance of continuing to study these often-overlooked microorganisms.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13010085/s1, Figure S1: Sequencing results of archaeal and bacterial abundance reads in negative controls used for Decontam; Figure S2: Microbial co-occurrence network in the neonatal stool; Figure S3: Microbial co-occurrence network comparison in human colostrum; Table S1: Sequencing summary; Table S2: Decontam results.; Table S3: Summary of numerical values of Phyla Relative Abundances (%); Table S4: Summary of numerical values of relative abundances of genera; Table S5: Statistical test for Alpha Diversity Indexes.
Author Contributions
M.S.-L.: Conceptualization, Investigation, Methodology, Visualization, Writing—original draft, reviewing and editing. J.M.V.-I.: Data curation, Formal analysis, Visualization, Software. D.L.R.-G.: Conceptualization, Investigation, Methodology, Visualization. A.P.-E.: Methodology, Supervision. J.M.H.-H.: Validation, Writing—reviewing and editing. M.N.R.-C.: Conceptualization, Methodology, Funding Acquisition. C.P.-C.: Conceptualization, Validation, Writing—reviewing and editing. K.C.-C.: Data curation, Methodology, Writing—reviewing and editing, Investigation. C.J.J.-C.: Methodology, Investigation, Supervision, J.G.-M.: Writing—original draft, reviewing and editing, Resources, Project administration, and Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Consejo Nacional de Humanidades, Ciencias y Tecnologías (CONAHCYT) Mexico: CONACYT-163235, INFR-2011-01; Consejo Nacional de Humanidades, Ciencias y Tecnologías (CONAHCYT) Mexico: CONACYT FORDECYT-PRONACES/6669/2020_Programa Presupuestario F003-Ciencia de Frontera 2019. The funding body was not involved in study design; collection, management, analysis, and interpretation of data; or the decision to submit for publication.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of the General Hospital “Dr José María Rodríguez” (Project identification code: 217B560002018006).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated for this study can be found in the NCBI BioProject ID PRJNA1018680 Link https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1018680 (accessed on 11 December 2023).
Acknowledgments
We are grateful to all families who agreed to participate in the study, Rodrigo García-Gutiérrez for technical support in the laboratory, and Viridiana Rosas-Ocegueda for administrative assistance. C.P.-C. (47399), J.M.H.-H. (225832), and J.G.-M. (19815) are Fellows from the Sistema Nacional de Investigadores, Mexico. We thank CONAHCyT for Doctoral Fellowships 997152 to J.M.V.-I., and 777953 to K.C.-C.; Master Fellowships 1140881 to M.S.-L., 1140600 to D.L.R.-G., and Estancias-Posdoctorales-por-México Fellowship 321600 to C.J.J.-C.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pace, R.M.; Williams, J.E.; Robertson, B.; Lackey, K.A.; Meehan, C.L.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. Variation in Human Milk Composition Is Related to Differences in Milk and Infant Fecal Microbial Communities. Microorganisms 2021, 9, 1153. [Google Scholar] [CrossRef] [PubMed]
- Gila-Diaz, A.; Arribas, S.M.; Algara, A.; Martín-Cabrejas, M.A.; López de Pablo, Á.L.; Sáenz de Pipaón, M.; Ramiro-Cortijo, D. A Review of Bioactive Factors in Human Breastmilk: A Focus on Prematurity. Nutrients 2019, 11, 1307. [Google Scholar] [CrossRef]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Langa, S.; Martín, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The Human Milk Microbiota: Origin and Potential Roles in Health and Disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Selma-Royo, M.; Calvo-Lerma, J.; Bäuerl, C.; Esteban-Torres, M.; Cabrera-Rubio, R.; Collado, M.C. Human Milk Microbiota: What Did We Learn in the Last 20 Years? Microbiome Res. Rep. 2022, 1, 19. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human Milk Composition: Nutrients and Bioactive Factors. Pediatr. Clin. 2013, 60, 49–74. [Google Scholar] [CrossRef]
- Thapa, B.R. Health Factors in Colostrum. Indian J. Pediatr. 2005, 72, 579–581. [Google Scholar] [CrossRef]
- Uruakpa, F.O.; Ismond, M.A.H.; Akobundu, E.N.T. Colostrum and Its Benefits: A Review. Nutr. Res. 2002, 22, 755–767. [Google Scholar] [CrossRef]
- Stinson, L.F.; Sindi, A.S.M.; Cheema, A.S.; Lai, C.T.; Mühlhäusler, B.S.; Wlodek, M.E.; Payne, M.S.; Geddes, D.T. The Human Milk Microbiome: Who, What, When, Where, Why, and How? Nutr. Rev. 2021, 79, 529–543. [Google Scholar] [CrossRef]
- Fitzstevens, J.L.; Smith, K.C.; Hagadorn, J.I.; Caimano, M.J.; Matson, A.P.; Brownell, E.A. Systematic Review of the Human Milk Microbiota. Nutr. Clin. Pract. 2017, 32, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Notarbartolo, V.; Giuffrè, M.; Montante, C.; Corsello, G.; Carta, M. Composition of Human Breast Milk Microbiota and Its Role in Children’s Health. Pediatr. Gastroenterol. Hepatol. Nutr. 2022, 25, 194–210. [Google Scholar] [CrossRef]
- Rodríguez, J.M. The Origin of Human Milk Bacteria: Is There a Bacterial Entero-Mammary Pathway during Late Pregnancy and Lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef]
- Ruiz, L.; García-Carral, C.; Rodriguez, J.M. Unfolding the Human Milk Microbiome Landscape in the Omics Era. Front. Microbiol. 2019, 10, 1378. [Google Scholar] [CrossRef]
- Moossavi, S.; Azad, M.B. Origins of Human Milk Microbiota: New Evidence and Arising Questions. Gut Microbes 2020, 12, 1667722. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.; Ricciardi-Castagnoli, P. Dendritic Cells Express Tight Junction Proteins and Penetrate Gut Epithelial Monolayers to Sample Bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.A. Human Milk Microbiota and Oligosaccharides: A Glimpse into Benefits, Diversity, and Correlations. Nutrients 2021, 13, 1123. [Google Scholar] [CrossRef]
- Jiménez, E.; de Andrés, J.; Manrique, M.; Pareja-Tobes, P.; Tobes, R.; Martínez-Blanch, J.F.; Codoñer, F.M.; Ramón, D.; Fernández, L.; Rodríguez, J.M. Metagenomic Analysis of Milk of Healthy and Mastitis-Suffering Women. J. Hum. Lact. 2015, 31, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Togo, A.H.; Grine, G.; Khelaifia, S.; des Robert, C.; Brevaut, V.; Caputo, A.; Baptiste, E.; Bonnet, M.; Levasseur, A.; Drancourt, M.; et al. Culture of Methanogenic Archaea from Human Colostrum and Milk. Sci. Rep. 2019, 9, 18653. [Google Scholar] [CrossRef]
- Palmer, C.; Bik, E.M.; Giulio, D.D.; Realman, D.A.; Brown, P.O. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007, 5, e117. Available online: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0050177 (accessed on 11 July 2022). [CrossRef]
- Grine, G.; Boualam, M.A.; Drancourt, M. Methanobrevibacter Smithii, a Methanogen Consistently Colonising the Newborn Stomach. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 2449–2455. [Google Scholar] [CrossRef] [PubMed]
- Sagheddu, V.; Patrone, V.; Miragoli, F.; Morelli, L. Abundance and Diversity of Hydrogenotrophic Microorganisms in the Infant Gut before the Weaning Period Assessed by Denaturing Gradient Gel Electrophoresis and Quantitative PCR. Front. Nutr. 2017, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Wampach, L.; Heintz-Buschart, A.; Hogan, A.; Muller, E.E.L.; Narayanasamy, S.; Laczny, C.C.; Hugerth, L.W.; Bindl, L.; Bottu, J.; Andersson, A.F.; et al. Colonization and Succession within the Human Gut Microbiome by Archaea, Bacteria, and Microeukaryotes during the First Year of Life. Front. Microbiol. 2017, 8, 738. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Henry, M.; El Khéchine, A.; Raoult, D.; Drancourt, M. High Prevalence of Methanobrevibacter Smithii and Methanosphaera Stadtmanae Detected in the Human Gut Using an Improved DNA Detection Protocol. PLoS ONE 2009, 4, e7063. [Google Scholar] [CrossRef] [PubMed]
- Borrel, G.; Brugère, J.-F.; Gribaldo, S.; Schmitz, R.A.; Moissl-Eichinger, C. The Host-Associated Archaeome. Nat. Rev. Microbiol. 2020, 18, 622–636. [Google Scholar] [CrossRef] [PubMed]
- Chibani, C.M.; Mahnert, A.; Borrel, G.; Almeida, A.; Werner, A.; Brugère, J.-F.; Gribaldo, S.; Finn, R.D.; Schmitz, R.A.; Moissl-Eichinger, C. A Catalogue of 1,167 Genomes from the Human Gut Archaeome. Nat. Microbiol. 2022, 7, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, R.; Mahnert, A.; Duller, S.; Moissl-Eichinger, C. Archaeal Key-Residents within the Human Microbiome: Characteristics, Interactions and Involvement in Health and Disease. Curr. Opin. Microbiol. 2022, 67, 102146. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Whitman, W.B. Methanogens. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 602–606. ISBN 978-0-12-384733-1. [Google Scholar]
- van de Pol, J.A.A.; van Best, N.; Mbakwa, C.A.; Thijs, C.; Savelkoul, P.H.; Arts, I.C.W.; Hornef, M.W.; Mommers, M.; Penders, J. Gut Colonization by Methanogenic Archaea Is Associated with Organic Dairy Consumption in Children. Front. Microbiol. 2017, 8, 355. [Google Scholar] [CrossRef] [PubMed]
- Buan, N.R. Methanogens: Pushing the Boundaries of Biology. Emerg. Top. Life Sci. 2018, 2, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Meier, D.; van Grinsven, S.; Michel, A.; Eickenbusch, P.; Glombitza, C.; Han, X.; Fiskal, A.; Bernasconi, S.; Schubert, C.J.; Lever, M.A. Hydrogen–Independent CO2 Reduction Dominates Methanogenesis in Five Temperate Lakes That Differ in Trophic States. ISME Commun. 2024, 4, ycae089. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.P.; Conway, P.L.; Schlundt, J. Methanogens in Humans: Potentially Beneficial or Harmful for Health. Appl. Microbiol. Biotechnol. 2018, 102, 3095–3104. [Google Scholar] [CrossRef]
- Samuel, B.S.; Gordon, J.I. A Humanized Gnotobiotic Mouse Model of Host–Archaeal–Bacterial Mutualism. Proc. Natl. Acad. Sci. USA 2006, 103, 10011–10016. [Google Scholar] [CrossRef]
- Weaver, G.A.; Krause, J.A.; Miller, T.L.; Wolin, M.J. Incidence of Methanogenic Bacteria in a Sigmoidoscopy Population: An Association of Methanogenic Bacteria and Diverticulosis. Gut 1986, 27, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Yazici, C.; Arslan, D.C.; Abraham, R.; Cushing, K.; Keshavarzian, A.; Mutlu, E.A. Breath Methane Levels Are Increased Among Patients with Diverticulosis. Dig. Dis. Sci. 2016, 61, 2648–2654. [Google Scholar] [CrossRef] [PubMed]
- Lecours, P.B.; Marsolais, D.; Cormier, Y.; Berberi, M.; Haché, C.; Bourdages, R.; Duchaine, C. Increased Prevalence of Methanosphaera Stadtmanae in Inflammatory Bowel Diseases. PLoS ONE 2014, 9, e87734. [Google Scholar] [CrossRef]
- Ghavami, S.B.; Rostami, E.; Sephay, A.A.; Shahrokh, S.; Balaii, H.; Aghdaei, H.A.; Zali, M.R. Alterations of the Human Gut Methanobrevibacter Smithii as a Biomarker for Inflammatory Bowel Diseases. Microb. Pathog. 2018, 117, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Brugère, J.-F.; Borrel, G.; Gaci, N.; Tottey, W.; O’Toole, P.W.; Malpuech-Brugère, C. Archaebiotics. Gut Microbes 2014, 5, 5–10. [Google Scholar] [CrossRef]
- Ramezani, A.; Nolin, T.D.; Barrows, I.R.; Serrano, M.G.; Buck, G.A.; Regunathan-Shenk, R.; West, R.E.; Latham, P.S.; Amdur, R.; Raj, D.S. Gut Colonization with Methanogenic Archaea Lowers Plasma Trimethylamine N-Oxide Concentrations in Apolipoprotein E−/− Mice. Sci. Rep. 2018, 8, 14752. [Google Scholar] [CrossRef] [PubMed]
- Sereme, Y.; Mezouar, S.; Grine, G.; Mege, J.L.; Drancourt, M.; Corbeau, P.; Vitte, J. Methanogenic Archaea: Emerging Partners in the Field of Allergic Diseases. Clin. Rev. Allergy Immunol. 2019, 57, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Camara, A.; Konate, S.; Tidjani Alou, M.; Kodio, A.; Togo, A.H.; Cortaredona, S.; Henrissat, B.; Thera, M.A.; Doumbo, O.K.; Raoult, D.; et al. Clinical Evidence of the Role of Methanobrevibacter Smithii in Severe Acute Malnutrition. Sci. Rep. 2021, 11, 5426. [Google Scholar] [CrossRef]
- Zhang, H.; DiBaise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human Gut Microbiota in Obesity and after Gastric Bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef]
- Maya-Lucas, O.; Murugesan, S.; Nirmalkar, K.; Alcaraz, L.D.; Hoyo-Vadillo, C.; Pizano-Zárate, M.L.; García-Mena, J. The Gut Microbiome of Mexican Children Affected by Obesity. Anaerobe 2019, 55, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Amabebe, E.; Robert, F.O.; Agbalalah, T.; Orubu, E.S.F. Microbial Dysbiosis-Induced Obesity: Role of Gut Microbiota in Homoeostasis of Energy Metabolism. Br. J. Nutr. 2020, 123, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, C.K. New-Found Link between Microbiota and Obesity. World J. Gastrointest. Pathophysiol. 2015, 6, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Ignacio, A.; Fernandes, M.R.; Rodrigues, V.A.A.; Groppo, F.C.; Cardoso, A.L.; Avila-Campos, M.J.; Nakano, V. Correlation between Body Mass Index and Faecal Microbiota from Children. Clin. Microbiol. Infect. 2016, 22, e1–e258. [Google Scholar] [CrossRef] [PubMed]
- Djemai, K.; Drancourt, M.; Tidjani Alou, M. Bacteria and Methanogens in the Human Microbiome: A Review of Syntrophic Interactions. Microb. Ecol. 2022, 83, 536–554. [Google Scholar] [CrossRef] [PubMed]
- Lurie-Weinberger, M.N.; Gophna, U. Archaea in and on the Human Body: Health Implications and Future Directions. PLoS Pathog. 2015, 11, e1004833. [Google Scholar] [CrossRef] [PubMed]
- Nkamga, V.D.; Henrissat, B.; Drancourt, M. Archaea: Essential Inhabitants of the Human Digestive Microbiota. Hum. Microbiome J. 2017, 3, 1–8. [Google Scholar] [CrossRef]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-Specific Primer and Probe Sets to Detect Methanogenic Communities Using Quantitative Real-Time Polymerase Chain Reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Gállego-Bravo, A.K.; García-Mena, J.; Piña-Escobedo, A.; López-Jiménez, G.; Gutiérrez-Castillo, M.E.; Tovar-Gálvez, L.R. Monitoring of a Microbial Community during Bioaugmentation with Hydrogenotrophic Methanogens to Improve Methane Yield of an Anaerobic Digestion Process. Biotechnol. Lett. 2023, 45, 1339–1353. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 Unifies Microbial Data in a Single Reference Tree. Nat. Biotechnol. 2024, 42, 715–718. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2022. [Google Scholar]
- Bisanz, J.E. Qiime2r: Importing QIIME2 Artifacts and Associated Data into R Sessions. 2018. Available online: https://github.com/jbisanz/qiime2R (accessed on 13 December 2024).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Love, M.; Ahlmann-Eltze, C.; Forbes, K.; Anders, S.; Huber, W.; RADIANT EU FP7; NIH NHGRI; CZI. DESeq2: Differential Gene Expression Analysis Based on the Negative Binomial Distribution. 2023. Available online: https://bioc.r-universe.dev/DESeq2 (accessed on 13 December 2024).
- Nixon, M.P.; McGovern, K.C.; Letourneau, J.; David, L.A.; Lazar, N.A.; Mukherjee, S.; Silverman, J.D. Scale Reliant Inference. arXiv 2024, arXiv:2201.03616. [Google Scholar]
- Khleborodova, A. Lefser: R Implementation of the LEfSE Method for Microbiome Biomarker Discovery. 2024. Available online: https://www.bioconductor.org/packages/release/bioc/html/lefser.html (accessed on 13 December 2024).
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Auguie, B. GridExtra: Miscellaneous Functions for “Grid” Graphics. 2017. Available online: https://cran.r-project.org/package=gridExtra (accessed on 13 December 2024).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Owens, J.A.; Saeedi, B.; Cohen, C.E.; Bellissimo, M.P.; Naudin, C.; Darby, T.; Druzak, S.; Maner-Smith, K.; Orr, M.; et al. Microbial Metabolite Delta-Valerobetaine Is a Diet-Dependent Obesogen. Nat. Metab. 2021, 3, 1694–1705. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Chen, X.; Zhang, Y.; Zhang, H.; Xie, P. Gut Microbiota and Its Metabolites in Depression: From Pathogenesis to Treatment. eBioMedicine 2023, 90, 104527. [Google Scholar] [CrossRef] [PubMed]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple Statistical Identification and Removal of Contaminant Sequences in Marker-Gene and Metagenomics Data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Nava, G.M.; Carbonero, F.; Croix, J.A.; Greenberg, E.; Gaskins, H.R. Abundance and Diversity of Mucosa-Associated Hydrogenotrophic Microbes in the Healthy Human Colon. ISME J. 2012, 6, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hugon, P.; Lagier, J.-C.; Colson, P.; Bittar, F.; Raoult, D. Repertoire of Human Gut Microbes. Microb. Pathog. 2017, 106, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Mihajlovski, A.; Doré, J.; Levenez, F.; Alric, M.; Brugère, J.-F. Molecular Evaluation of the Human Gut Methanogenic Archaeal Microbiota Reveals an Age-Associated Increase of the Diversity. Environ. Microbiol. Rep. 2010, 2, 272–280. [Google Scholar] [CrossRef]
- Mihajlovski, A.; Alric, M.; Brugère, J.-F. A Putative New Order of Methanogenic Archaea Inhabiting the Human Gut, as Revealed by Molecular Analyses of the mcrA Gene. Res. Microbiol. 2008, 159, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, P.D.; Shanahan, F.; Marchesi, J.R. Human Methanogen Diversity and Incidence in Healthy and Diseased Colonic Groups Using mcrA Gene Analysis. BMC Microbiol. 2008, 8, 79. [Google Scholar] [CrossRef]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugère, J.-F. Archaea and the Human Gut: New Beginning of an Old Story. World J. Gastroenterol. 2014, 20, 16062–16078. [Google Scholar] [CrossRef]
- Mukhopadhyay, B.; Purwantini, E.; Conway de Macario, E.; Daniels, L. Characterization of aMethanosarcina Strain Isolated from Goat Feces, and That Grows on H2-CO2 Only after Adaptation. Curr. Microbiol. 1991, 23, 165–173. [Google Scholar] [CrossRef]
- Gomathi, V.; Ramasamy, K.; Ramalakshmi, A.; Ramanathan, A. Methan Emission by Gut Symbionts of Termites. Acad. J. Plant Sci. 2009, 2, 189–194. [Google Scholar]
- Xiong, X.; Rao, Y.; Tu, X.; Wang, Z.; Gong, J.; Yang, Y.; Wu, H.; Liu, X. Gut Archaea Associated with Bacteria Colonization and Succession during Piglet Weaning Transitions. BMC Vet. Res. 2022, 18, 243. [Google Scholar] [CrossRef] [PubMed]
- Robichaux, M.; Howell, M.; Boopathy, R. Methanogenic Activity in Human Periodontal Pocket. Curr. Microbiol. 2003, 46, 53–58. [Google Scholar] [CrossRef]
- Matarazzo, F.; Ribeiro, A.C.; Feres, M.; Faveri, M.; Mayer, M.P.A. Diversity and Quantitative Analysis of Archaea in Aggressive Periodontitis and Periodontally Healthy Subjects. J. Clin. Periodontol. 2011, 38, 621–627. [Google Scholar] [CrossRef]
- Nguyen-Hieu, T.; Khelaifia, S.; Aboudharam, G.; Drancourt, M. Methanogenic Archaea in Subgingival Sites: A Review. APMIS 2013, 121, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.J.; Auerbach, A.K.; Moissl-Eichinger, C. Archaea on Human Skin. PLoS ONE 2013, 8, e65388. [Google Scholar] [CrossRef] [PubMed]
- Huynh, H.T.T.; Pignoly, M.; Nkamga, V.D.; Drancourt, M.; Aboudharam, G. The Repertoire of Archaea Cultivated from Severe Periodontitis. PLoS ONE 2015, 10, e0121565. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.-L.; Szafrański, S.P.; Jarek, M.; Bhuju, S.; Wagner-Döbler, I. Dysbiosis in Chronic Periodontitis: Key Microbial Players and Interactions with the Human Host. Sci. Rep. 2017, 7, 3703. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N. Symbiotic Interactions of Archaea in Animal and Human Microbiomes. Curr. Clin. Microbiol. Rep. 2023, 10, 161–173. [Google Scholar] [CrossRef]
- Bang, C.; Weidenbach, K.; Gutsmann, T.; Heine, H.; Schmitz, R.A. The Intestinal Archaea Methanosphaera Stadtmanae and Methanobrevibacter Smithii Activate Human Dendritic Cells. PLoS ONE 2014, 9, e99411. [Google Scholar] [CrossRef]
- Di Simone, N.; Santamaria Ortiz, A.; Specchia, M.; Tersigni, C.; Villa, P.; Gasbarrini, A.; Scambia, G.; D’Ippolito, S. Recent Insights on the Maternal Microbiota: Impact on Pregnancy Outcomes. Front. Immunol. 2020, 11, 528202. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Murtaza, G.; Kalhoro, D.H.; Kalhoro, M.S.; Metwally, E.; Chughtai, M.I.; Mazhar, M.U.; Khan, S.A. Relationship between Gut Microbiota and Host-Metabolism: Emphasis on Hormones Related to Reproductive Function. Anim. Nutr. 2021, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.; Kim, N. Roles of Sex Hormones and Gender in the Gut Microbiota. J. Neurogastroenterol. Motil. 2021, 27, 314–325. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Merra, G.; Noce, A.; Marrone, G.; Cintoni, M.; Tarsitano, M.G.; Capacci, A.; De Lorenzo, A. Influence of Mediterranean Diet on Human Gut Microbiota. Nutrients 2021, 13, 7. [Google Scholar] [CrossRef]
- Ramos, S.; Martín, M.Á. Impact of Diet on Gut Microbiota. Curr. Opin. Food Sci. 2021, 37, 83–90. [Google Scholar] [CrossRef]
- Nova, E.; Gómez-Martinez, S.; González-Soltero, R. The Influence of Dietary Factors on the Gut Microbiota. Microorganisms 2022, 10, 1368. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.; Yang, H. Factors Affecting the Composition of the Gut Microbiota, and Its Modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Cheng, Y.; Selma-Royo, M.; Cao, X.; Calatayud, M.; Qi, Q.; Zhou, J.; Zeng, L.; Garcia-Mantrana, I.; Collado, M.C.; Han, B. Influence of Geographical Location on Maternal-Infant Microbiota: Study in Two Populations From Asia and Europe. Front. Cell. Infect. Microbiol. 2022, 11, 663513. [Google Scholar] [CrossRef] [PubMed]
- Corona-Cervantes, K.; García-González, I.; Villalobos-Flores, L.E.; Hernández-Quiroz, F.; Piña-Escobedo, A.; Hoyo-Vadillo, C.; Rangel-Calvillo, M.N.; García-Mena, J. Human Milk Microbiota Associated with Early Colonization of the Neonatal Gut in Mexican Newborns. PeerJ 2020, 8, e9205. [Google Scholar] [CrossRef]
- Koskinen, K.; Pausan, M.R.; Perras, A.K.; Beck, M.; Bang, C.; Mora, M.; Schilhabel, A.; Schmitz, R.; Moissl-Eichinger, C. First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio 2017, 8, e00824-17. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef]
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef] [PubMed]
- Ekiel, I.; Sprott, G.D.; Patel, G.B. Acetate and CO2 Assimilation by Methanothrix Concilii. J. Bacteriol. 1985, 162, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, A.; Raftery, M.; Saunders, N.F.W.; Guilhaus, M.; Cavicchioli, R. Biology of the Cold Adapted Archaeon, Methanococcoides Burtonii Determined by Proteomics Using Liquid Chromatography-Tandem Mass Spectrometry. J. Proteome Res. 2004, 3, 1164–1176. [Google Scholar] [CrossRef]
- Lyu, Z.; Shao, N.; Akinyemi, T.; Whitman, W.B. Methanogenesis. Curr. Biol. 2018, 28, R727–R732. [Google Scholar] [CrossRef] [PubMed]
- Shima, S.; Warkentin, E.; Thauer, R.K.; Ermler, U. Structure and Function of Enzymes Involved in the Methanogenic Pathway Utilizing Carbon Dioxide and Molecular Hydrogen. J. Biosci. Bioeng. 2002, 93, 519–530. [Google Scholar] [CrossRef]
- Drevland, R.M.; Waheed, A.; Graham, D.E. Enzymology and Evolution of the Pyruvate Pathway to 2-Oxobutyrate in Methanocaldococcus Jannaschii. J. Bacteriol. 2007, 189, 4391–4400. [Google Scholar] [CrossRef] [PubMed]
- Eikmanns, B.; Jaenchen, R.; Thauer, R.K. Propionate Assimilation by Methanogenic Bacteria. Arch. Microbiol. 1983, 136, 106–110. [Google Scholar] [CrossRef]
- Duller, S.; Moissl-Eichinger, C. Archaea in the Human Microbiome and Potential Effects on Human Infectious Disease. Emerg. Infect. Dis. 2024, 30, 1505–1513. [Google Scholar] [CrossRef]
- Drancourt, M.; Djemai, K.; Gouriet, F.; Grine, G.; Loukil, A.; Bedotto, M.; Levasseur, A.; Lepidi, H.; Bou-Khalil, J.; Khelaifia, S.; et al. Methanobrevibacter Smithii Archaemia in Febrile Patients With Bacteremia, Including Those With Endocarditis. Clin. Infect. Dis. 2021, 73, e2571–e2579. [Google Scholar] [CrossRef]
- Olomu, I.N.; Pena-Cortes, L.C.; Long, R.A.; Vyas, A.; Krichevskiy, O.; Luellwitz, R.; Singh, P.; Mulks, M.H. Elimination of “Kitome” and “Splashome” Contamination Results in Lack of Detection of a Unique Placental Microbiome. BMC Microbiol. 2020, 20, 157. [Google Scholar] [CrossRef]
- Mutuyemungu, E.; Singh, M.; Liu, S.; Rose, D.J. Intestinal Gas Production by the Gut Microbiota: A Review. J. Funct. Foods 2023, 100, 105367. [Google Scholar] [CrossRef]
- Mafra, D.; Ribeiro, M.; Fonseca, L.; Regis, B.; Cardozo, L.F.M.F.; Fragoso Dos Santos, H.; Emiliano de Jesus, H.; Schultz, J.; Shiels, P.G.; Stenvinkel, P.; et al. Archaea from the Gut Microbiota of Humans: Could Be Linked to Chronic Diseases? Anaerobe 2022, 77, 102629. [Google Scholar] [CrossRef]
- Dombrowska-Pali, A.; Wiktorczyk-Kapischke, N.; Chrustek, A.; Olszewska-Słonina, D.; Gospodarek-Komkowska, E.; Socha, M.W. Human Milk Microbiome-A Review of Scientific Reports. Nutrients 2024, 16, 1420. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).