Relationship Between the Host Plant Range of Insects and Symbiont Bacteria
Abstract
1. Introduction
2. Relationship Between the Host Plant Range of Tephritids and Symbiont Bacteria
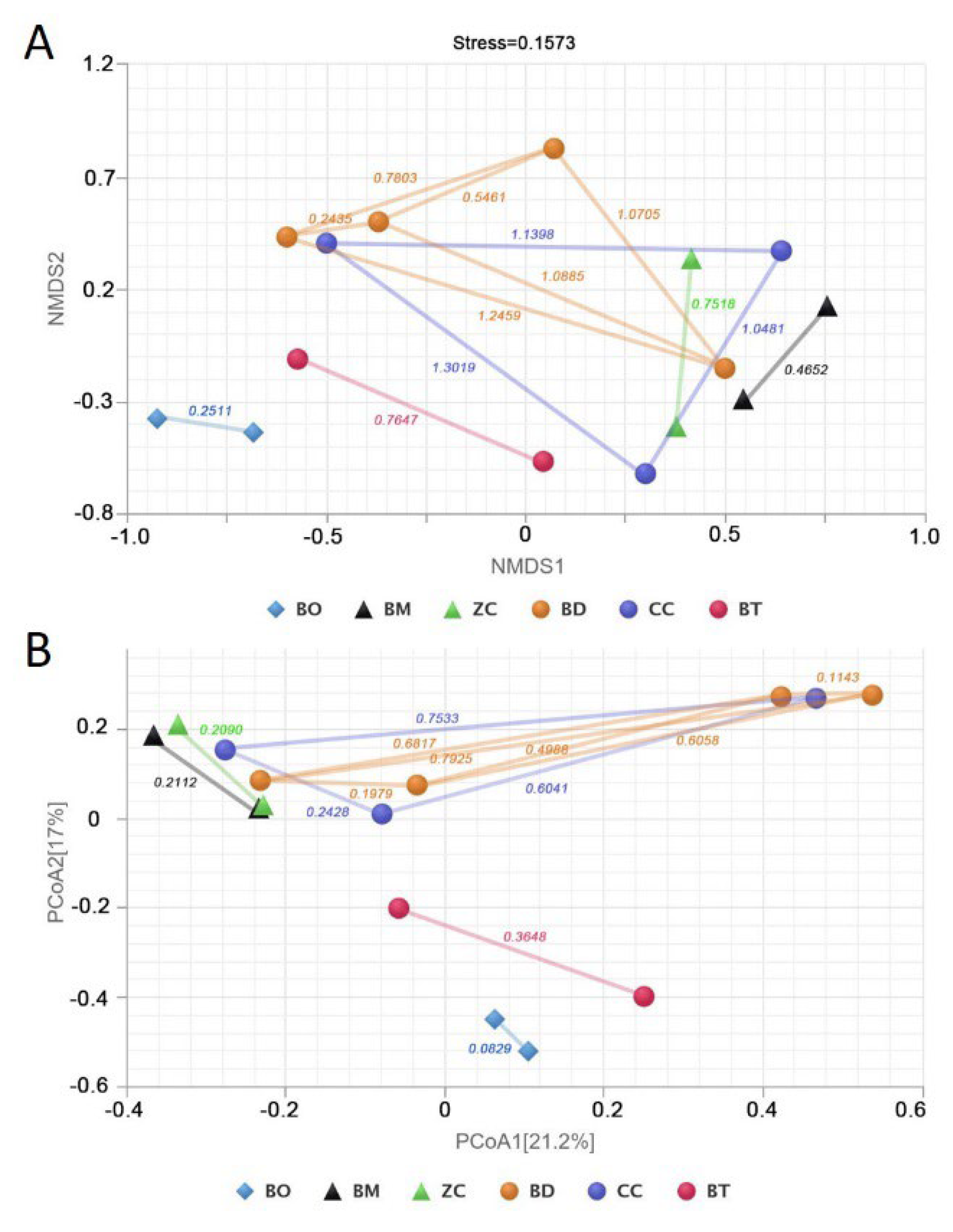
2.1. Is the Narrowing of the Insect’s Dietary Range Accompanied by a Reduction in the Diversity and Variation in Gut Bacteria?
| Name | Species | Sequencing Method | Source |
|---|---|---|---|
| 1AL | Anastrepha ludens (Loew, 1873) | Pyrosequencing | [44] |
| 1AO | Anastrepha obliqua (Macquart, 1835) | Pyrosequencing | [44] |
| 1ASE | Anastrepha serpentina (Wiedemann, 1830) | Pyrosequencing | [44] |
| 1AST | Anastrepha striata (Schiner, 1868) | Pyrosequencing | [44] |
| 2AF | Anastrepha fraterculus (Wiedemann, 1830) | Illumina | [45] |
| 3AO | Anastrepha obliqua (Macquart, 1835) | Metatranscriptomic | [37] |
| 4AG | Anastrepha grandis (Macquart, 1846) | Pyrosequencing | [46] |
| 4AL | Anastrepha ludens (Loew, 1873) | Pyrosequencing | [46] |
| 4BO | Bactrocera oleae (Rossi, 1790) | Pyrosequencing | [46] |
| 5CC | Ceratitis capitata (Wiedemann, 1824) | Illumina | [47] |
| 6CCW | Ceratitis capitata (Wiedemann, 1824) | Illumina | [48] |
| 6CCL | Ceratitis capitata (Wiedemann, 1824) | Illumina | [48] |
| 7BM | Bactrocera minax (Enderlein, 1920) | Pyrosequencing | [49] |
| 8BM | Bactrocera minax (Enderlein, 1920) | Metagenomic | [50] |
| 9BO | Bactrocera oleae (Rossi, 1790) | Illumina | [51] |
| 10ZC | Zeugodacus cucurbitae (Coquillett, 1899) | Illumina | [52] |
| 11BD1 | Bactrocera dorsalis (Hendel, 1912) | Illumina | [53] |
| 11BD2 | Bactrocera dorsalis (Hendel, 1912) | Illumina | [53] |
| 11BD3 | Bactrocera dorsalis (Hendel, 1912) | Illumina | [53] |
| 12BD | Bactrocera dorsalis (Hendel, 1912) | Pyrosequencing | [54] |
| 13BD | Bactrocera dorsalis (Hendel, 1912) | Illumina | [55] |
| 14BD | Bactrocera dorsalis (Hendel, 1912) | Illumina | [43] |
| 14ZC | Zeugodacus cucurbitae (Coquillett, 1899) | Illumina | [43] |
| 14BO | Bactrocera oleae (Rossi, 1790) | Illumina | [43] |
| 14BZ | Bactrocera zonata (Saunders, 1842) | Illumina | [43] |
| 14CC | Ceratitis capitata (Wiedemann, 1824) | Illumina | [43] |
| 14CQ | Ceratitis quilicii (De Meyer, Mwatawala and Virgilio, 2016) | Illumina | [43] |
| 14CR | Ceratitis rosa (Karsch, 1887) | Illumina | [43] |
| 14CCO | Ceratitis cosyra (Walker, 1849) | Illumina | [43] |
| 14CF | Ceratitis flexuosa (Walker, 1853) | Illumina | [43] |
| 14CP | Ceratitis podocarpi (Bezzi, 1924) | Illumina | [43] |
| 15BT | Bactrocera tryoni (Saunders, 1842) | Illumina | [56] |
| 16BT1 | Bactrocera tryoni (Saunders, 1842) | Illumina | [56] |
| 16BT2 | Bactrocera tryoni (Saunders, 1842) | Illumina | [56] |
| 16BT3 | Bactrocera tryoni (Saunders, 1842) | Illumina | [56] |
| 16BT4 | Bactrocera tryoni (Saunders, 1842) | Illumina | [56] |
| 16BT5 | Bactrocera tryoni (Saunders, 1842) | Illumina | [56] |
| 17ZT | Bactrocera tau (Saunders, 1842) | Illumina | [57] |
| 18BZ | Bactrocera zonata (Saunders, 1842) | Illumina | [58] |
| 19BC | Bactrocera carambolae (Drew and Hancock, 1994) | Illumina | [59] |
2.2. Factors Contributing to Differences in Intestinal Commensal Bacteria in the Same Species of Tephritidae
2.3. Core Flora Was Present in Monophagous and Oligophagous Insects
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Friend, W.G. Nutritional requirements of phytophagous insects. Annu. Rev. Entomol. 1958, 3, 57–74. [Google Scholar] [CrossRef]
- Mitter, C.; Farrell, B.; Wiegmann, B. The Phylogenetic Study of Adaptive Zones: Has Phytophagy Promoted Insect Diversification? Am. Nat. 1988, 132, 107–128. [Google Scholar] [CrossRef]
- Cates, R.G. Host plant predictability and the feeding patterns of monophagous, oligophagous, and polyphagous insect herbivores. Oecologia 1981, 48, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Bjorndal, K.A. Flexibility of digestive responses in two generalist herbivores, the tortoises Geochelone carbonaria and Geochelone denticulata. Oecologia 1989, 78, 317–321. [Google Scholar] [CrossRef]
- Reymond, P.; Bodenhausen, N.; Van Poecke, R.M.; Krishnamurthy, V.; Dicke, M.; Farmer, E.E. A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant. Cell 2004, 16, 3132–3147. [Google Scholar] [CrossRef]
- Jurenka, R.; Russell, K.; O’Neal, M. Phytoecdysteroids as antifeedants towards several beetles that include polyphagous and monophagous feeding guilds. Pest Manag. Sci. 2017, 73, 1633–1637. [Google Scholar] [CrossRef]
- White, I.M.; Elson-Harris, M.M. Fruit Flies of Economic Significance: Their Identification and Bionomics; CAB International: Wallingford, UK, 1992. [Google Scholar]
- Qin, Y.; Paini, D.R.; Wang, C.; Fang, Y.; Li, Z. Global establishment risk of economically important fruit fly species (Tephritidae). PLoS ONE 2015, 10, e0116424. [Google Scholar] [CrossRef]
- Brues, C.T. Choice of Food and Numerical Abundance Among Insects. J. Econ. Entomol. 1923, 1, 46–51. [Google Scholar] [CrossRef]
- Cates, R.G. Feeding patterns of monophagous, oligophagous, and polyphagous insect herbivores: The effect of resource abundance and plant chemistry. Oecologia 1980, 46, 22–31. [Google Scholar] [CrossRef]
- Hafsi, A.; Facon, B.; Ravigne, V.; Chiroleu, F.; Quilici, S.; Chermiti, B.; Duyck, P.F. Host plant range of a fruit fly community (Diptera: Tephritidae): Does fruit composition influence larval performance? BMC Ecol. 2016, 16, 40. [Google Scholar] [CrossRef]
- Prokopy, R.J.; Owens, E.D. Visual Generalist with Visual Specialist Phytophagous Insects: Host Selection Behaviour and Application to Management. Entomol. Exp. Appl. 1978, 3, 609–620. [Google Scholar] [CrossRef]
- Steffan-Dewenter, I.; Tscharntke, T. Butterfly community structure in fragmented habitats. Ecol. Lett. 2004, 5, 449–456. [Google Scholar]
- Clarke, A.R.; Armstrong, K.F.; Carmichael, A.E.; Milne, J.R.; Raghu, S.; Roderick, G.K.; Yeates, D.K. Invasive phytophagous pests arising through a recent tropical evolutionary radiation: The Bactrocera dorsalis complex of fruit flies. Annu. Rev. Entomol. 2005, 50, 293–319. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.F.; Yao, Z.; Bai, S.; Cai, Z.; Zhang, H. Tephritidae fruit fly gut microbiome diversity, function and potential for applications. Bull. Entomol. Res. 2020, 110, 423–437. [Google Scholar] [CrossRef]
- Werren, J.H.; O’Neill, S.L. The evolution of heritable symbionts. In Influential Passengers: Inherited Microorganisms and Arthropod Reproduction; Oxford University Press: Oxford, UK, 1997. [Google Scholar]
- Margulis, L.; Fester, R. Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis; Biology, Environmental Science; MIT Press: Cambridge, MA, USA, 1991. [Google Scholar]
- Thomas, S.; Izard, J.; Walsh, E.; Batich, K.; Chongsathidkiet, P.; Clarke, G.; Sela, D.A.; Muller, A.J.; Mullin, J.M.; Albert, K.; et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res. 2017, 77, 1783–1812. [Google Scholar] [CrossRef]
- Ofek-Lalzar, M.; Sela, N.; Goldman-Voronov, M.; Green, S.J.; Hadar, Y.; Minz, D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014, 5, 4950. [Google Scholar] [CrossRef]
- Harman, G.E.; Uphoff, N. Symbiotic Root-Endophytic Soil Microbes Improve Crop Productivity and Provide Environmental Benefits. Scientifica 2019, 2019, 9106395. [Google Scholar] [CrossRef]
- Glick, B.R.; Gamalero, E. Recent Developments in the Study of Plant Microbiomes. Microorganisms 2021, 9, 1533. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Majumder, R.; Sutcliffe, B.; Taylor, P.W.; Chapman, T.A. Next-Generation Sequencing reveals relationship between the larval microbiome and food substrate in the polyphagous Queensland fruit fly. Sci. Rep. 2019, 9, 14292. [Google Scholar] [CrossRef]
- Singh, S.; Singh, A.; Baweja, V.; Roy, A.; Chakraborty, A.; Singh, I.K. Molecular Rationale of Insect-Microbes Symbiosis—From Insect Behaviour to Mechanism. Microorganisms 2021, 9, 2422. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. The microbial dimension in insect nutritional ecology. Funct. Ecol. 2009, 1, 38–47. [Google Scholar] [CrossRef]
- Baumann, P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Degnan, P.H.; Santos, S.R.; Ochman, H. The players in a mutualistic symbiosis: Insects, bacteria, viruses, and virulence genes. Proc. Natl. Acad. Sci. USA 2005, 47, 16919–16926. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Russell, J.A.; Moran, N.A.; Hunter, M.S. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 2003, 100, 1803–1807. [Google Scholar] [CrossRef]
- Oliver, K.M.; Moran, N.A.; Hunter, M.S. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. Biol. Sci. 2006, 273, 1273–1280. [Google Scholar] [CrossRef]
- Montllor, C.B.; Maxmen, A.; Purcell, A.H. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 2002, 2, 189–195. [Google Scholar] [CrossRef]
- Asplen, M.K.; Bano, N.; Brady, C.M.; Desneux, N.; Hopper, K.R.; Malouines, C.; Oliver, K.M.; White, J.A.; Heimpel, G.E. Specialisation of bacterial endosymbionts that protect aphids from parasitoids. Ecol. Entomol. 2014, 39, 736–739. [Google Scholar] [CrossRef]
- Oliver, K.M.; Degnan, P.H.; Burke, G.R.; Moran, N.A. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 2010, 55, 247–266. [Google Scholar] [CrossRef]
- Baumann, P.; Moran, N.A.; Baumann, L. The evolution and genetics of aphid endosymbionts. Bioscience 1997, 1, 12–20. [Google Scholar] [CrossRef]
- Hansen, A.K.; Moran, N.A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2014, 23, 1473–1496. [Google Scholar] [CrossRef] [PubMed]
- Bigiotti, G.; Sacchetti, P.; Pastorelli, R.; Lauzon, C.R.; Belcari, A. Bacterial symbiosis in Bactrocera oleae, an Achilles’ heel for its pest control. Insect Sci. 2021, 28, 874–884. [Google Scholar] [CrossRef]
- Estes, A.M.; Hearn, D.J.; Bronstein, J.L.; Pierson, E.A. The olive fly endosymbiont, “Candidatus Erwinia dacicola,” switches from an intracellular existence to an extracellular existence during host insect development. Appl. Environ. Microbiol. 2009, 75, 7097–7106. [Google Scholar] [CrossRef]
- Cardenas-Hernandez, V.; Lemos-Lucumi, C.A.; Toro-Perea, N. Comparative metatranscriptomics reveals effect of host plant on microbiota gene expression of Anastrepha obliqua (Diptera: Tephritidae) larvae. Environ. Entomol. 2024, 53, 157–167. [Google Scholar] [CrossRef]
- Su, Q.; Zhou, X.; Zhang, Y. Symbiont-mediated functions in insect hosts. Commun. Integr. Biol. 2013, 6, e23804. [Google Scholar] [CrossRef]
- Douglas, A.E. Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 1998, 43, 17–37. [Google Scholar] [CrossRef]
- Morrow, J.L.; Frommer, M.; Shearman, D.C.; Riegler, M. The Microbiome of Field-Caught and Laboratory-Adapted Australian Tephritid Fruit Fly Species with Different Host Plant Use and Specialisation. Microb. Ecol. 2015, 70, 498–508. [Google Scholar] [CrossRef]
- Guidolin, A.S.; Cônsoli, F.L. Symbiont Diversity of Aphis (Toxoptera) citricidus (Hemiptera: Aphididae) as Influenced by Host Plants. Invertebr. Microbiol. 2017, 73, 201–210. [Google Scholar] [CrossRef]
- Russell, J.A.; Moran, N.A. Costs and benefits of symbiont infection in aphids: Variation among symbionts and across temperatures. Proc. Biol. Sci. 2006, 273, 603–610. [Google Scholar] [CrossRef]
- De Cock, M.; Virgilio, M.; Vandamme, P.; Bourtzis, K.; De Meyer, M.; Willems, A. Comparative Microbiomics of Tephritid Frugivorous Pests (Diptera: Tephritidae) From the Field: A Tale of High Variability Across and Within Species. Front. Microbiol. 2020, 11, 1890. [Google Scholar] [CrossRef]
- Ventura, C.; Briones-Roblero, C.I.; Hernandez, E.; Rivera-Orduna, F.N.; Zuniga, G. Comparative Analysis of the Gut Bacterial Community of Four Anastrepha Fruit Flies (Diptera: Tephritidae) Based on Pyrosequencing. Curr. Microbiol. 2018, 75, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Salgueiro, J.; Nussenbaum, A.L.; Milla, F.H.; Asimakis, E.; Goane, L.; Ruiz, M.J.; Bachmann, G.E.; Vera, M.T.; Stathopoulou, P.; Bourtzis, K.; et al. Analysis of the Gut Bacterial Community of Wild Larvae of Anastrepha fraterculus sp. 1: Effect of Host Fruit, Environment, and Prominent Stable Associations of the Genera Wolbachia, Tatumella, and Enterobacter. Front. Microbiol. 2022, 13, 822990. [Google Scholar] [CrossRef] [PubMed]
- Augustinos, A.A.; Tsiamis, G.; Cáceres, C.; Abd-Alla, A.M.M.; Bourtzis, K. Taxonomy, Diet, and Developmental Stage Contribute to the Structuring of Gut-Associated Bacterial Communities in Tephritid Pest Species. Front. Microbiol. 2019, 10, 2004. [Google Scholar] [CrossRef] [PubMed]
- Malacrinò, A.; Campolo, O.; Medina, R.F.; Palmeri, V. Instar- and host-associated differentiation of bacterial communities in the Mediterranean fruit fly Ceratitis capitata. PLoS ONE 2018, 13, e0194131. [Google Scholar] [CrossRef]
- Bel Mokhtar, N.; Catalá-Oltra, M.; Stathopoulou, P.; Asimakis, E.; Remmal, I.; Remmas, N.; Maurady, A.; Britel, M.R.; García De Oteyza, J.; Tsiamis, G.; et al. Dynamics of the Gut Bacteriome During a Laboratory Adaptation Process of the Mediterranean Fruit Fly, Ceratitis capitata. Front. Microbiol. 2022, 13, 919760. [Google Scholar] [CrossRef]
- Andongma, A.A.; Dong, L.W.Y. Assessment of the Bacteria community structure across life stages of the Chinese Citrus Fly, Bactrocera minax (Diptera: Tephritidae). BMC Microbiol. 2019, 19, 285. [Google Scholar] [CrossRef]
- Cao, S.; Ren, X.; Zhang, G.; Wang, H.; Wei, B.; Niu, C. Gut microbiota metagenomics and mediation of phenol degradation in Bactrocera minax (Diptera, Tephritidae). Pest Manag. Sci. 2024, 80, 3935–3944. [Google Scholar] [CrossRef]
- Koskinioti, P.; Ras, E.; Augustinos, A.A.; Tsiamis, G.; Beukeboom, L.W.; Caceres, C.; Bourtzis, K. The effects of geographic origin and antibiotic treatment on the gut symbiotic communities of Bactrocera oleae populations. Entomol. Exp. Appl. 2019, 3, 197–208. [Google Scholar] [CrossRef]
- Choudhary, J.S.; Naaz, N.; Prabhakar, C.S.; Das, B.; Singh, A.K.; Bhatt, B.P. High Taxonomic and Functional Diversity of Bacterial Communities Associated with Melon Fly, Zeugodacus cucurbitae (Diptera: Tephritidae). Curr. Microbiol. 2021, 78, 611–623. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, X.; Chen, Z.; Wang, Z.; Lu, Y.; Cheng, D. The Divergence in Bacterial Components Associated with Bactrocera dorsalis across Developmental Stages. Front. Microbiol. 2018, 9, 114. [Google Scholar] [CrossRef]
- Andongma, A.A.; Wan, L.; Dong, Y.; Li, P.; Desneux, N.; White, J.A.; Niu, C. Pyrosequencing reveals a shift in symbiotic bacteria populations across life stages of Bactrocera dorsalis. Sci. Rep. 2015, 5, 9470. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, H.; Ren, L.; Cheng, D. Microbial Communities in Different Developmental Stages of the Oriental Fruit Fly, Bactrocera dorsalis, Are Associated with Differentially Expressed Peptidoglycan Recognition Protein-Encoding Genes. Appl. Environ. Microbiol. 2019, 85, e00803-19. [Google Scholar] [CrossRef] [PubMed]
- Majumder, R.; Sutcliffe, B.; Taylor, P.W.; Chapman, T.A. Microbiome of the Queensland Fruit Fly through Metamorphosis. Microorganisms 2020, 8, 795. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.S.; Shi, G.; Liu, L.J.; Li, Z.H. Diversity of bacteria in different life stages and their impact on the development and reproduction of Zeugodacus tau (Diptera: Tephritidae). Insect Sci. 2021, 28, 363–376. [Google Scholar] [CrossRef]
- Naaz, N.; Choudhary, J.S.; Choudhary, A.; Dutta, A.; Das, B. Developmental stage-associated microbiota profile of the peach fruit fly, Bactrocera zonata (Diptera: Tephritidae) and their functional prediction using 16S rRNA gene metabarcoding sequencing. 3 Biotech 2020, 10, 390. [Google Scholar] [CrossRef]
- Yong, H.S.; Song, S.L.; Chua, K.O.; Lim, P.E. High Diversity of Bacterial Communities in Developmental Stages of Bactrocera carambolae (Insecta: Tephritidae) Revealed by Illumina MiSeq Sequencing of 16S rRNA Gene. Curr. Microbiol. 2017, 74, 1076–1082. [Google Scholar] [CrossRef]
- Girard, M.; Luis, P.; Valiente, M.C.; Minard, G. Crosstalk between the microbiota and insect postembryonic development. Trends Microbiol. 2023, 31, 181–196. [Google Scholar] [CrossRef]
- Feldhaar, H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 2011, 5, 533–543. [Google Scholar] [CrossRef]
- Yang, Z.W.; Luo, J.Y.; Men, Y.; Liu, Z.H.; Zheng, Z.K.; Wang, Y.H.; Xie, Q. Different roles of host and habitat in determining the microbial communities of plant-feeding true bugs. Microbiome 2023, 11, 244. [Google Scholar] [CrossRef]
- Liu, S.H.; Chen, Y.; Li, W.; Tang, G.H.; Yang, Y.; Jiang, H.B.; Dou, W.; Wang, J.J. Diversity of Bacterial Communities in the Intestinal Tracts of Two Geographically Distant Populations of Bactrocera dorsalis (Diptera: Tephritidae). J. Econ. Entomol. 2018, 111, 2861–2868. [Google Scholar] [CrossRef]
- Yong, H.S.; Song, S.L.; Eamsobhana, P.; Pasartvit, A.; Lim, P.E. Differential abundance and core members of the bacterial community associated with wild male Zeugodacus cucurbitae fruit flies (Insecta: Tephritidae) from three geographical regions of Southeast Asia. Mol. Biol. Rep. 2019, 46, 3765–3776. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chen, J.; Qin, M.; Jiang, L.; Qiao, G. Geography-dependent symbiont communities in two oligophagous aphid species. FEMS Microbiol. Ecol. 2021, 97, fiab132. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Koga, R.; Shibao, H.; Matsumoto, T.; Fukatsu, T. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 2002, 11, 2123–2135. [Google Scholar] [CrossRef]
- Najar-Rodriguez, A.J.; Mcgraw, E.A.; Mensah, R.K.; Pittman, G.W.; Walter, G.H. The microbial flora of Aphis gossypii: Patterns across host plants and geographical space. J. Invertebr. Pathol. 2009, 100, 123–126. [Google Scholar] [CrossRef]
- Zytynska, S.E.; Weisser, W.W. The natural occurrence of secondary bacterial symbionts in aphids. Ecol. Entomol. 2015, 1, 13–26. [Google Scholar] [CrossRef]
- Jones, R.T.; Bressan, A.; Greenwell, A.M.; Fierer, N. Bacterial communities of two parthenogenetic aphid species cocolonizing two host plants across the Hawaiian Islands. Appl. Environ. Microbiol. 2011, 77, 8345–8349. [Google Scholar] [CrossRef]
- Gomes, S.I.F.; Kielak, A.M.; Hannula, S.E.; Heinen, R.; Jongen, R.; Keesmaat, I.; De Long, J.R.; Bezemer, T.M. Microbiomes of a specialist caterpillar are consistent across different habitats but also resemble the local soil microbial communities. Anim. Microbiome 2020, 2, 37. [Google Scholar] [CrossRef]
- Gallo-Franco, J.J.; Toro-Perea, N. Variations in the Bacterial Communities in Anastrepha obliqua (Diptera: Tephritidae) According to the Insect Life Stage and Host Plant. Curr. Microbiol. 2020, 77, 1283–1291. [Google Scholar] [CrossRef]
- Akami, M.; Ren, X.; Wang, Y.; Mansour, A.; Cao, S.; Qi, X.; Ngakou, A.; Ngane, R.A.N.; Niu, C. Host fruits shape the changes in the gut microbiota and development of Bactrocera dorsalis (Diptera: Tephritidae) larvae. Int. J. Trop. Insect Sci. 2022, 42, 2127–2141. [Google Scholar] [CrossRef]
- Ferrari, J.; West, J.A.; Via, S.; Godfray, H.C. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 2012, 66, 375–390. [Google Scholar] [CrossRef]
- Russell, J.A.; Weldon, S.; Smith, A.H.; Kim, K.L.; Hu, Y.; Lukasik, P.; Doll, S.; Anastopoulos, I.; Novin, M.; Oliver, K.M. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol. Ecol. 2013, 22, 2045–2059. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, J.P.; Outreman, Y.; Mieuzet, L.; Simon, J.C. Bacterial communities associated with host-adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS ONE 2015, 10, e0120664. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.M.; Asplen, M.K.; Desneux, N.; Heimpel, G.E.; Hopper, K.R.; Linnen, C.R.; Oliver, K.M.; Wulff, J.A.; White, J.A. Worldwide populations of the aphid Aphis craccivora are infected with diverse facultative bacterial symbionts. Microb. Ecol. 2014, 67, 195–204. [Google Scholar] [CrossRef]
- Guidolin, A.S.; Consoli, F.L. Diversity of the Most Commonly Reported Facultative Symbionts in Two Closely-Related Aphids with Different Host Ranges. Neotrop. Entomol. 2018, 47, 440–446. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Behar, A.; Yuval, B.; Jurkevitch, E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol. Ecol. 2005, 14, 2637–2643. [Google Scholar] [CrossRef]
- Ben-Yosef, M.; Aharon, Y.; Jurkevitch, E.; Yuval, B. Give us the tools and we will do the job: Symbiotic bacteria affect olive fly fitness in a diet-dependent fashion. Proc. R. Soc. B Biol. Sci. 2010, 277, 1545–1552. [Google Scholar] [CrossRef]
- Ben-Yosef, M.; Pasternak, Z.; Jurkevitch, E.; Yuval, B. Symbiotic bacteria enable olive flies (Bactrocera oleae) to exploit intractable sources of nitrogen. J. Evol. Biol. 2014, 27, 2695–2705. [Google Scholar] [CrossRef]
- Ben-Yosef, M.; Pasternak, Z.; Jurkevitch, E.; Yuval, B. Symbiotic bacteria enable olive fly larvae to overcome host defences. R. Soc. Open Sci. 2015, 2, 150170. [Google Scholar] [CrossRef]
- Szklarzewicz, T.; Michalik, A. Transovarial Transmission of Symbionts in Insects. Results Probl. Cell Differ. 2017, 63, 43–67. [Google Scholar] [CrossRef]
- Rashid, M.A.; Andongma, A.A.; Dong, Y.C.; Ren, X.M.; Niu, C.Y. Effect of gut bacteria on fitness of the Chinese citrus fly, Bactrocera minax (Diptera: Tephritidae). Symbiosis 2018, 76, 63–69. [Google Scholar] [CrossRef]
- Wang, A.; Yao, Z.; Zheng, W.; Zhang, H. Bacterial communities in the gut and reproductive organs of Bactrocera minax (Diptera: Tephritidae) based on 454 pyrosequencing. PLoS ONE 2014, 9, e106988. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Phloem-sap feeding by animals: Problems and solutions. J. Exp. Bot. 2006, 57, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.K.; Moran, N.A. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. USA 2011, 108, 2849–2854. [Google Scholar] [CrossRef] [PubMed]
- Perez-Brocal, V.; Gil, R.; Ramos, S.; Lamelas, A.; Postigo, M.; Michelena, J.M.; Silva, F.J.; Moya, A.; Latorre, A. A small microbial genome: The end of a long symbiotic relationship? Science 2006, 314, 312–313. [Google Scholar] [CrossRef]
- Mccutcheon, J.P.; Moran, N.A. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2011, 10, 13–26. [Google Scholar] [CrossRef]
- Shigenobu, S.; Watanabe, H.; Hattori, M.; Sakaki, Y.; Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 2000, 407, 81–86. [Google Scholar] [CrossRef]
- van Ham, R.C.; Kamerbeek, J.; Palacios, C.; Rausell, C.; Abascal, F.; Bastolla, U.; Fernandez, J.M.; Jimenez, L.; Postigo, M.; Silva, F.J.; et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 2003, 100, 581–586. [Google Scholar] [CrossRef]
- Michalik, A.; Szwedo, J.; Stroinski, A.; Swierczewski, D.; Szklarzewicz, T. Symbiotic cornucopia of the monophagous planthopper Ommatidiotus dissimilis (Fallen, 1806) (Hemiptera: Fulgoromorpha: Caliscelidae). Protoplasma 2018, 255, 1317–1329. [Google Scholar] [CrossRef]
- Xue, J.; Zhou, X.; Zhang, C.X.; Yu, L.L.; Fan, H.W.; Wang, Z.; Xu, H.J.; Xi, Y.; Zhu, Z.R.; Zhou, W.W.; et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014, 15, 521. [Google Scholar] [CrossRef]
- Sinno, M.; Bezier, A.; Vinale, F.; Giron, D.; Laudonia, S.; Garonna, A.P.; Pennacchio, F. Symbiosis disruption in the olive fruit fly, Bactrocera oleae (Rossi), as a potential tool for sustainable control. Pest Manag. Sci. 2020, 76, 3199–3207. [Google Scholar] [CrossRef] [PubMed]
- Nobre, T. Monophagous olive moth and its bacterial microbiota: Unveiling the holobiont towards pest management. Biocontrol Sci. Technol. 2021, 31, 107–111. [Google Scholar] [CrossRef]
- Estes, A.M.; Nestel, D.; Belcari, A.; Jessup, A.; Rempoulakis, P.; Economopoulos, A.P. A basis for the renewal of sterile insect technique for the olive fly, Bactrocera oleae (Rossi). J. Appl. Entomol. 2011, 1–2, 1–16. [Google Scholar] [CrossRef]
- Sacchetti, P.; Pastorelli, R.; Bigiotti, G.; Guidi, R.; Ruschioni, S.; Viti, C.; Belcari, A. Olive fruit fly rearing procedures affect the vertical transmission of the bacterial symbiont Candidatus Erwinia dacicola. BMC Biotechnol. 2019, 19, 91. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, M.M.; More, R.P.; Rangasamy, A.; Gandhi, G.R.; Muthugounder, M.; Thiruvengadam, V.; Samaddar, S.; Jalali, S.K.; Sa, T. Gut Bacterial Diversity of Insecticide-Susceptible and -Resistant Nymphs of the Brown Planthopper Nilaparvata lugens Stal (Hemiptera: Delphacidae) and Elucidation of Their Putative Functional Roles. J. Microbiol. Biotechnol. 2018, 28, 976–986. [Google Scholar] [CrossRef]

| Name | Wild/Laboratory | Host Plant | Geography |
|---|---|---|---|
| 1AL | Wild | Bitter orange (Citrus aurantium) | Soconusco Region, Chiapas State, Mexico |
| 1AO | Wild | Mango (Mangifera indica), | Soconusco Region, Chiapas State, Mexico |
| 1ASE | Wild | Mamey sapote (Pouteria sapota) | Soconusco Region, Chiapas State, Mexico |
| 1AST | Wild | Guava (Psidium guajava) | Soconusco Region, Chiapas State, Mexico |
| 2AF | Wild | Peaches and guavas | Horco Molle, Tucuman Province, Argentina; Concordia, Entre Rios Province, Argentina |
| 3AO | Wild | Spondias purpurea, Mangifera indica, and Averrhoa carambola | Valle del Cauca, southwestern Colombia |
| 4AG | Laboratory | / | / |
| 4AL | Laboratory | / | / |
| 4BO | Laboratory | / | / |
| 5CC | Wild | Orange fruits (Citrus sinensis) | Reggio Calabria, Italy |
| 6CCW | Wild | Mandarin orange (Citrus reticulada and Citrus unshiu) | Valencia, Spain |
| 6CCL | Laboratory | / | / |
| 7BM | Wild | Citrus | Yichang, Hubei, China |
| 8BM | Wild | Citrus | Yichang, Hubei, China |
| 9BO | Wild | Olives | Greece |
| 10ZC | Wild | Cucumber (Cucumis sativus) | Farming Systems Research Center for Hill and Plateau Region, Ranchi, India |
| 11BD1 | Laboratory | / | / |
| 11BD2 | Wild | Carambola (Averrhoa carambola) | Huizhou, Guangdong, China |
| 11BD3 | Wild | Carambola (Averrhoa carambola) | Nansha, Guangdong, China |
| 12BD | Wild | Unknown | Wuhan, Hubei, China |
| 13BD | Wild | Carambola (Averrhoa carambola) | Guangzhou, Guangdong, China |
| 14BD | Wild | Eriobotrya japonica, Mangifera indica, Annona muricata, and Psidium guajava | South Africa; Tanzania |
| 14ZC | Wild | Coccinia grandis, Momordica charantia, Citrullus lanatus, and Cucumis sativus | Reunion; Tanzania |
| 14BO | Wild | Olea europea | Greece; Italy |
| 14BZ | Wild | Terminalia catappa | Reunion |
| 14CC | Wild | Citrus reticulata, Ficus carica; Malus pumila, Ficus carica; Pyrus communis | Greece; Italy; South Africa |
| 14CQ | Wild | Eriobotrya japonica, Psidium catlleyanum, Psidium guajava, Eriobotrya japonica, and Harpephyllum caffrum | Reunion; South Africa |
| 14CR | Wild | Citrus sinensis | Mozambique |
| 14CCO | Wild | Sclerocarya birrea and Annona muricata | Tanzania; South Africa |
| 14CF | Wild | Antiaris toxicaria | Kenya |
| 14CP | Wild | Afrocarpus falcatus | South Africa |
| 15BT | Wild | Pomegranates (Punica granatum), green apples (Malus pumila), and quinces (Cydonia oblonga) | New South Wales and Victoria, Australia |
| 16BT1 | Wild | Hog plum | Nambour |
| 16BT2 | Wild | Sapodilla | Whiteside |
| 16BT3 | Wild | Sapodilla | Nambour |
| 16BT4 | Wild | Pomegranate | Commealla |
| 16BT5 | Wild | Green apple | Echuca |
| 17ZT | Laboratory | / | / |
| 18BZ | Wild | Wood apple (Aegle marmelos) | Research farm of ICAR Research Complexfor Eastern Region, Ranchi, India |
| 19BC | Wild | Carambola (Averrhoa carambola) | Wilayah Persekutuan Kuala Lumpur, Malaysia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, D.; Yin, C.; Jing, J.; Li, Z.; Liu, L. Relationship Between the Host Plant Range of Insects and Symbiont Bacteria. Microorganisms 2025, 13, 189. https://doi.org/10.3390/microorganisms13010189
Ge D, Yin C, Jing J, Li Z, Liu L. Relationship Between the Host Plant Range of Insects and Symbiont Bacteria. Microorganisms. 2025; 13(1):189. https://doi.org/10.3390/microorganisms13010189
Chicago/Turabian StyleGe, Doudou, Chongwen Yin, Jiayu Jing, Zhihong Li, and Lijun Liu. 2025. "Relationship Between the Host Plant Range of Insects and Symbiont Bacteria" Microorganisms 13, no. 1: 189. https://doi.org/10.3390/microorganisms13010189
APA StyleGe, D., Yin, C., Jing, J., Li, Z., & Liu, L. (2025). Relationship Between the Host Plant Range of Insects and Symbiont Bacteria. Microorganisms, 13(1), 189. https://doi.org/10.3390/microorganisms13010189






