Improving Geldanamycin Production in Streptomyces geldanamycininus Through UV Mutagenesis of Protoplast
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Growth Conditions
2.2. Preparation and Optimization of Protoplasts
2.3. Determination of Protoplast Preparation Rate and Regeneration Rate
2.4. Mutagenesis of Protoplasts
2.5. Screening of High-Yielding Strains
2.6. Detection and Quantification of Geldanamycin Titer
2.7. DNA Extraction and Whole-Genome Sequencing
2.8. RNA Extraction and Transcriptome Sequencing
2.9. Transcriptome Data Analysis
2.10. Reverse Transcription and qRT-PCR Analysis
2.11. Statistical Analysis
3. Results
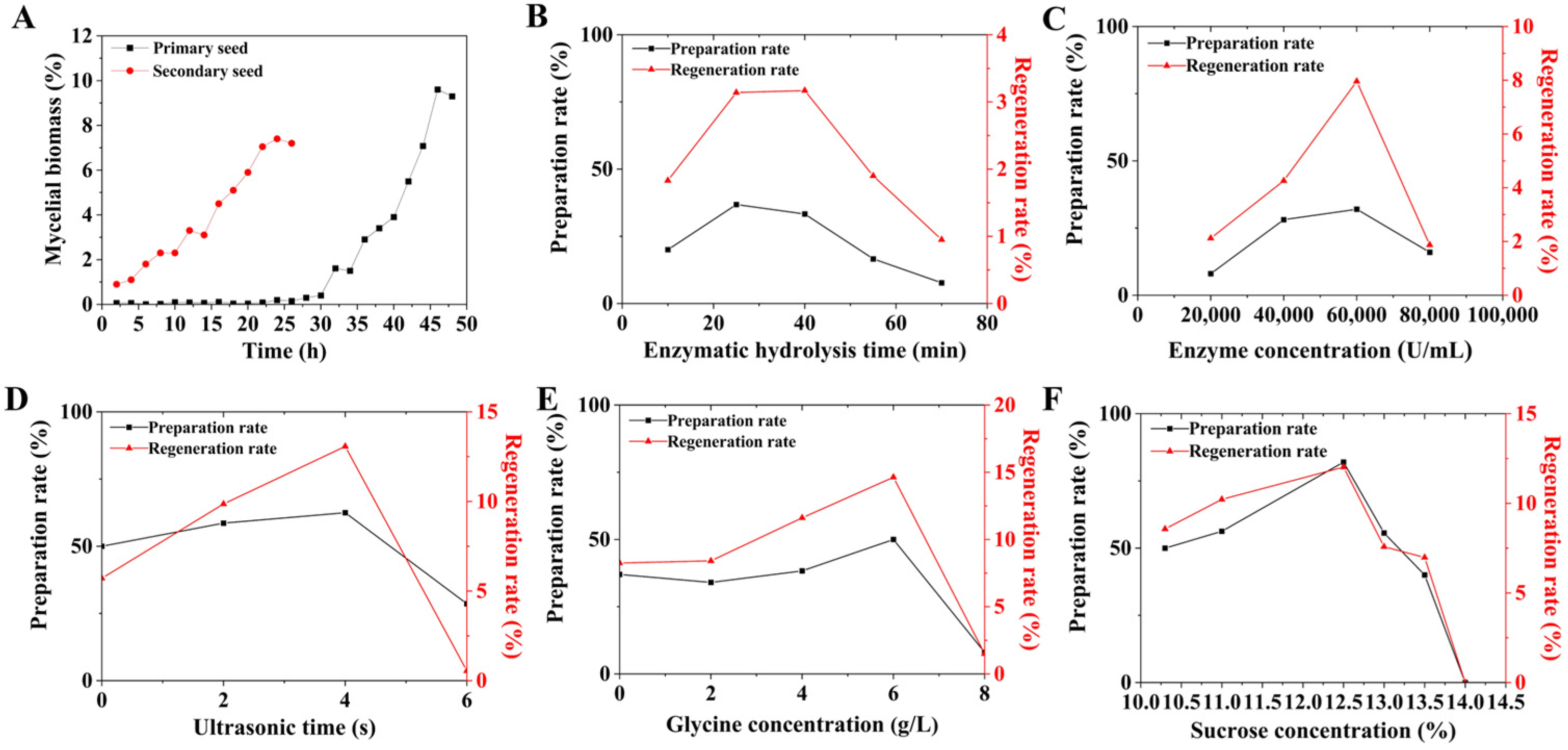
3.1. Effects of Various Factors on the Preparation Rate and Regeneration Rate of FIM18-0592 Protoplasts
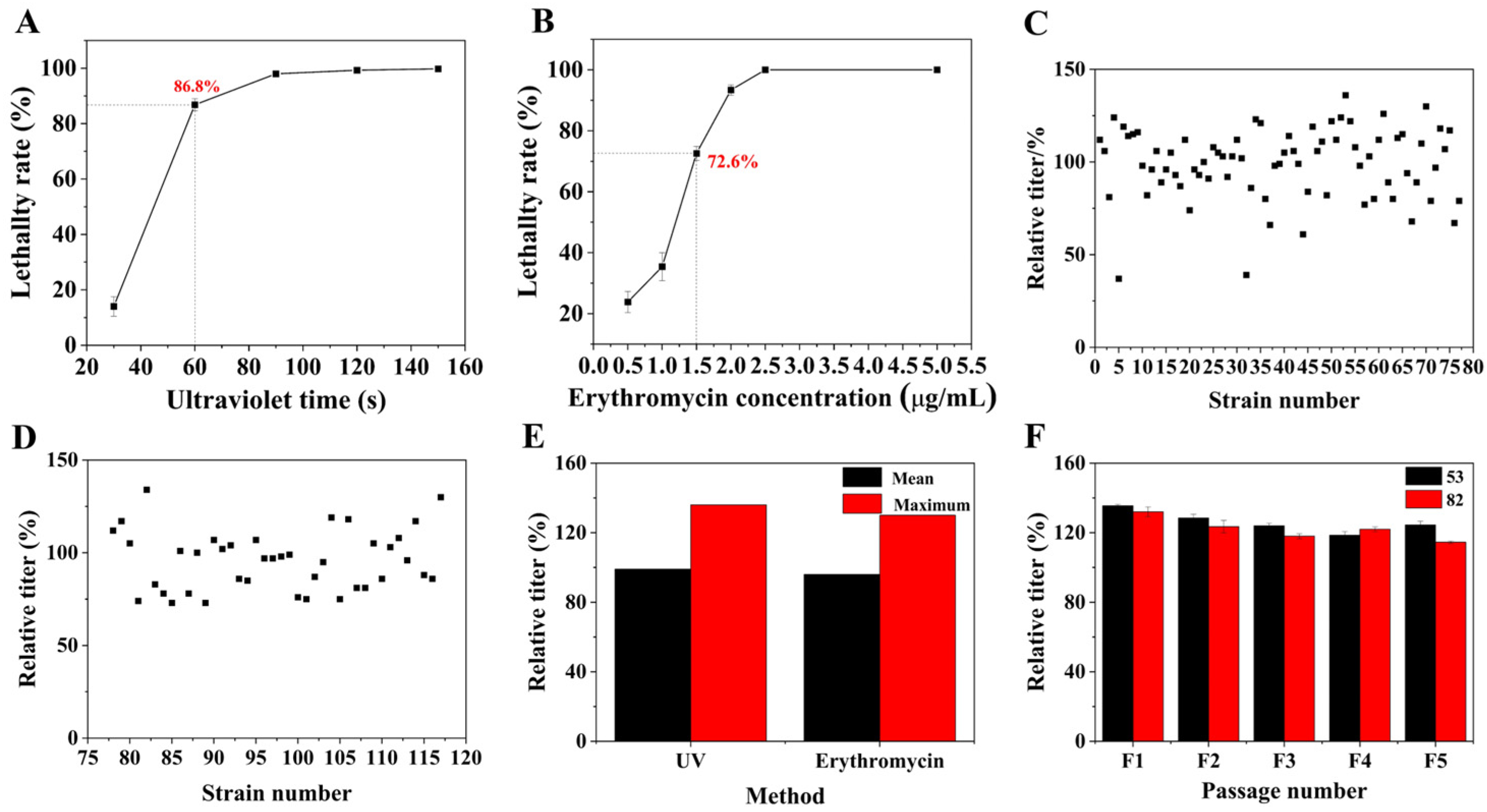
3.2. Analysis of the Mutagenesis Results for FIM 18-0592 Protoplasts
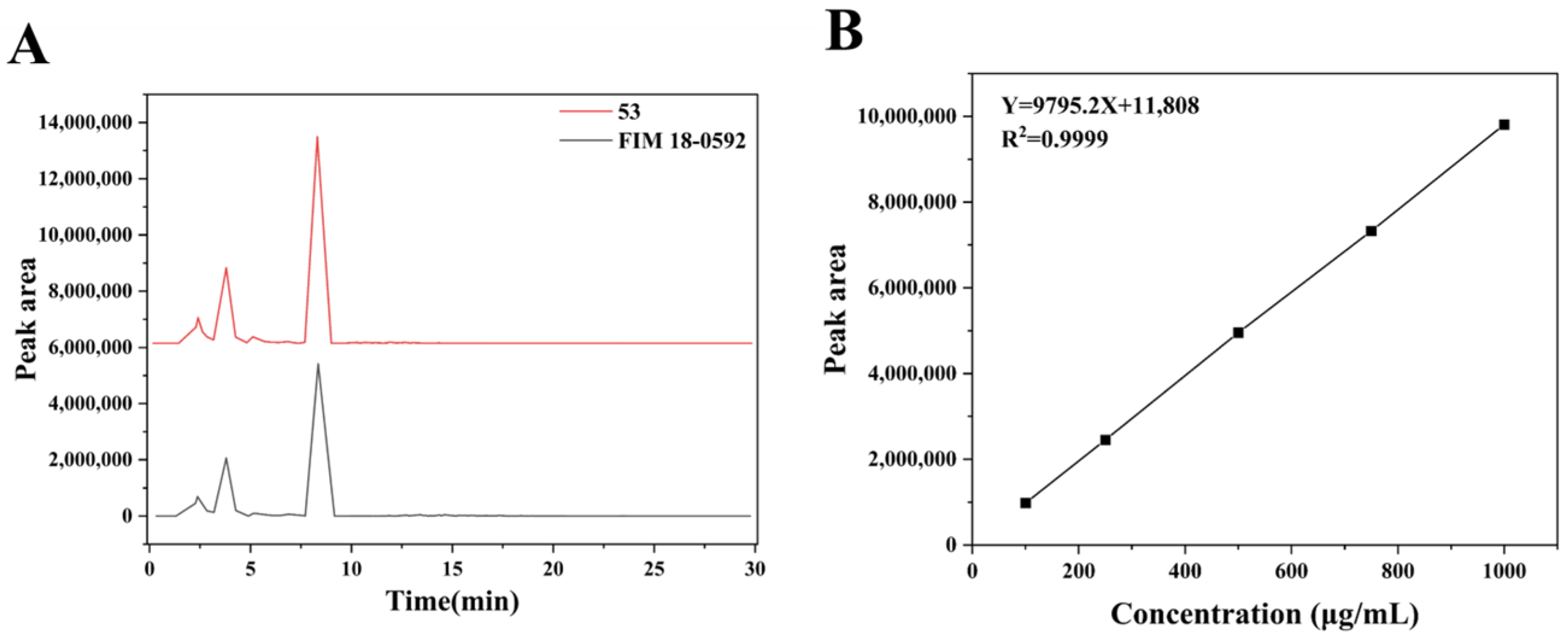
3.3. Detection and Quantification of Geldanamycin Between FIM 18-0592 and High-Yielding Strain 53
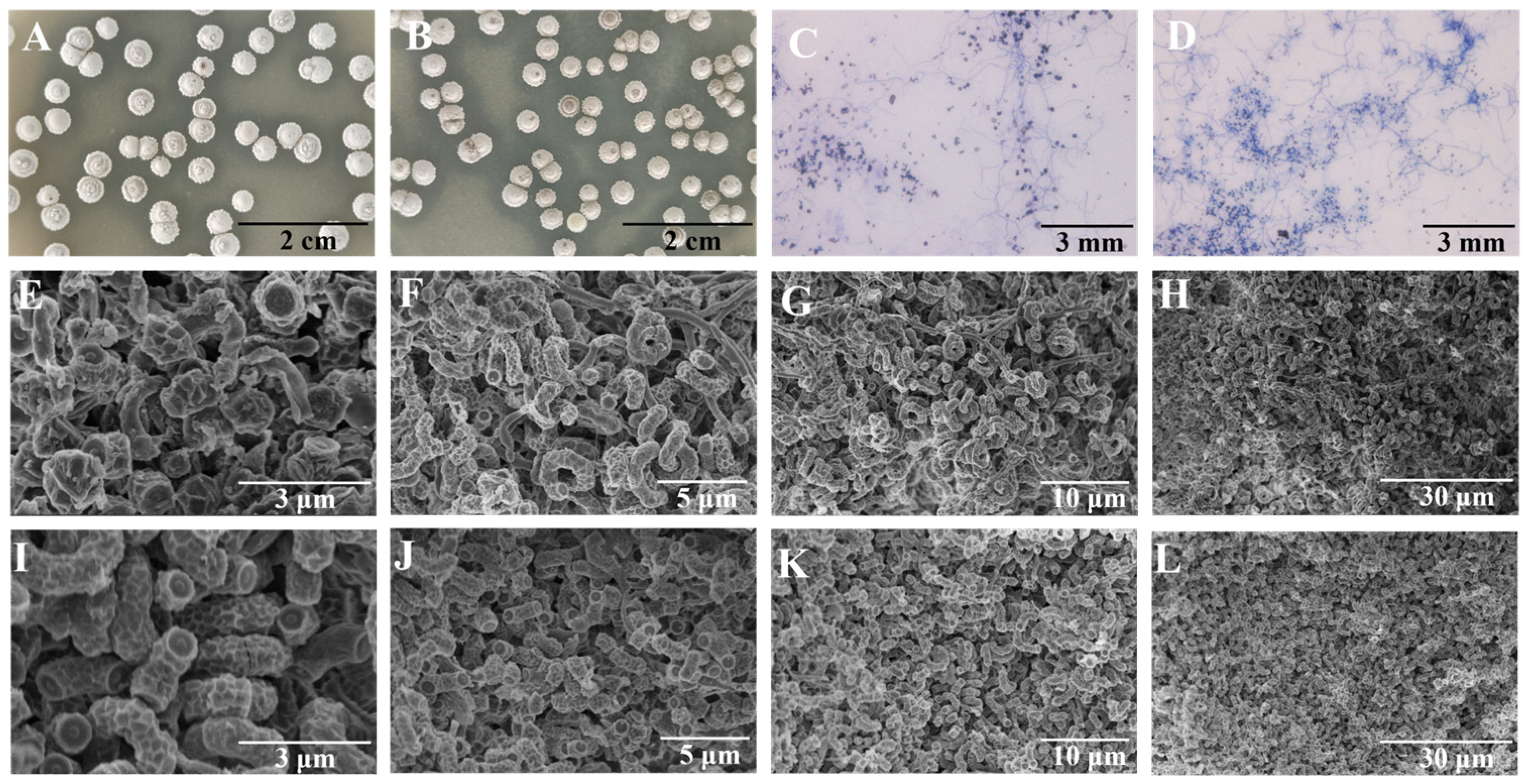
3.4. Comparative Analysis of Morphological Characteristics Between FIM 18-0592 and High-Yielding Strain 53
3.5. Whole-Genome Sequencing Analysis of FIM18-0592
3.6. Resequencing Analysis of High-Yielding Strain 53
3.7. Transcriptome Sequencing Analysis of FIM18-0592 and High-Yielding Strain 53
3.7.1. Analysis of DEGs Between High-Yielding and Original Strains
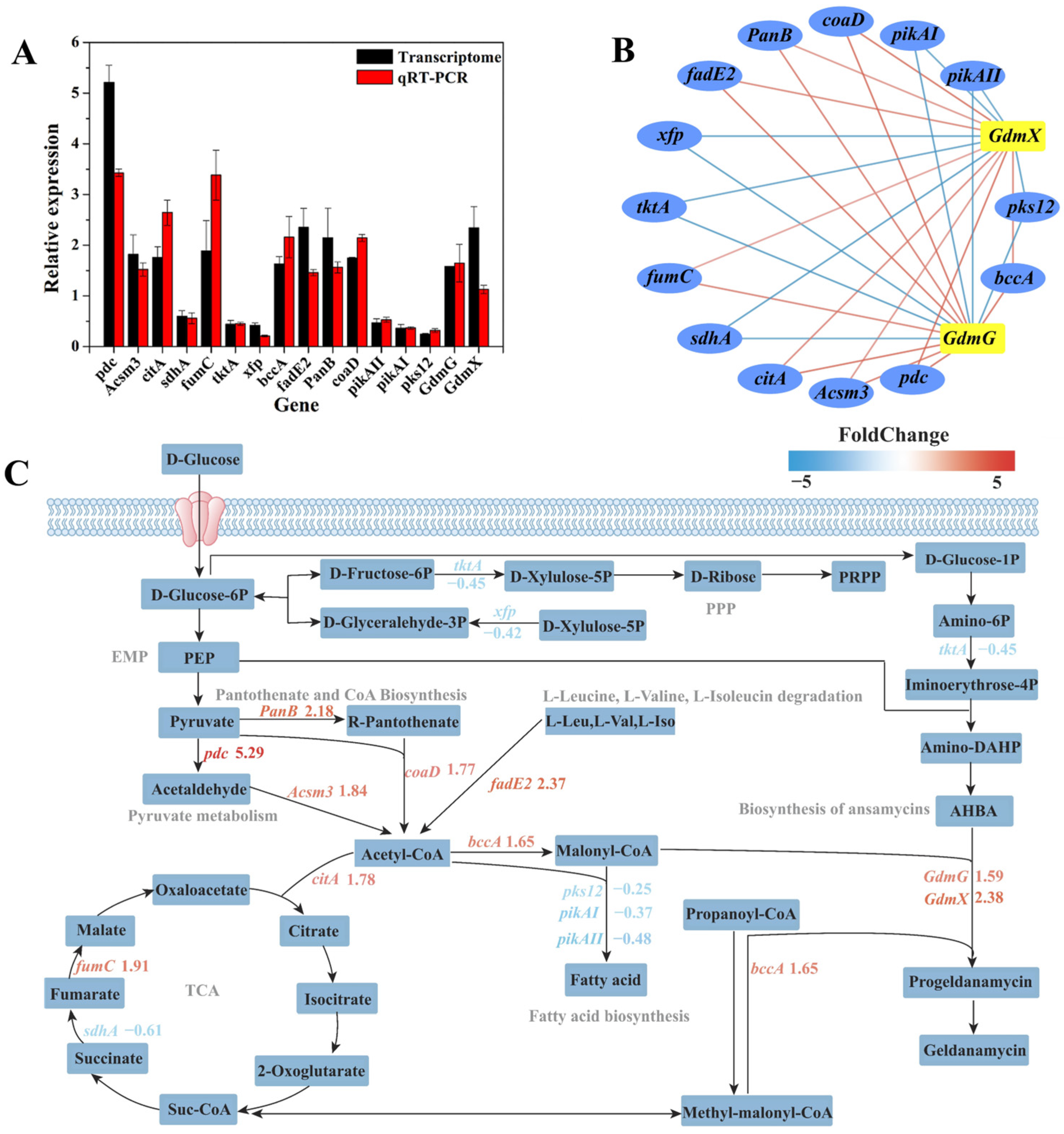
3.7.2. Analysis of Metabolic Pathway of Key Genes with Significant Differential Expression
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- DeBoer, C.; Meulman, P.A.; Wnuk, R.J.; Peterson, D.H. Geldanamycin, a new antibiotic. J. Antibiot. 1970, 23, 442–447. [Google Scholar] [CrossRef]
- Gorska, M.; Popowska, U.; Sielicka-Dudzin, A.; Kuban-Jankowska, A.; Sawczuk, W.; Knap, N.; Cicero, G.; Wozniak, F. Geldanamycin and its derivatives as Hsp90 inhibitors. Front. Biosci. 2012, 7, 2269–2277. [Google Scholar] [CrossRef]
- Trendowski, M. PU-H71: An improvement on nature’s solutions to oncogenic Hsp90 addiction. Pharmacol. Res. 2015, 99, 202–216. [Google Scholar] [CrossRef]
- Petersen, A.L.O.A.; Cull, B.; Dias, B.R.S.; Palma, L.C.; Luz, Y.D.S.; de Menezes, J.P.B.; Mottram, J.C.; Veras, P.S.T. 17-AAG-induced activation of the autophagic pathway in Leishmania is associated with parasite death. Microorganisms 2021, 9, 1089. [Google Scholar] [CrossRef]
- Dowall, S.D.; Bewley, K.; Watson, R.J.; Vasan, S.S.; Ghosh, C.; Konai, M.M.; Gausdal, G.; Lorens, J.B.; Long, J.; Barclay, W.; et al. Antiviral screening of multiple compounds against Ebola virus. Viruses 2016, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, W.; Tan, J.; Sun, Y.; Wan, Z.; Li, R. Antifungal activity of geldanamycin alone or in combination with fluconazole against Candida species. Mycopathologia 2013, 175, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Xia, T.; Wu, W.; Wang, R.; Qi, M.; Zhang, S.; Li, J.; Yang, Y.; Zheng, D.; Lin, S.; et al. Critical secondary metabolites confer the broad-spectrum pathogenic fungi resistance property of a marine-originating Streptomyces sp. HNBCa1. J. Agric. Food Chem. 2024, 72, 13164–13174. [Google Scholar] [CrossRef]
- Heisey, R.M.; Putnam, A.R. Herbicidal effects of geldanamycin and nigericin, antibiotics from Streptomyces hygroscopicus. J. Nat. Prod. 1986, 49, 859–865. [Google Scholar] [CrossRef]
- Kozeko, L.; Talalaiev, O.; Neimash, V.; Povarchuk, V. A protective role of HSP90 chaperone in gamma-irradiated Arabidopsis thaliana seeds. Life Sci. Space Res. 2015, 6, 51–58. [Google Scholar] [CrossRef]
- Kozeko, L.E. Phenotypic variability of Arabidopsis thaliana seedlings as a result of inhibition of Hsp90 chaperones. Tsitol Genet 2013, 47, 18–33. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, Y.; Liu, D.; Zhang, C.; Zhang, F. Geldanamycin (GDA) induce the increase of genomic DNA polymorphism and DNA methylation in Triticum aestivum L. Seed 2016, 35, 1–4. [Google Scholar]
- Díaz-Cruz, G.A.; Liu, J.Y.; Tahlan, K.; Bignell, D.R.D. Nigericin and geldanamycin are phytotoxic specialized metabolites produced by the plant pathogen Streptomyces sp. 11-1-2. Microbiol. Spectr. 2022, 10, e0231421. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Zhang, F.; Zhou, K.; Zhao, Q.; Sun, H.; Wang, S.; Zhao, Y.; Fu, J. Breeding of high protein Chlorella sorokiniana using protoplast fusion. Bioresour. Technol. 2020, 313, 123624. [Google Scholar] [CrossRef]
- Meng, S.; Wu, H.; Wang, L.; Zhang, B.; Bai, L. Enhancement of antibiotic productions by engineered nitrate utilization in Actinomycetes. Appl. Microbiol. Biotechnol. 2017, 101, 5341–5352. [Google Scholar] [CrossRef]
- Yang, L.; Yuan, Y.; Fang, Z.; Lin, R.; Jiang, H.; Zhou, J. Identification of a Streptomyces strain and study on the fermentation process of geldanamycin production. Biotechnol. Bull. 2024, 40, 299–309. [Google Scholar]
- Dobson, L.F.; O’Cleirigh, C.C.; O’Shea, D.G. The influence of morphology on geldanamycin production in submerged fermentations of Streptomyces hygroscopicus var. geldanus. Appl. Microbiol. Biotechnol. 2008, 79, 859–866. [Google Scholar] [CrossRef]
- Hospet, R.; Thangadurai, D.; Cruz-Martins, N.; Sangeetha, J.; Anu Appaiah, K.A.; Chowdhury, Z.Z.; Bedi, N.; Soytong, K.; Al Tawaha, A.R.M.; Jabeen, S.; et al. Genome shuffling for phenotypic improvement of industrial strains through recursive protoplast fusion technology. Crit. Rev. Food Sci. Nutr. 2023, 63, 2960–2969. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.M.; Li, J.S.; Liu, D.; Liu, F.; Wang, Y.T.; Song, X.R.; Wang, M. Genome shuffling of Streptomyces gilvosporeus for improving natamycin production. J. Agric. Food Chem. 2012, 60, 6026–6036. [Google Scholar] [CrossRef]
- Khaliq, S.; Akhtar, K.; Ghauri, M.A.; Iqbal, R.; Khalid, A.M.; Muddassar, M. Change in colony morphology and kinetics of tylosin production after UV and gamma irradiation mutagenesis of Streptomyces fradiae NRRL-2702. Microbiol. Res. 2009, 164, 469–477. [Google Scholar] [CrossRef]
- Jelić, D.; Antolović, R. From erythromycin to azithromycin and new potential ribosome-binding antimicrobials. Antibiotics 2016, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Ye, Z.; Lu, L. Enhancement of angucycline production by combined UV mutagenesis and ribosome engineering and fermentation optimization in Streptomyces dengpaensis XZHG99T. Prep. Biochem. Biotechnol. 2021, 51, 173–182. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kasahara, K.; Izawa, M.; Ochi, K. Applicability of ribosome engineering to vitamin B12 production by Propionibacterium shermanii. Biosci. Biotechnol. Biochem. 2017, 81, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Yue, X.R.; Wang, C.X.; Wang, J.R.; Zhao, J.N.; Yang, Z.P.; Fu, Q.K.; Wu, C.S.; Hu, W.; Li, Y.Z.; et al. Ribosome engineering of Myxococcus xanthus for enhancing the heterologous production of epothilones. Microb. Cell Factories 2024, 23, 346. [Google Scholar] [CrossRef] [PubMed]
- Hua, L.; Shi, H.; Lin, Q.; Wang, H.; Gao, Y.; Zeng, J.; Lou, K.; Huo, X. Selection and genetic analysis of high polysaccharide-producing mutants in Inonotus obliquus. Microorganisms 2024, 12, 1335. [Google Scholar] [CrossRef]
- Wan, H.M.; Chen, C.C.; Chang, T.S.; Giridhar, R.N.; Wu, W.T. Combining induced mutation and protoplasting for strain improvement of Aspergillus oryzae for kojic acid production. Biotechnol. Lett. 2004, 26, 1163–1166. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.Q.; Zhou, Y.H.; Chen, X.S.; Wu, J.Y.; Wei, P.; Yuan, L.X.; Yao, J.M. Genome shuffling and ribosome engineering of Streptomyces virginiae for improved virginiamycin production. Bioprocess Biosyst. Eng. 2018, 41, 729–738. [Google Scholar] [CrossRef]
- Pareek, C.S.; Smoczynski, R.; Tretyn, A. Sequencing technologies and genome sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar] [CrossRef]
- Wang, G.; Shi, T.; Chen, T.; Wang, X.; Wang, Y.; Liu, D.; Guo, J.; Fu, J.; Feng, L.; Wang, Z.; et al. Integrated whole-genome and transcriptome sequence analysis reveals the genetic characteristics of a riboflavin-overproducing Bacillus subtilis. Metab. Eng. 2018, 48, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.Q.; Xu, Z.W.; Men, X.H.; Zhang, B.; Liu, Z.Q.; Zheng, Y.G. Enhancement of protoplast preparation and regeneration of Hirsutella sinensis based on process optimization. Biotechnol. Lett. 2020, 42, 2357–2366. [Google Scholar] [CrossRef]
- Li, N.; Lu, J.; Wang, Z.; Du, P.; Li, P.; Su, J.; Xiao, J.; Wang, M.; Wang, J.; Wang, R. Improving the regeneration rate of deep lethal mutant protoplasts by fusion to promote efficient L-lysine fermentation. BMC Biotechnol. 2023, 23, 22. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Gao, H.; Tang, Y.; Lv, R.; Jiang, W.; Jiang, Y.; Zhang, W.; Xin, F.; Jiang, M. Transcriptomic analysis reveals the potential mechanisms for improving carotenoid production in Rhodosporidium toruloides Z11 under light stress. J. Agric. Food Chem. 2024, 72, 3793–3799. [Google Scholar] [CrossRef]
- Suryadi, H.; Irianti, M.I.; Septiarini, T.H. Methods of random mutagenesis of Aspergillus strain for increasing kojic acid production. Curr. Pharm. Biotechnol. 2022, 23, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Kollerov, V.; Donova, M. Ursodeoxycholic acid production by Gibberella zeae mutants. AMB Express 2022, 12, 105. [Google Scholar] [CrossRef]
- Zhu, Z.; Wu, X.; Lv, B.; Wu, G.; Wang, J.; Jiang, W.; Li, P.; He, J.; Chen, J.; Chen, M.; et al. A new approach for breeding low-temperature-resistant Volvariella volvacea strains: Genome shuffling in edible fungi. Biotechnol. Appl. Biochem. 2016, 63, 605–615. [Google Scholar] [CrossRef]
- Wang, X.; Ning, X.; Zhao, Q.; Kang, Q.; Bai, L. Improved PKS gene expression with strong endogenous promoter resulted in geldanamycin yield increase. Biotechnol. J. 2017, 12, 1700268. [Google Scholar] [CrossRef]
- Martín, J.F.; Ramos, A.; Liras, P. Regulation of geldanamycin biosynthesis by cluster-situated transcription factors and the master regulator PhoP. Antibiotics 2019, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Vetcher, L.; Tian, Z.Q.; McDaniel, R.; Rascher, A.; Revill, W.P.; Hutchinson, C.R.; Hu, Z. Rapid engineering of the geldanamycin biosynthesis pathway by Red/ET recombination and gene complementation. Appl. Environ. Microbiol. 2005, 71, 1829–1835. [Google Scholar] [CrossRef]
- Allen, I.W.; Ritchie, D.A. Cloning and analysis of DNA sequences from Streptomyces hygroscopicus encoding geldanamycin biosynthesis. Mol. Genet. Genom. 1994, 243, 593–599. [Google Scholar] [CrossRef]
- Rascher, A.; Hu, Z.; Viswanathan, N.; Schirmer, A.; Reid, R.; Nierman, W.C.; Lewis, M.; Hutchinson, C.R. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol. Lett. 2003, 218, 223–230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Favoino, G.; Krink, N.; Schwanemann, T.; Wierckx, N.; Nikel, P.I. Enhanced biosynthesis of poly(3-hydroxybutyrate) in engineered strains of Pseudomonas putida via increased malonyl-CoA availability. Microb. Biotechnol. 2024, 17, e70044. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.L.; Du, F.; Nong, F.T.; Li, J.; Huang, P.W.; Ma, W.; Gu, Y.; Sun, X.M. Function of the polyketide synthase domains of Schizochytrium sp. on fatty acid synthesis in Yarrowia lipolytica. J. Agric. Food Chem. 2023, 71, 2446–2454. [Google Scholar] [CrossRef]
- Xie, X.; Meesapyodsuk, D.; Qiu, X. Ketoacylsynthase domains of a polyunsaturated fatty acid synthase in Thraustochytrium sp. strain ATCC 26185 can effectively function as stand-alone enzymes in Escherichia coli. Appl. Environ. Microbiol. 2017, 83, e03133-16. [Google Scholar] [CrossRef]
- Salie, M.J.; Thelen, J.J. Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim. Biophys. Acta 2016, 1861, 1207–1213. [Google Scholar] [CrossRef]
- Chen, Y.; Siewers, V.; Nielsen, J. Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e42475. [Google Scholar] [CrossRef] [PubMed]

| Order | Name | Gene ID | Primer |
|---|---|---|---|
| 1 | pdc | chr_1968-F1 | TGCCAGGAAATCTCCACTCA |
| chr_1968-R1 | CGTAGAAGTGTCCGTTGTCCA | ||
| 2 | Acsm3 | chr_107-F1 | ATTCACCACCGTGGACACCG |
| chr_107-R1 | CCGCGCCGTAGAATACGTCT | ||
| 3 | citA | chr_4649-F1 | TCGTACCCGGACTCGAAGGA |
| chr_4649-R1 | AGGCACCGTCGACCAGCA | ||
| 4 | sdhA | chr_4555-F1 | ACAAGGGCAAGCGGTTCA |
| chr_4555-R1 | TTGTAGTCGAGGCGGATGG | ||
| 5 | fumC | chr_6221-F | GCGTGGTGGACAAGGACATC |
| chr_6221-R | AGGACTGGCTGGCGTTGAC | ||
| 6 | tktA | chr_10051-F | GACCGCAACGACCACTCAC |
| chr_10051-R | GAACGCCCGCCACTTGTC | ||
| 7 | xfp | chr_8692-F1 | AAATGGCTGAAGGTGTCCC |
| chr_8692-R1 | GTGCGAGAAGCCGTTGTG | ||
| 8 | bccA | chr_4470-F1 | GTGCTTCGTCGAGCGCTAC |
| chr_4470-R1 | CCTTGAGGATGGCTTTGGAG | ||
| 9 | fadE2 | chr_2830-F1 | CGTGATGGGCAAGACCG |
| chr_2830-R1 | CCGTGGTAGTGGTCCTCGTA | ||
| 10 | PanB | chr_3428-F | CGCAGAAGCAGTCCGAGAAG |
| chr_3428-R | TCGTAGGCGGTGAGCATCG | ||
| 11 | coaD | chr_6777-F | CGCCAGACCACCGAGGAG |
| chr_6777-R | CTGGAGGAGAGGAAGCTGTAGG | ||
| 12 | pikAII | chr_1226-F1 | GGGACCTCGACTCGCTCTAT |
| chr_1226-R1 | GGCGAGATCCCGAAGAAA | ||
| 13 | pikAI | chr_1222-F | TGGAGGCGATTCACGAGGAG |
| chr_1222-R | ACGGTCTGGCGGAGGTTG | ||
| 14 | pks12 | chr_1219-F | ACCAACGGCGTCGGAGTG |
| chr_1219-R | GCTTGTGCTGGAAGGCGTAG | ||
| 15 | GdmG | chr_1816-F1 | AGTCGATGTGACCGAAGAAATT |
| chr_1816-R1 | GAAGGTGCCGATCTCCAAC | ||
| 16 | GdmX | chr_1803-F1 | GCAGCATCACATCCTCAACA |
| chr_1803-R1 | GCGGAATTCCCAGTCGTAG | ||
| 17 | rpoA | chr_5758-F | GTCGCACGCTCCTCTCCTC |
| chr_5758-R | ACGACGAGCTGCTTGATGTTG | ||
| 18 | hrdB | chr_6978-F1 | GATTCCGCCAACCCAGTG |
| chr_6978-R1 | CTTCTGCGGCACTGACCA |
| Order | Mutant Gene | Name | Normal Base | Variant Base | Variation Type | Encoded Protein |
|---|---|---|---|---|---|---|
| 1 | chr-3081 | PpSQ1 00400 | C | T | nonsynonymous | dihydroxy-acid dehydratase |
| 2 | chr-1222 | pikAI | CA | C | frameshift deletion | type I polyketide synthase |
| 3 | chr-1226 | pikAII | C | CC | frameshift insertion | type I polyketide synthase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Y.; Yang, L.; Fang, Z.; Chen, H.; Sun, F.; Jiang, H.; Zhou, J. Improving Geldanamycin Production in Streptomyces geldanamycininus Through UV Mutagenesis of Protoplast. Microorganisms 2025, 13, 186. https://doi.org/10.3390/microorganisms13010186
Yuan Y, Yang L, Fang Z, Chen H, Sun F, Jiang H, Zhou J. Improving Geldanamycin Production in Streptomyces geldanamycininus Through UV Mutagenesis of Protoplast. Microorganisms. 2025; 13(1):186. https://doi.org/10.3390/microorganisms13010186
Chicago/Turabian StyleYuan, Yuan, Lu Yang, Zhikai Fang, Haimin Chen, Fei Sun, Hong Jiang, and Jian Zhou. 2025. "Improving Geldanamycin Production in Streptomyces geldanamycininus Through UV Mutagenesis of Protoplast" Microorganisms 13, no. 1: 186. https://doi.org/10.3390/microorganisms13010186
APA StyleYuan, Y., Yang, L., Fang, Z., Chen, H., Sun, F., Jiang, H., & Zhou, J. (2025). Improving Geldanamycin Production in Streptomyces geldanamycininus Through UV Mutagenesis of Protoplast. Microorganisms, 13(1), 186. https://doi.org/10.3390/microorganisms13010186








