Changes in RNA Splicing: A New Paradigm of Transcriptional Responses to Probiotic Action in the Mammalian Brain
Abstract
1. Introduction
2. Method
2.1. Description of Datasets Used
2.2. Gene Alternative Splicing and Expression Analysis
2.3. Functional Enrichment Analysis
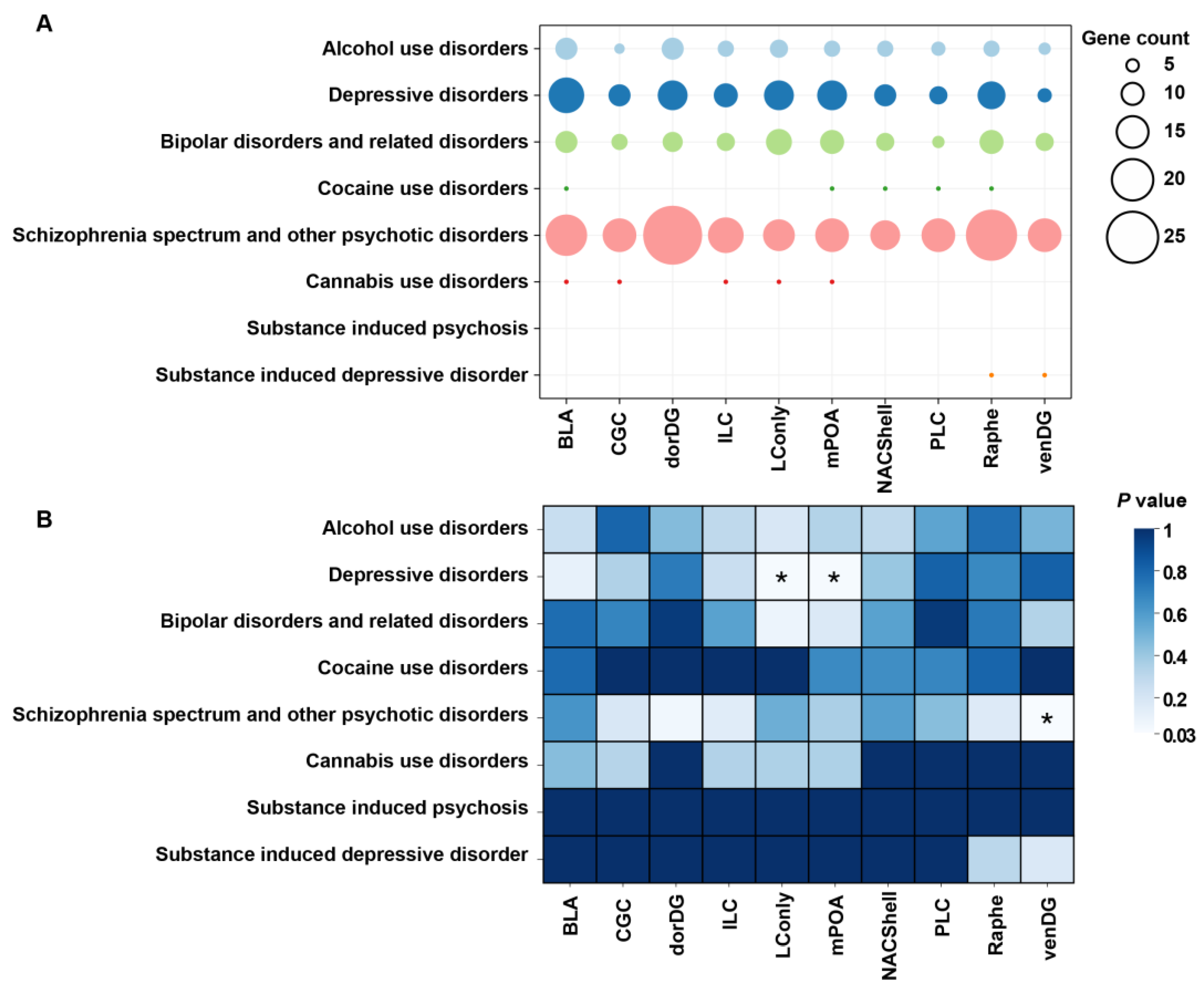
2.4. Psychiatric Disorder Association Analysis
3. Results
3.1. Probiotics Promote Extensive Alternative Splicing Changes Across the Brain
3.2. Probiotics Modulate Brain Neural Signaling Pathways via Alternative Splicing
3.3. Probiotic DSGs Are Associated with Psychiatric Disorders and Their Risk Genes
3.4. Limited Overlap Between Probiotic-Induced Splicing Events and Differential Gene Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liang, S.; Wu, X.; Jin, F. Gut-Brain Psychology: Rethinking Psychology From the Microbiota-Gut-Brain Axis. Front. Integr. Neurosci. 2018, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Shin, C. The Microbiota-Gut-Brain Axis in Neuropsychiatric Disorders: Pathophysiological Mechanisms and Novel Treatments. Curr. Neuropharmacol. 2018, 16, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Groussin, M.; Mazel, F.; Alm, E.J. Co-evolution and Co-speciation of Host-Gut Bacteria Systems. Cell Host Microbe 2020, 28, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Longo, S.; Rizza, S.; Federici, M. Microbiota-gut-brain axis: Relationships among the vagus nerve, gut microbiota, obesity, and diabetes. Acta Diabetol. 2023, 60, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, J.; Chen, Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients 2021, 13, 2099. [Google Scholar] [CrossRef] [PubMed]
- Holzer, P. Gut Signals and Gut Feelings: Science at the Interface of Data and Beliefs. Front. Behav. Neurosci. 2022, 16, 929332. [Google Scholar] [CrossRef]
- Galland, L. The gut microbiome and the brain. J. Med. Food 2014, 17, 1261–1272. [Google Scholar] [CrossRef]
- Madison, A.; Kiecolt-Glaser, J.K. Stress, depression, diet, and the gut microbiota: Human-bacteria interactions at the core of psychoneuroimmunology and nutrition. Curr. Opin. Behav. Sci. 2019, 28, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Collaborators, G.B.D.N. Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 2019, 18, 459–480. [Google Scholar] [CrossRef]
- Sabayan, B.; Doyle, S.; Rost, N.S.; Sorond, F.A.; Lakshminarayan, K.; Launer, L.J. The role of population-level preventive care for brain health in ageing. Lancet Healthy Longev. 2023, 4, e274–e283. [Google Scholar] [CrossRef] [PubMed]
- Popa-Wagner, A.; Dumitrascu, D.I.; Capitanescu, B.; Petcu, E.B.; Surugiu, R.; Fang, W.H.; Dumbrava, D.A. Dietary habits, lifestyle factors and neurodegenerative diseases. Neural Regen. Res. 2020, 15, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.C.; Schulte-Mecklenbeck, A.; Madireddy, L.; Pawlitzki, M.; Strippel, C.; Rauber, S.; Kramer, J.; Rolfes, L.; Ruck, T.; Beuker, C.; et al. Classification of neurological diseases using multi-dimensional CSF analysis. Brain 2021, 144, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Siuly, S.; Zhang, Y. Medical Big Data: Neurological Diseases Diagnosis Through Medical Data Analysis. Data Sci. Eng. 2016, 1, 54–64. [Google Scholar] [CrossRef]
- Person, H.; Keefer, L. Psychological comorbidity in gastrointestinal diseases: Update on the brain-gut-microbiome axis. Prog. Neuropsychopharmacol. Biol. Psychiatry 2021, 107, 110209. [Google Scholar] [CrossRef]
- Staudacher, H.M.; Mikocka-Walus, A.; Ford, A.C. Common mental disorders in irritable bowel syndrome: Pathophysiology, management, and considerations for future randomised controlled trials. Lancet Gastroenterol. Hepatol. 2021, 6, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Westfall, S.; Lomis, N.; Prakash, S. Longevity extension in Drosophila through gut-brain communication. Sci. Rep. 2018, 8, 8362. [Google Scholar] [CrossRef]
- Lee, J.G.; Cho, H.J.; Jeong, Y.M.; Lee, J.S. Genetic Approaches Using Zebrafish to Study the Microbiota-Gut-Brain Axis in Neurological Disorders. Cells 2021, 10, 566. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Cruz, N.J.; Kang, D.W.; Gandal, M.J.; Wang, B.; Kim, Y.M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 2019, 177, 1600–1618.e1617. [Google Scholar] [CrossRef]
- Huang, T.T.; Lai, J.B.; Du, Y.L.; Xu, Y.; Ruan, L.M.; Hu, S.H. Current Understanding of Gut Microbiota in Mood Disorders: An Update of Human Studies. Front. Genet. 2019, 10, 98. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Khan, A.; Seban, N.; Littlejohn, N.; Shah, A.; Srinivasan, S. A homeostatic gut-to-brain insulin antagonist restrains neuronally stimulated fat loss. Nat. Commun. 2024, 15, 6869. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Mandhare, A.; Tryphena, K.P.; Srivastava, S.; Shaikh, M.F.; Singh, S.B.; Khatri, D.K. Epigenetics in depression and gut-brain axis: A molecular crosstalk. Front. Aging Neurosci. 2022, 14, 1048333. [Google Scholar] [CrossRef] [PubMed]
- Bourassa, M.W.; Alim, I.; Bultman, S.J.; Ratan, R.R. Butyrate, neuroepigenetics and the gut microbiome: Can a high fiber diet improve brain health? Neurosci. Lett. 2016, 625, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balazsi, S.; Hajnady, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef]
- Shock, T.; Badang, L.; Ferguson, B.; Martinez-Guryn, K. The interplay between diet, gut microbes, and host epigenetics in health and disease. J. Nutr. Biochem. 2021, 95, 108631. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Zeng, B.H.; Li, W.W.; Zhou, C.J.; Fan, S.H.; Cheng, K.; Zeng, L.; Zheng, P.; Fang, L.; Wei, H.; et al. Effects of gut microbiota on the microRNA and mRNA expression in the hippocampus of mice. Behav. Brain Res. 2017, 322, 34–41. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative splicing and related RNA binding proteins in human health and disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Naro, C.; Cesari, E.; Sette, C. Splicing regulation in brain and testis: Common themes for highly specialized organs. Cell Cycle 2021, 20, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Nikom, D.; Zheng, S. Alternative splicing in neurodegenerative disease and the promise of RNA therapies. Nat. Rev. Neurosci. 2023, 24, 457–473. [Google Scholar] [CrossRef]
- Martinez, F.J.; Pratt, G.A.; Van Nostrand, E.L.; Batra, R.; Huelga, S.C.; Kapeli, K.; Freese, P.; Chun, S.J.; Ling, K.; Gelboin-Burkhart, C.; et al. Protein-RNA Networks Regulated by Normal and ALS-Associated Mutant HNRNPA2B1 in the Nervous System. Neuron 2016, 92, 780–795. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.H.; Liu, S.P.; Huang, S.J.; Chen, H.J.; Chen, P.R.; Lin, Y.H.; Ho, Y.C.; Chang, W.L.; Tsai, C.H.; Shyu, W.C.; et al. Aberrant alternative splicing events in Parkinson’s disease. Cell Transplant. 2013, 22, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Mu, X.; Shi, Y.; Zheng, H. Distinct Roles of Honeybee Gut Bacteria on Host Metabolism and Neurological Processes. Microbiol. Spectr. 2022, 10, e02438-21. [Google Scholar] [CrossRef] [PubMed]
- Rayan, N.A.; Aow, J.; Lim, M.G.L.; Arcego, D.M.; Ryan, R.; Nourbakhsh, N.; de Lima, R.M.S.; Craig, K.; Zhang, T.Y.; Goh, Y.T.; et al. Shared and unique transcriptomic signatures of antidepressant and probiotics action in the mammalian brain. Mol. Psychiatry 2024, 29, 3653–3668. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Gutierrez-Sacristan, A.; Grosdidier, S.; Valverde, O.; Torrens, M.; Bravo, A.; Pinero, J.; Sanz, F.; Furlong, L.I. PsyGeNET: A knowledge platform on psychiatric disorders and their genes. Bioinformatics 2015, 31, 3075–3077. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Sacristan, A.; Hernandez-Ferrer, C.; Gonzalez, J.R.; Furlong, L.I. psygenet2r: A R/Bioconductor package for the analysis of psychiatric disease genes. Bioinformatics 2017, 33, 4004–4006. [Google Scholar] [CrossRef] [PubMed]
- Kiattiweerasak, A.; Aumpan, N.; Chonprasertsuk, S.; Pornthisarn, B.; Siramolpiwat, S.; Bhanthumkomol, P.; Nunanan, P.; Issariyakulkarn, N.; Mahachai, V.; Yamaoka, Y.; et al. Efficacy and safety of Lacticaseibacillus rhamnosus R0011 and Lactobacillus helveticus R0052 as an adjuvant for Helicobacter pylori eradication: A double-blind, randomized, placebo-controlled study. Front. Gastroenterol. 2023, 2, 1245993. [Google Scholar] [CrossRef]
- Salapa, H.E.; Thibault, P.A.; Libner, C.D.; Ding, Y.; Clarke, J.W.E.; Denomy, C.; Hutchinson, C.; Abidullah, H.M.; Austin Hammond, S.; Pastushok, L.; et al. hnRNP A1 dysfunction alters RNA splicing and drives neurodegeneration in multiple sclerosis (MS). Nat. Commun. 2024, 15, 356. [Google Scholar] [CrossRef]
- Fujii, T.; Nagamori, S.; Wiriyasermkul, P.; Zheng, S.; Yago, A.; Shimizu, T.; Tabuchi, Y.; Okumura, T.; Fujii, T.; Takeshima, H.; et al. Parkinson’s disease-associated ATP13A2/PARK9 functions as a lysosomal H+,K+-ATPase. Nat. Commun. 2023, 14, 2174. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Cencic, R.; Cai, R.; Schmeing, T.M.; Pelletier, J. RNA-tethering assay and eIF4G:eIF4A obligate dimer design uncovers multiple eIF4F functional complexes. Nucleic Acids Res. 2020, 48, 8562–8575. [Google Scholar] [CrossRef]
- Paul, M.S.; Duncan, A.R.; Genetti, C.A.; Pan, H.; Jackson, A.; Grant, P.E.; Shi, J.; Pinelli, M.; Brunetti-Pierri, N.; Garza-Flores, A.; et al. Rare EIF4A2 variants are associated with a neurodevelopmental disorder characterized by intellectual disability, hypotonia, and epilepsy. Am. J. Hum. Genet. 2023, 110, 120–145. [Google Scholar] [CrossRef]
- Janin, J.; Bahadur, R.P.; Chakrabarti, P. Protein-protein interaction and quaternary structure. Q. Rev. Biophys. 2008, 41, 133–180. [Google Scholar] [CrossRef]
- Kawamura, N.; Sun-Wada, G.H.; Wada, Y. Loss of G2 subunit of vacuolar-type proton transporting ATPase leads to G1 subunit upregulation in the brain. Sci. Rep. 2015, 5, 14027. [Google Scholar] [CrossRef]
- Falace, A.; Volpedo, G.; Scala, M.; Zara, F.; Striano, P.; Fassio, A. V-ATPase Dysfunction in the Brain: Genetic Insights and Therapeutic Opportunities. Cells 2024, 13, 1441. [Google Scholar] [CrossRef] [PubMed]
- Twine, N.A.; Janitz, K.; Wilkins, M.R.; Janitz, M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS ONE 2011, 6, e16266. [Google Scholar] [CrossRef]
- Li, D.; McIntosh, C.S.; Mastaglia, F.L.; Wilton, S.D.; Aung-Htut, M.T. Neurodegenerative diseases: A hotbed for splicing defects and the potential therapies. Transl. Neurodegener. 2021, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Jakubauskiene, E.; Kanopka, A. Alternative Splicing and Hypoxia Puzzle in Alzheimer’s and Parkinson’s Diseases. Genes 2021, 12, 1272. [Google Scholar] [CrossRef]
- Montagna, E.; Dorostkar, M.M.; Herms, J. The Role of APP in Structural Spine Plasticity. Front. Mol. Neurosci. 2017, 10, 136. [Google Scholar] [CrossRef]
- Turner, P.R.; O’Connor, K.; Tate, W.P.; Abraham, W.C. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog. Neurobiol. 2003, 70, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Gegelashvili, G.; Bock, E.; Schousboe, A.; Linnemann, D. Two types of amyloid precursor protein (APP) mRNA in rat glioma cell lines: Upregulation via a cyclic AMP-dependent pathway. Brain Res. Mol. Brain Res. 1996, 37, 151–156. [Google Scholar] [CrossRef]
- Sandbrink, R.; Masters, C.L.; Beyreuther, K. APP gene family. Alternative splicing generates functionally related isoforms. Ann. N. Y Acad. Sci. 1996, 777, 281–287. [Google Scholar] [CrossRef]
- Sawmiller, D.; Koyama, N.; Fujiwara, M.; Segawa, T.; Maeda, M.; Mori, T. Targeting apolipoprotein E and N-terminal amyloid beta-protein precursor interaction improves cognition and reduces amyloid pathology in Alzheimer’s mice. J. Biol. Chem. 2023, 299, 104846. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Walker, D.; Bernardo, A.; Brodbeck, J.; Balestra, M.E.; Huang, Y. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J. Neurosci. 2008, 28, 1452–1459. [Google Scholar] [CrossRef]
- Strang, K.H.; Golde, T.E.; Giasson, B.I. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Investig. 2019, 99, 912–928. [Google Scholar] [CrossRef] [PubMed]
- Shehadeh, L.A.; Yu, K.; Wang, L.; Guevara, A.; Singer, C.; Vance, J.; Papapetropoulos, S. SRRM2, a potential blood biomarker revealing high alternative splicing in Parkinson’s disease. PLoS ONE 2010, 5, e9104. [Google Scholar] [CrossRef] [PubMed]
- Karlebach, G.; Steinhaus, R.; Danis, D.; Devoucoux, M.; Anczukow, O.; Sheynkman, G.; Seelow, D.; Robinson, P.N. Alternative splicing is coupled to gene expression in a subset of variably expressed genes. NPJ Genom. Med. 2024, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Rosonina, E.; Blencowe, B.J. Gene expression: The close coupling of transcription and splicing. Curr. Biol. 2002, 12, R319–R321. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, Z.M.; Zheng, M.J.; Liu, X. Phosphatidylethanolamine-binding protein 1 (PEBP1) as a potential target for the treatment for depression. CNS Neurosci. Ther. 2013, 19, 982–983. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, D. The Role of RNA Splicing in Liver Function and Disease: A Focus on Metabolic Dysfunction-Associated Steatotic Liver Disease. Genes 2024, 15, 1181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, Z.; Castle, J.; Sun, S.; Johnson, J.; Krainer, A.R.; Zhang, M.Q. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes. Dev. 2008, 22, 2550–2563. [Google Scholar] [CrossRef] [PubMed]
- Bercik, P.; Denou, E.; Collins, J.; Jackson, W.; Lu, J.; Jury, J.; Deng, Y.; Blennerhassett, P.; Macri, J.; McCoy, K.D.; et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 2011, 141, 599–609.e3. [Google Scholar] [CrossRef] [PubMed]
- Damiani, F.; Cornuti, S.; Tognini, P. The gut-brain connection: Exploring the influence of the gut microbiota on neuroplasticity and neurodevelopmental disorders. Neuropharmacology 2023, 231, 109491. [Google Scholar] [CrossRef]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Bjorkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Grau-Bove, X.; Ruiz-Trillo, I.; Irimia, M. Origin of exon skipping-rich transcriptomes in animals driven by evolution of gene architecture. Genome Biol. 2018, 19, 135. [Google Scholar] [CrossRef]
- Hatje, K.; Rahman, R.U.; Vidal, R.O.; Simm, D.; Hammesfahr, B.; Bansal, V.; Rajput, A.; Mickael, M.E.; Sun, T.; Bonn, S.; et al. The landscape of human mutually exclusive splicing. Mol. Syst. Biol. 2017, 13, 959. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.D.; Babu, M.M.; Lees, J.; Orengo, C.A. Biological impact of mutually exclusive exon switching. PLoS Comput. Biol. 2021, 17, e1008708. [Google Scholar] [CrossRef] [PubMed]
- Van Booven, D.; Mengying, L.; Sunil Rao, J.; Blokhin, I.O.; Dayne Mayfield, R.; Barbier, E.; Heilig, M.; Wahlestedt, C. Alcohol use disorder causes global changes in splicing in the human brain. Transl. Psychiatry 2021, 11, 2. [Google Scholar] [CrossRef]
- Ota, V.K.; Santoro, M.L.; Spindola, L.M.; Pan, P.M.; Simabucuro, A.; Xavier, G.; Vieira-Fonseca, T.; Zanardo, E.A.; Dos Santos, F.R.C.; Schafer, J.L.; et al. Gene expression changes associated with trajectories of psychopathology in a longitudinal cohort of children and adolescents. Transl. Psychiatry 2020, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-β Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Suzuki, H.; Tsukahara, T. Alternative splicing regulation of APP exon 7 by RBFox proteins. Neurochem. Int. 2014, 78, 7–17. [Google Scholar] [CrossRef]
- Jacobs, A.; Elmer, K.R. Alternative splicing and gene expression play contrasting roles in the parallel phenotypic evolution of a salmonid fish. Mol. Ecol. 2021, 30, 4955–4969. [Google Scholar] [CrossRef]
- Verta, J.P.; Jacobs, A. The role of alternative splicing in adaptation and evolution. Trends Ecol. Evol. 2022, 37, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Grantham, M.E.; Brisson, J.A. Extensive Differential Splicing Underlies Phenotypically Plastic Aphid Morphs. Mol. Biol. Evol. 2018, 35, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Spellman, R.; Llorian, M.; Smith, C.W. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell 2007, 27, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Su, C.J.; Edmonds, K.K.; Liang, G.; Elowitz, M.B. Dynamics and functional roles of splicing factor autoregulation. Cell Rep. 2022, 39, 110985. [Google Scholar] [CrossRef] [PubMed]
- Anko, M.L.; Muller-McNicoll, M.; Brandl, H.; Curk, T.; Gorup, C.; Henry, I.; Ule, J.; Neugebauer, K.M. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012, 13, R17. [Google Scholar] [CrossRef]
- Yau, J.O.; Chaichim, C.; Power, J.M.; McNally, G.P. The Roles of Basolateral Amygdala Parvalbumin Neurons in Fear Learning. J. Neurosci. 2021, 41, 9223–9234. [Google Scholar] [CrossRef] [PubMed]
- Suthard, R.L.; Senne, R.A.; Buzharsky, M.D.; Pyo, A.Y.; Dorst, K.E.; Diep, A.H.; Cole, R.H.; Ramirez, S. Basolateral Amygdala Astrocytes Are Engaged by the Acquisition and Expression of a Contextual Fear Memory. J. Neurosci. 2023, 43, 4997–5013. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, S.; Musso, F.; Mobascher, A.; Warbrick, T.; Winterer, G.; Gallinat, J. Hippocampal subfields predict positive symptoms in schizophrenia: First evidence from brain morphometry. Transl. Psychiatry 2012, 2, e127. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, S.; Turner, J.A.; Calhoun, V.D.; Lim, K.O.; Mueller, B.; Bustillo, J.R.; O’Leary, D.S.; McEwen, S.; Voyvodic, J.; Belger, A.; et al. Dentate gyrus volume deficit in schizophrenia. Psychol. Med. 2020, 50, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
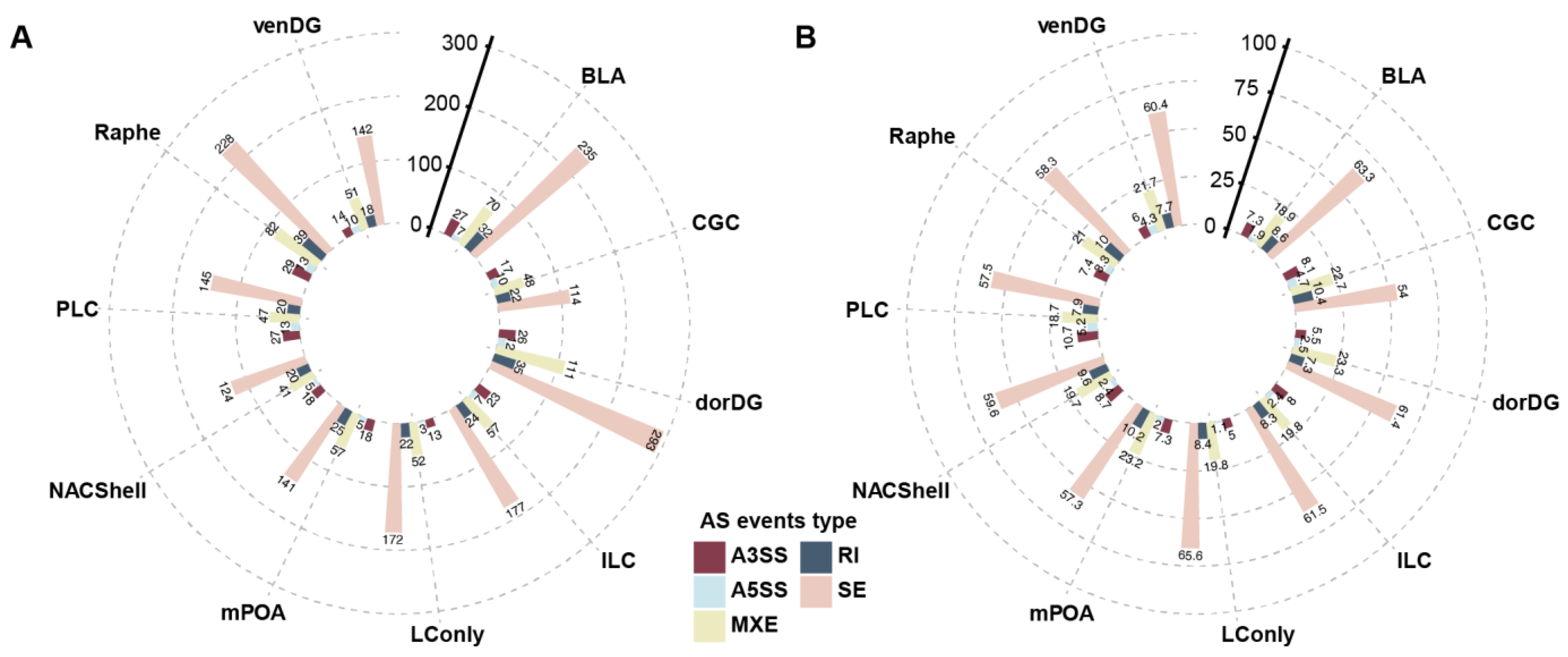
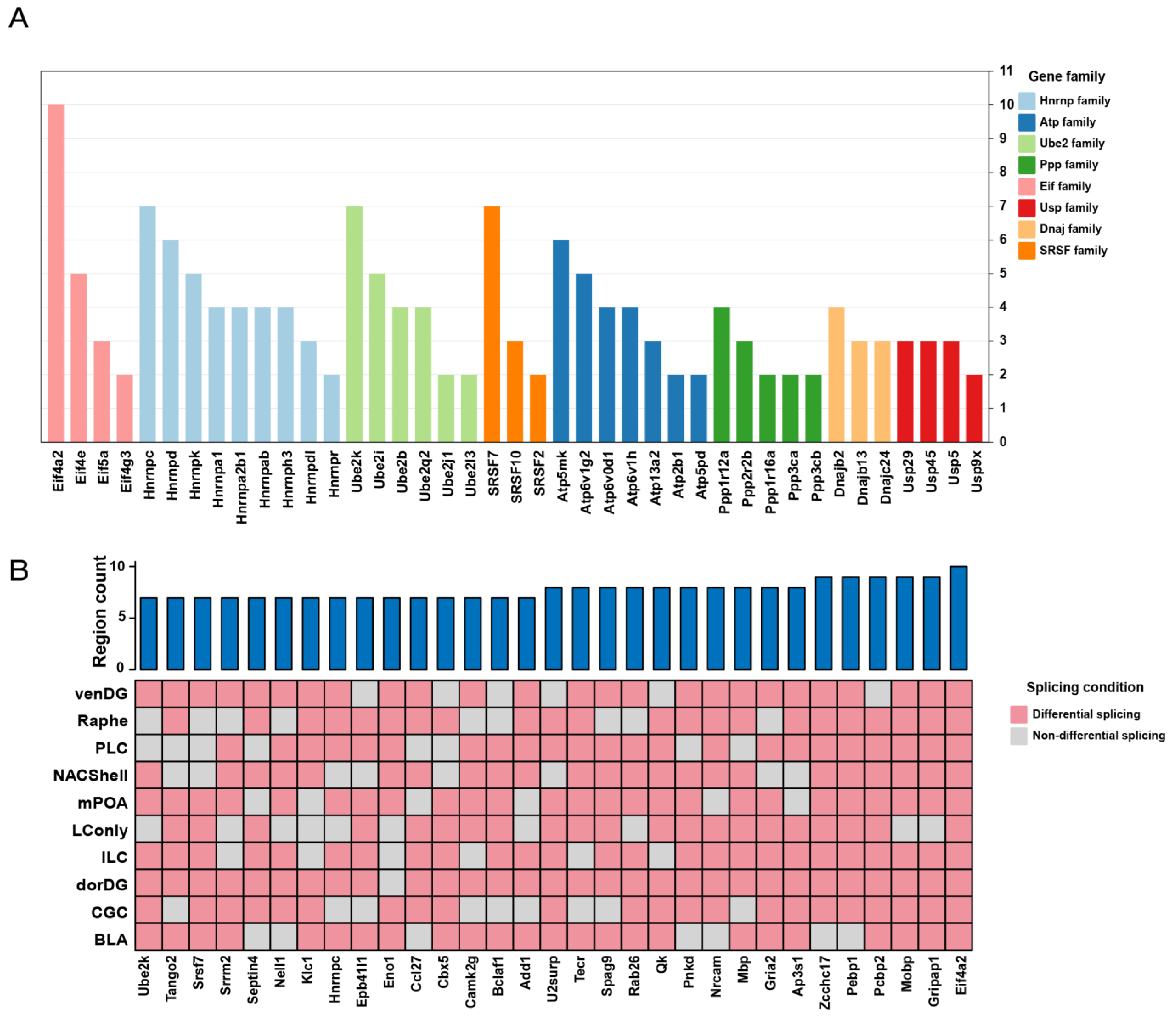

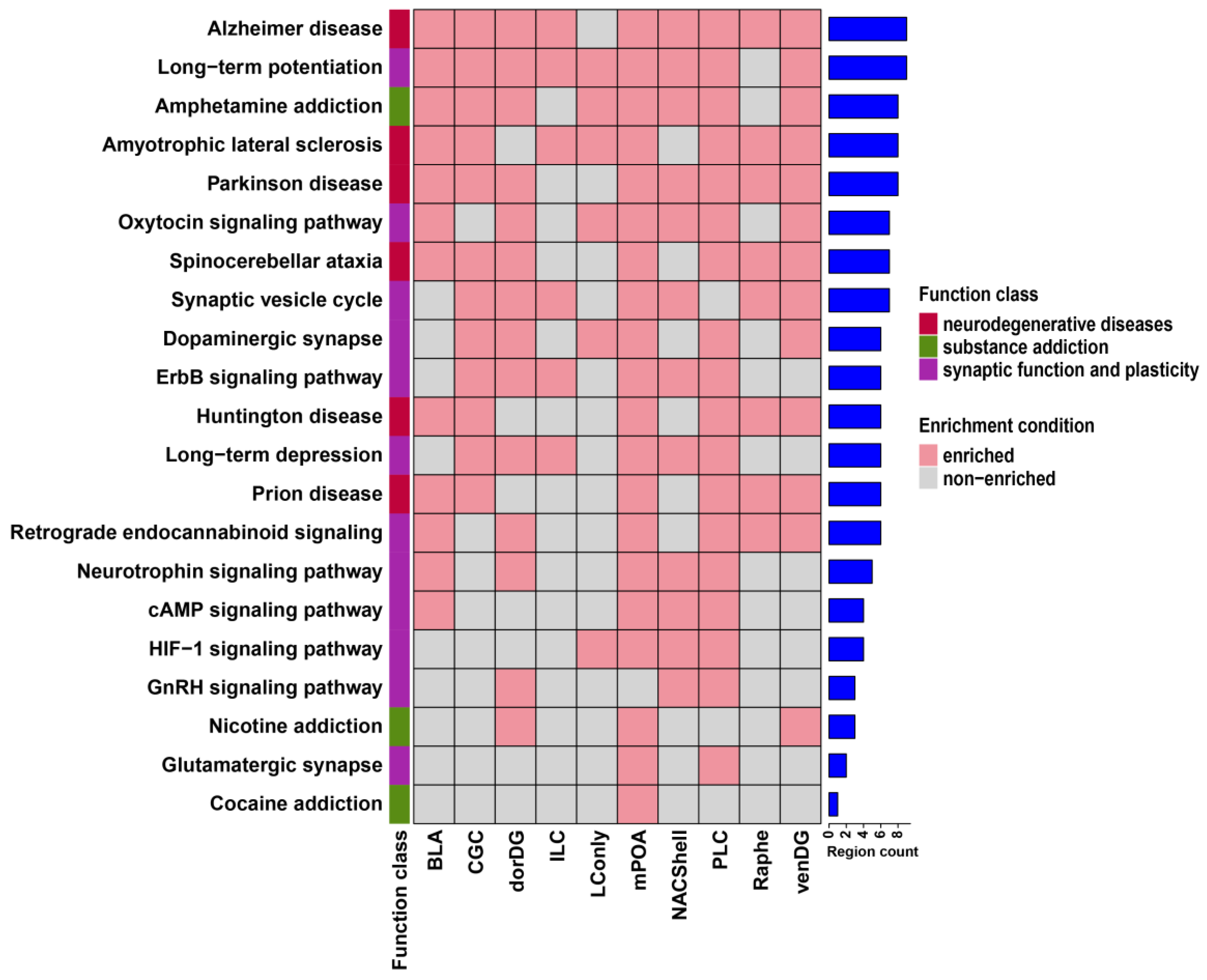
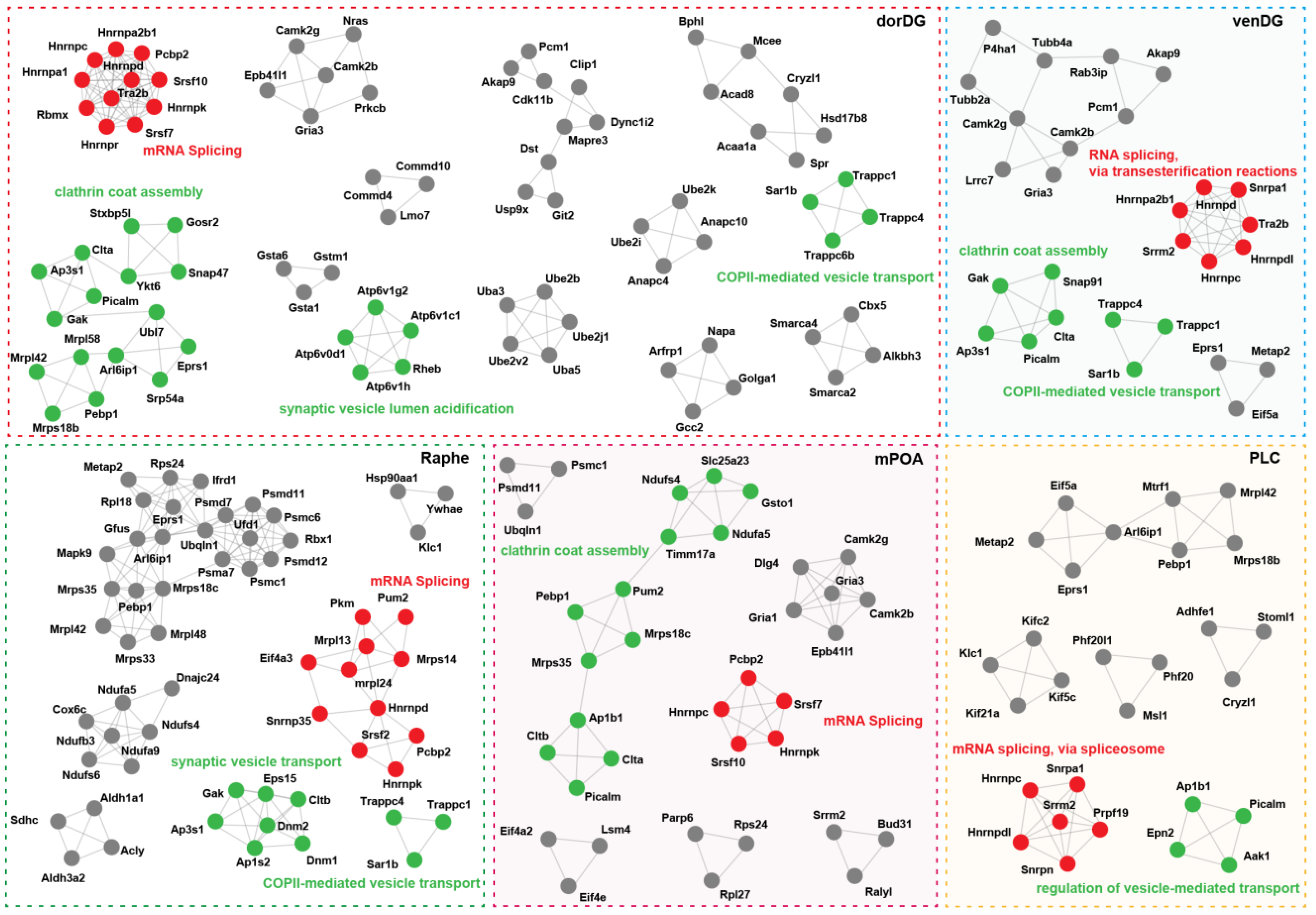


| BLA | CGC | dorDG | ILC | LConly | mPOA | NACShell | PLC | Raphe | venDG | |
|---|---|---|---|---|---|---|---|---|---|---|
| DSGs | 314 | 186 | 369 | 247 | 225 | 200 | 174 | 221 | 309 | 190 |
| DSGs vs. DEGs | 42 | 20 | 220 | 154 | 43 | 39 | 17 | 38 | 37 | 49 |
| Overlap Ratio | 13% | 11% | 60% | 62% | 19% | 20% | 10% | 17% | 12% | 26% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, X.; Zhu, L.; Zhang, Z. Changes in RNA Splicing: A New Paradigm of Transcriptional Responses to Probiotic Action in the Mammalian Brain. Microorganisms 2025, 13, 165. https://doi.org/10.3390/microorganisms13010165
Yue X, Zhu L, Zhang Z. Changes in RNA Splicing: A New Paradigm of Transcriptional Responses to Probiotic Action in the Mammalian Brain. Microorganisms. 2025; 13(1):165. https://doi.org/10.3390/microorganisms13010165
Chicago/Turabian StyleYue, Xiaojie, Lei Zhu, and Zhigang Zhang. 2025. "Changes in RNA Splicing: A New Paradigm of Transcriptional Responses to Probiotic Action in the Mammalian Brain" Microorganisms 13, no. 1: 165. https://doi.org/10.3390/microorganisms13010165
APA StyleYue, X., Zhu, L., & Zhang, Z. (2025). Changes in RNA Splicing: A New Paradigm of Transcriptional Responses to Probiotic Action in the Mammalian Brain. Microorganisms, 13(1), 165. https://doi.org/10.3390/microorganisms13010165






