From the Cytoplasm into the Nucleus—Hepatitis B Virus Travel and Genome Repair
Abstract
1. Preface
Overview of the HBV Life Cycle
2. Core Protein, Capsids, and Their Phosphorylation
2.1. Core Protein
2.2. Capsids
2.3. Cytoplasmic Transport
2.3.1. Principles of Active Cytoplasmic Transport
2.3.2. Retrograde Transport of Other Viruses
2.3.3. HBV: Cytosolic Genome Release Versus Genome Transport in the Capsid
2.4. Nuclear Transport and Genome Release
2.4.1. Principles of Active Nuclear Import
2.4.2. Nuclear Import of Genomes of Other Viruses
2.4.3. Nuclear Import of HBV Genomes
2.5. Repair of the HBV Genome
3. Summary
Therapeutic Implications
Funding
Conflicts of Interest
References
- Mason, W.S. Animal models and the molecular biology of hepadnavirus infection. Cold Spring Harb. Perspect. Med. 2015, 5, a021352. [Google Scholar] [CrossRef]
- Allweiss, L.; Strick-Marchand, H. In-vitro and in-vivo models for hepatitis B cure research. Curr. Opin. HIV AIDS 2020, 15, 173–179. [Google Scholar] [CrossRef]
- Gerlich, W.H. Medical virology of hepatitis B: How it began and where we are now. Virol. J. 2013, 10, 239. [Google Scholar] [CrossRef]
- Liu, H.; Hong, X.; Xi, J.; Menne, S.; Hu, J.; Wang, J.C. Cryo-EM structures of human hepatitis B and woodchuck hepatitis virus small spherical subviral particles. Sci. Adv. 2022, 8, eabo4184. [Google Scholar] [CrossRef] [PubMed]
- Blondot, M.L.; Bruss, V.; Kann, M. Intracellular transport and egress of hepatitis B virus. J. Hepatol. 2016, 64, S49–S59. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Mason, W.S.; Summers, J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J. Virol. 1996, 70, 4567–4575. [Google Scholar] [CrossRef]
- Zhao, X.L.; Yang, J.R.; Lin, S.Z.; Ma, H.; Guo, F.; Yang, R.F.; Zhang, H.H.; Han, J.C.; Wei, L.; Pan, X.B. Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut 2016, 65, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Lindh, M.; Rydell, G.E.; Larsson, S.B. Impact of integrated viral DNA on the goal to clear hepatitis B surface antigen with different therapeutic strategies. Curr. Opin. Virol. 2018, 30, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Caminiti, G. HBV-Integration Studies in the Clinic: Role in the Natural History of Infection. Viruses 2021, 13, 368. [Google Scholar] [CrossRef]
- Ringlander, J.; Malmstrom, S.; Eilard, A.; Stromberg, L.G.; Stenback, J.B.; Andersson, M.E.; Larsson, S.B.; Kann, M.; Nilsson, S.; Hellstrand, K.; et al. Hepatitis B virus particles in serum contain minus strand DNA and degraded pregenomic RNA of variable and inverse lengths. Liver Int. 2024, 44, 1775–1780. [Google Scholar] [CrossRef]
- Luckenbaugh, L.; Kitrinos, K.M.; Delaney, W.E.t.; Hu, J. Genome-free hepatitis B virion levels in patient sera as a potential marker to monitor response to antiviral therapy. J. Viral Hepat. 2015, 22, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Herrscher, C.; Pastor, F.; Burlaud-Gaillard, J.; Dumans, A.; Seigneuret, F.; Moreau, A.; Patient, R.; Eymieux, S.; de Rocquigny, H.; Hourioux, C.; et al. Hepatitis B virus entry into HepG2-NTCP cells requires clathrin-mediated endocytosis. Cell. Microbiol. 2020, 22, e13205. [Google Scholar] [CrossRef]
- Huang, H.C.; Chen, C.C.; Chang, W.C.; Tao, M.H.; Huang, C. Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J. Virol. 2012, 86, 9443–9453. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Hildt, E. Intracellular Trafficking of HBV Particles. Cells 2020, 9, 2023. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Somiya, M.; Shimada, N.; Sakamoto, W.; Yoshimoto, N.; Iijima, M.; Tatematsu, K.; Nakai, T.; Okajima, T.; Maruyama, A.; et al. Mutational analysis of hepatitis B virus pre-S1 (9–24) fusogenic peptide. Biochem. Biophys. Res. Commun. 2016, 474, 406–412. [Google Scholar] [CrossRef]
- Perez-Vargas, J.; Teppa, E.; Amirache, F.; Boson, B.; Pereira de Oliveira, R.; Combet, C.; Bockmann, A.; Fusil, F.; Freitas, N.; Carbone, A.; et al. A fusion peptide in preS1 and the human protein disulfide isomerase ERp57 are involved in hepatitis B virus membrane fusion process. eLife 2021, 10, e64507. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, R.A. “Lifespan” of liver cells. Autoradio-graphic study using tritiated thymidine in normal, cirrhotic, and partially hepatectomized rats. Arch. Intern. Med. 1961, 107, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Magami, Y.; Azuma, T.; Inokuchi, H.; Kokuno, S.; Moriyasu, F.; Kawai, K.; Hattori, T. Cell proliferation and renewal of normal hepatocytes and bile duct cells in adult mouse liver. Liver 2002, 22, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Moreno, A.; Ploss, A. Mechanisms of Hepatitis B Virus cccDNA and Minichromosome Formation and HBV Gene Transcription. Viruses 2024, 16, 609. [Google Scholar] [CrossRef]
- Bacher, J.; Allweiss, L.; Dandri, M. SMC5/6-Mediated Transcriptional Regulation of Hepatitis B Virus and Its Therapeutic Potential. Viruses 2024, 16, 1667. [Google Scholar] [CrossRef] [PubMed]
- Cougot, D.; Wu, Y.; Cairo, S.; Caramel, J.; Renard, C.A.; Levy, L.; Buendia, M.A.; Neuveut, C. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J. Biol. Chem. 2007, 282, 4277–4287. [Google Scholar] [CrossRef] [PubMed]
- Dandri, M. Epigenetic modulation in chronic hepatitis B virus infection. Semin. Immunopathol. 2020, 42, 173–185. [Google Scholar] [CrossRef]
- Zhong, Y.; Wu, C.; Xu, Z.; Teng, Y.; Zhao, L.; Zhao, K.; Wang, J.; Wang, W.; Zhan, Q.; Zhu, C.; et al. Hepatitis B Virus Core Protein Is Not Required for Covalently Closed Circular DNA Transcriptional Regulation. J. Virol. 2022, 96, e0136222. [Google Scholar] [CrossRef]
- Ding, J.; Yi, Z.; Zai, W.; Wu, M.; Chen, B.; Cai, Q.; Zhang, X.; Yuan, Z. Illuminating the Live-Cell Dynamics of Hepatitis B Virus Covalently Closed Circular DNA Using the CRISPR-Tag System. mBio 2023, 14, e0355022. [Google Scholar] [CrossRef]
- Kremsdorf, D.; Lekbaby, B.; Bablon, P.; Sotty, J.; Augustin, J.; Schnuriger, A.; Pol, J.; Soussan, P. Alternative splicing of viral transcripts: The dark side of HBV. Gut 2021, 70, 2373–2382. [Google Scholar] [CrossRef] [PubMed]
- Bartenschlager, R.; Junker-Niepmann, M.; Schaller, H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 1990, 64, 5324–5332. [Google Scholar] [CrossRef] [PubMed]
- Tavis, J.E.; Ganem, D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 1996, 70, 5741–5750. [Google Scholar] [CrossRef] [PubMed]
- Tavis, J.E.; Massey, B.; Gong, Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, ε. J. Virol. 1998, 72, 5789–5796. [Google Scholar] [CrossRef] [PubMed]
- Selzer, L.; Zlotnick, A. Assembly and Release of Hepatitis B Virus. Cold Spring Harb. Perspect. Med. 2015, 5, a021394. [Google Scholar] [CrossRef] [PubMed]
- Prange, R. Hepatitis B virus movement through the hepatocyte: An update. Biol. Cell 2022, 114, 325–348. [Google Scholar] [CrossRef]
- Clark, D.N.; Tajwar, R.; Hu, J.; Tavis, J.E. The hepatitis B virus polymerase. Enzymes 2021, 50, 195–226. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Seeger, C. Hepadnavirus Genome Replication and Persistence. Cold Spring Harb. Perspect. Med. 2015, 5, a021386. [Google Scholar] [CrossRef] [PubMed]
- Lott, L.; Beames, B.; Notvall, L.; Lanford, R.E. Interaction between hepatitis B virus core protein and reverse transcriptase. J. Virol. 2000, 74, 11479–11489. [Google Scholar] [CrossRef] [PubMed]
- Bruss, V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 1997, 71, 9350–9357. [Google Scholar] [CrossRef] [PubMed]
- Bruss, V.; Thomssen, R. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J. Virol. 1994, 68, 1643–1650. [Google Scholar] [CrossRef] [PubMed]
- Niklasch, M.; Zimmermann, P.; Nassal, M. The Hepatitis B Virus Nucleocapsid-Dynamic Compartment for Infectious Virus Production and New Antiviral Target. Biomedicines 2021, 9, 1577. [Google Scholar] [CrossRef]
- Ko, C.; Chakraborty, A.; Chou, W.M.; Hasreiter, J.; Wettengel, J.M.; Stadler, D.; Bester, R.; Asen, T.; Zhang, K.; Wisskirchen, K.; et al. Hepatitis B virus genome recycling and de novo secondary infection events maintain stable cccDNA levels. J. Hepatol. 2018, 69, 1231–1241. [Google Scholar] [CrossRef]
- Nair, S.; Zlotnick, A. HBV Core Protein Is in Flux between Cytoplasmic, Nuclear, and Nucleolar Compartments. mBio 2021, 12, e03514-20. [Google Scholar] [CrossRef] [PubMed]
- Gerlich, W.H.; Goldmann, U.; Muller, R.; Stibbe, W.; Wolff, W. Specificity and localization of the hepatitis B virus-associated protein kinase. J. Virol. 1982, 42, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, L.G.; Martinez, V.; Loh, Y.T.; Rogler, C.E.; Chisari, F.V. Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice. J. Virol. 1994, 68, 5469–5475. [Google Scholar] [CrossRef] [PubMed]
- Li, H.C.; Huang, E.Y.; Su, P.Y.; Wu, S.Y.; Yang, C.C.; Lin, Y.S.; Chang, W.C.; Shih, C. Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathog. 2010, 6, e1001162. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.C.; Su, I.J.; Lai, M.Y.; Chen, D.S.; Chang, M.H.; Chuang, S.M.; Sung, J.L. Biologic and prognostic significance of hepatocyte hepatitis B core antigen expressions in the natural course of chronic hepatitis B virus infection. J. Hepatol. 1987, 5, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.B.; Han, J.C.; Wei, L.; Peng, D.D.; Gao, Y. Subcellular distribution and translocation of hepatitis B virus core protein in HepG2.2.15 cells. Zhonghua Gan Zang Bing. Za Zhi 2008, 16, 29–32. [Google Scholar] [PubMed]
- Cao, F.; Tavis, J.E. Detection and characterization of cytoplasmic hepatitis B virus reverse transcriptase. J. Gen. Virol. 2004, 85, 3353–3360. [Google Scholar] [CrossRef] [PubMed]
- Unchwaniwala, N.; Sherer, N.M.; Loeb, D.D. Hepatitis B Virus Polymerase Localizes to the Mitochondria, and Its Terminal Protein Domain Contains the Mitochondrial Targeting Signal. J. Virol. 2016, 90, 8705–8719. [Google Scholar] [CrossRef] [PubMed]
- Wynne, S.A.; Crowther, R.A.; Leslie, A.G. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 1999, 3, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, P.T.; Stahl, S.J.; Williams, R.W.; Steven, A.C. Hepatitis core antigen produced in Escherichia coli: Subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 1995, 34, 4919–4932. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, B.; Wynne, S.A.; Crowther, R.A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 1997, 386, 88–91. [Google Scholar] [CrossRef]
- Kann, M.; Sodeik, B.; Vlachou, A.; Gerlich, W.H.; Helenius, A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 1999, 145, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.T.; Liaw, Y.F.; Ou, J.H. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J. Virol. 1990, 64, 6141–6147. [Google Scholar] [CrossRef] [PubMed]
- Su, P.Y.; Yang, C.J.; Chu, T.H.; Chang, C.H.; Chiang, C.; Tang, F.M.; Lee, C.Y.; Shih, C. HBV maintains electrostatic homeostasis by modulating negative charges from phosphoserine and encapsidated nucleic acids. Sci. Rep. 2016, 6, 38959. [Google Scholar] [CrossRef]
- Luo, J.; Xi, J.; Gao, L.; Hu, J. Role of Hepatitis B virus capsid phosphorylation in nucleocapsid disassembly and covalently closed circular DNA formation. PLoS Pathog. 2020, 16, e1008459. [Google Scholar] [CrossRef]
- Zlotnick, A.; Cheng, N.; Stahl, S.J.; Conway, J.F.; Steven, A.C.; Wingfield, P.T. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: Implications for morphogenesis and organization of encapsidated RNA. Proc. Natl. Acad. Sci. USA 1997, 94, 9556–9561. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, M.; Sawano, Y.; Kosuge, S.; Yamano, Y.; Kuroki, K.; Ohtsuki, K. High phosphorylation of HBV core protein by two α-type CK2-activated cAMP-dependent protein kinases in vitro. FEBS Lett. 2006, 580, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Heger-Stevic, J.; Zimmermann, P.; Lecoq, L.; Bottcher, B.; Nassal, M. Hepatitis B virus core protein phosphorylation: Identification of the SRPK1 target sites and impact of their occupancy on RNA binding and capsid structure. PLoS Pathog. 2018, 14, e1007488. [Google Scholar] [CrossRef]
- Lubyova, B.; Tikalova, E.; Krulova, K.; Hodek, J.; Zabransky, A.; Hirsch, I.; Weber, J. ATM-Dependent Phosphorylation of Hepatitis B Core Protein in Response to Genotoxic Stress. Viruses 2021, 13, 2438. [Google Scholar] [CrossRef] [PubMed]
- Diab, A.; Foca, A.; Fusil, F.; Lahlali, T.; Jalaguier, P.; Amirache, F.; N’Guyen, L.; Isorce, N.; Cosset, F.L.; Zoulim, F.; et al. Polo-like-kinase 1 is a proviral host factor for hepatitis B virus replication. Hepatology 2017, 66, 1750–1765. [Google Scholar] [CrossRef] [PubMed]
- Wittkop, L.; Schwarz, A.; Cassany, A.; Grun-Bernhard, S.; Delaleau, M.; Rabe, B.; Cazenave, C.; Gerlich, W.; Glebe, D.; Kann, M. Inhibition of protein kinase C phosphorylation of hepatitis B virus capsids inhibits virion formation and causes intracellular capsid accumulation. Cell. Microbiol. 2010, 12, 962–975. [Google Scholar] [CrossRef] [PubMed]
- de Rocquigny, H.; Rat, V.; Pastor, F.; Darlix, J.L.; Hourioux, C.; Roingeard, P. Phosphorylation of the Arginine-Rich C-Terminal Domains of the Hepatitis B Virus (HBV) Core Protein as a Fine Regulator of the Interaction between HBc and Nucleic Acid. Viruses 2020, 12, 738. [Google Scholar] [CrossRef] [PubMed]
- Yip, R.P.H.; Kwok, D.C.Y.; Lai, L.T.F.; Ho, S.M.; Wong, I.C.K.; Chan, C.P.; Lau, W.C.Y.; Ngo, J.C.K. SRPK2 Mediates HBV Core Protein Phosphorylation and Capsid Assembly via Docking Interaction. PLoS Pathog. 2024, 20, e1011978. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J.C.; Zlotnick, A. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS Pathog. 2011, 7, e1002388. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; White, S.J.; Thompson, R.F.; Bingham, R.; Weiss, E.U.; Maskell, D.P.; Zlotnick, A.; Dykeman, E.; Tuma, R.; Twarock, R.; et al. HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Nat. Microbiol. 2017, 2, 17098. [Google Scholar] [CrossRef]
- Gazina, E.V.; Fielding, J.E.; Lin, B.; Anderson, D.A. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 2000, 74, 4721–4728. [Google Scholar] [CrossRef]
- Romero, S.; Unchwaniwala, N.; Evans, E.L.; Eliceiri, K.W.; Loeb, D.D.; Sherer, N.M. Live Cell Imaging Reveals HBV Capsid Translocation from the Nucleus To the Cytoplasm Enabled by Cell Division. mBio 2023, 14, e0330322. [Google Scholar] [CrossRef] [PubMed]
- Osseman, Q.; Gallucci, L.; Au, S.; Cazenave, C.; Berdance, E.; Blondot, M.L.; Cassany, A.; Begu, D.; Ragues, J.; Aknin, C.; et al. The chaperone dynein LL1 mediates cytoplasmic transport of empty and mature hepatitis B virus capsids. J. Hepatol. 2018, 68, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fan, G.; Wang, Z.; Chen, H.S.; Yin, C.C. Allosteric conformational changes of human HBV core protein transform its assembly. Sci. Rep. 2017, 7, 1404. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Tsuda, S.; Ogata, N.; Kataoka, M.; Sasaki, J.; Inuki, S.; Ohno, H.; Watashi, K.; Yoshiya, T.; Oishi, S. Synthesis of the full-length hepatitis B virus core protein and its capsid formation. Org. Biomol. Chem. 2024, 22, 2218–2225. [Google Scholar] [CrossRef]
- Scotto, J.; Hadchouel, M.; Wain-Hobson, S.; Sonigo, P.; Courouce, A.M.; Tiollais, P.; Brechot, C. Hepatitis B virus DNA in Dane particles: Evidence for the presence of replicative intermediates. J. Infect. Dis. 1985, 151, 610–617. [Google Scholar] [CrossRef]
- Khaykelson, D.; Asor, R.; Zhao, Z.; Schlicksup, C.J.; Zlotnick, A.; Raviv, U. Guanidine Hydrochloride-Induced Hepatitis B Virus Capsid Disassembly Hysteresis. Biochemistry 2024, 63, 1543–1552. [Google Scholar] [CrossRef]
- Hadden, J.A.; Perilla, J.R.; Schlicksup, C.J.; Venkatakrishnan, B.; Zlotnick, A.; Schulten, K. All-atom molecular dynamics of the HBV capsid reveals insights into biological function and cryo-EM resolution limits. eLife 2018, 7, e32478. [Google Scholar] [CrossRef] [PubMed]
- Venkatakrishnan, B.; Katen, S.P.; Francis, S.; Chirapu, S.; Finn, M.G.; Zlotnick, A. Hepatitis B Virus Capsids Have Diverse Structural Responses to Small-Molecule Ligands Bound to the Heteroaryldihydropyrimidine Pocket. J. Virol. 2016, 90, 3994–4004. [Google Scholar] [CrossRef] [PubMed]
- Rabe, B.; Delaleau, M.; Bischof, A.; Foss, M.; Sominskaya, I.; Pumpens, P.; Cazenave, C.; Castroviejo, M.; Kann, M. Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids. PLoS Pathog. 2009, 5, e1000563. [Google Scholar] [CrossRef]
- Bourne, C.R.; Katen, S.P.; Fulz, M.R.; Packianathan, C.; Zlotnick, A. A mutant hepatitis B virus core protein mimics inhibitors of icosahedral capsid self-assembly. Biochemistry 2009, 48, 1736–1742. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J.C.; Pierson, E.E.; Keifer, D.Z.; Delaleau, M.; Gallucci, L.; Cazenave, C.; Kann, M.; Jarrold, M.F.; Zlotnick, A. Importin β Can Bind Hepatitis B Virus Core Protein and Empty Core-Like Particles and Induce Structural Changes. PLoS Pathog. 2016, 12, e1005802. [Google Scholar] [CrossRef] [PubMed]
- Selzer, L.; Kant, R.; Wang, J.C.; Bothner, B.; Zlotnick, A. Hepatitis B Virus Core Protein Phosphorylation Sites Affect Capsid Stability and Transient Exposure of the C-terminal Domain. J. Biol. Chem. 2015, 290, 28584–28593. [Google Scholar] [CrossRef]
- Kenney, J.M.; von Bonsdorff, C.H.; Nassal, M.; Fuller, S.D. Evolutionary conservation in the hepatitis B virus core structure: Comparison of human and duck cores. Structure 1995, 3, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Dryden, K.A.; Wieland, S.F.; Whitten-Bauer, C.; Gerin, J.L.; Chisari, F.V.; Yeager, M. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol. Cell 2006, 22, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Roseman, A.M.; Berriman, J.A.; Wynne, S.A.; Butler, P.J.; Crowther, R.A. A structural model for maturation of the hepatitis B virus core. Proc. Natl. Acad. Sci. USA 2005, 102, 15821–15826. [Google Scholar] [CrossRef]
- Hu, Z.; Ban, H.; Zheng, H.; Liu, M.; Chang, J.; Guo, J.T. Protein phosphatase 1 catalyzes HBV core protein dephosphorylation and is co-packaged with viral pregenomic RNA into nucleocapsids. PLoS Pathog. 2020, 16, e1008669. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Shih, C. Significance of hepatitis B virus capsid dephosphorylation via polymerase. J. Biomed. Sci. 2024, 31, 34. [Google Scholar] [CrossRef]
- Hofmann, S.; Plank, V.; Groitl, P.; Skvorc, N.; Hofmann, K.; Luther, J.; Ko, C.; Zimmerman, P.; Bruss, V.; Stadler, D.; et al. SUMO Modification of Hepatitis B Virus Core Mediates Nuclear Entry, Promyelocytic Leukemia Nuclear Body Association, and Efficient Formation of Covalently Closed Circular DNA. Microbiol. Spectr. 2023, 11, e0044623. [Google Scholar] [CrossRef]
- Zimmerman, S.B.; Trach, S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991, 222, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, R.; Hoang, C.P.; Verkman, A.S. Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: Cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys. J. 1997, 72, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Potma, E.O.; de Boeij, W.P.; Bosgraaf, L.; Roelofs, J.; van Haastert, P.J.; Wiersma, D.A. Reduced protein diffusion rate by cytoskeleton in vegetative and polarized dictyostelium cells. Biophys. J. 2001, 81, 2010–2019. [Google Scholar] [CrossRef]
- Akhmanova, A.; Steinmetz, M.O. Control of microtubule organization and dynamics: Two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015, 16, 711–726. [Google Scholar] [CrossRef]
- Sallee, M.D.; Feldman, J.L. Microtubule organization across cell types and states. Curr. Biol. 2021, 31, R506–R511. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, T.; Volkman, L.E.; Welch, M.D. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 2010, 190, 187–195. [Google Scholar] [CrossRef]
- Trivedi, D.V.; Nag, S.; Spudich, A.; Ruppel, K.M.; Spudich, J.A. The Myosin Family of Mechanoenzymes: From Mechanisms to Therapeutic Approaches. Annu. Rev. Biochem. 2020, 89, 667–693. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Yang, W.C. The functions of kinesin and kinesin-related proteins in eukaryotes. Cell Adhes. Migr. 2020, 14, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, A.; Zhao, Y. Dyneins. Curr. Biol. 2023, 33, R1274–R1279. [Google Scholar] [CrossRef] [PubMed]
- Apcarian, A.; Cunningham, A.L.; Diefenbach, R.J. Identification of binding domains in the herpes simplex virus type 1 small capsid protein pUL35 (VP26). J. Gen. Virol. 2010, 91, 2659–2663. [Google Scholar] [CrossRef] [PubMed]
- Radtke, K.; Kieneke, D.; Wolfstein, A.; Michael, K.; Steffen, W.; Scholz, T.; Karger, A.; Sodeik, B. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010, 6, e1000991. [Google Scholar] [CrossRef]
- Lupberger, J.; Schaedler, S.; Peiran, A.; Hildt, E. Identification and characterization of a novel bipartite nuclear localization signal in the hepatitis B virus polymerase. World J. Gastroenterol. 2013, 19, 8000–8010. [Google Scholar] [CrossRef]
- Kann, M.; Bischof, A.; Gerlich, W.H. In vitro model for the nuclear transport of the hepadnavirus genome. J. Virol. 1997, 71, 1310–1316. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Hu, J. Formation of hepatitis B virus covalently closed circular DNA: Removal of genome-linked protein. J. Virol. 2007, 81, 6164–6174. [Google Scholar] [CrossRef]
- Guo, H.; Jiang, D.; Zhou, T.; Cuconati, A.; Block, T.M.; Guo, J.T. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: An intermediate of covalently closed circular DNA formation. J. Virol. 2007, 81, 12472–12484. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Zheng, X.; Cong, J.; Liu, Y.; Li, J.; Sun, R.; Tian, Z.G.; Wei, H.M. Cytoplasm-Translocated Ku70/80 Complex Sensing of HBV DNA Induces Hepatitis-Associated Chemokine Secretion. Front. Immunol. 2016, 7, 569. [Google Scholar] [CrossRef]
- Wieland, S.F.; Chisari, F.V. Stealth and cunning: Hepatitis B and hepatitis C viruses. J. Virol. 2005, 79, 9369–9380. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Zhang, T.; Tang, L.; Li, Y. Cytokines and Chemokines in HBV Infection. Front. Mol. Biosci. 2021, 8, 805625. [Google Scholar] [CrossRef]
- Osseman, Q.; Kann, M. Intracytoplasmic Transport of Hepatitis B Virus Capsids. Methods Mol. Biol. 2017, 1540, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Daniels, R.; Rusan, N.M.; Wadsworth, P.; Hebert, D.N. SV40 VP2 and VP3 insertion into ER membranes is controlled by the capsid protein VP1: Implications for DNA translocation out of the ER. Mol. Cell 2006, 24, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Rainey-Barger, E.K.; Magnuson, B.; Tsai, B. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J. Virol. 2007, 81, 12996–13004. [Google Scholar] [CrossRef] [PubMed]
- Gallucci, L.; Kann, M. Nuclear Import of Hepatitis B Virus Capsids and Genome. Viruses 2017, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Cautain, B.; Hill, R.; de Pedro, N.; Link, W. Components and regulation of nuclear transport processes. FEBS J. 2015, 282, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Yamada, K.; Yoneda, Y. Importin α: A key molecule in nuclear transport and non-transport functions. J. Biochem. 2016, 160, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.N.; Jin, H. The RGG motif proteins: Interactions, functions, and regulations. Wiley Interdiscip. Rev. RNA 2023, 14, e1748. [Google Scholar] [CrossRef]
- Jans, D.A.; Ackermann, M.J.; Bischoff, J.R.; Beach, D.H.; Peters, R. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J. Cell Biol. 1991, 115, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Rout, M.P.; Aitchison, J.D.; Magnasco, M.O.; Chait, B.T. Virtual gating and nuclear transport: The hole picture. Trends Cell Biol. 2003, 13, 622–628. [Google Scholar] [CrossRef]
- Rout, M.P.; Aitchison, J.D.; Suprapto, A.; Hjertaas, K.; Zhao, Y.; Chait, B.T. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 2000, 148, 635–651. [Google Scholar] [CrossRef]
- Lim, R.Y.; Huang, N.P.; Koser, J.; Deng, J.; Lau, K.H.; Schwarz-Herion, K.; Fahrenkrog, B.; Aebi, U. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 2006, 103, 9512–9517. [Google Scholar] [CrossRef] [PubMed]
- Stankunas, E.; Kohler, A. Docking a flexible basket onto the core of the nuclear pore complex. Nat. Cell Biol. 2024, 26, 1504–1519. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, M.E.; Plafker, K.; Smith, A.E.; Clurman, B.E.; Macara, I.G. Npap60/Nup50 is a tri-stable switch that stimulates importin-α:β-mediated nuclear protein import. Cell 2002, 110, 349–360. [Google Scholar] [CrossRef]
- Tsujii, A.; Miyamoto, Y.; Moriyama, T.; Tsuchiya, Y.; Obuse, C.; Mizuguchi, K.; Oka, M.; Yoneda, Y. Retinoblastoma-binding Protein 4-regulated Classical Nuclear Transport Is Involved in Cellular Senescence. J. Biol. Chem. 2015, 290, 29375–29388. [Google Scholar] [CrossRef] [PubMed]
- Gorlich, D.; Pante, N.; Kutay, U.; Aebi, U.; Bischoff, F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996, 15, 5584–5594. [Google Scholar] [CrossRef]
- Pante, N.; Kann, M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef]
- Ojala, P.M.; Sodeik, B.; Ebersold, M.W.; Kutay, U.; Helenius, A. Herpes simplex virus type 1 entry into host cells: Reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 2000, 20, 4922–4931. [Google Scholar] [CrossRef]
- Rollenhagen, C.; Muhlhausser, P.; Kutay, U.; Pante, N. Importin β-depending nuclear import pathways: Role of the adapter proteins in the docking and releasing steps. Mol. Biol. Cell 2003, 14, 2104–2115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brandariz-Nunez, A.; Liu, T.; Du, T.; Evilevitch, A. Pressure-driven release of viral genome into a host nucleus is a mechanism leading to herpes infection. eLife 2019, 8, e47212. [Google Scholar] [CrossRef] [PubMed]
- Cassany, A.; Ragues, J.; Guan, T.; Begu, D.; Wodrich, H.; Kann, M.; Nemerow, G.R.; Gerace, L. Nuclear import of adenovirus DNA involves direct interaction of hexon with an N-terminal domain of the nucleoporin Nup214. J. Virol. 2015, 89, 1719–1730. [Google Scholar] [CrossRef] [PubMed]
- San Martin, C. Latest insights on adenovirus structure and assembly. Viruses 2012, 4, 847–877. [Google Scholar] [CrossRef] [PubMed]
- Lagadec, F.; Carlon-Andres, I.; Ragues, J.; Port, S.; Wodrich, H.; Kehlenbach, R.H. CRM1 Promotes Capsid Disassembly and Nuclear Envelope Translocation of Adenovirus Independently of Its Export Function. J. Virol. 2022, 96, e0127321. [Google Scholar] [CrossRef]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715–11720. [Google Scholar] [CrossRef] [PubMed]
- Wiethoff, C.M.; Wodrich, H.; Gerace, L.; Nemerow, G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 2005, 79, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gonzalez, N.; Gomez-Gonzalez, A.; Hernando-Perez, M.; Bauer, M.; Greber, U.F.; San Martin, C.; de Pablo, P.J. Adenovirus core protein V reinforces the capsid and enhances genome release from disrupted particles. Sci. Adv. 2023, 9, eade9910. [Google Scholar] [CrossRef] [PubMed]
- Paci, G.; Zheng, T.; Caria, J.; Zilman, A.; Lemke, E.A. Molecular determinants of large cargo transport into the nucleus. eLife 2020, 9, e55963. [Google Scholar] [CrossRef]
- Liu, P.; Chen, S.; Wang, M.; Cheng, A. The role of nuclear localization signal in parvovirus life cycle. Virol. J. 2017, 14, 80. [Google Scholar] [CrossRef]
- Vihinen-Ranta, M.; Wang, D.; Weichert, W.S.; Parrish, C.R. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 2002, 76, 1884–1891. [Google Scholar] [CrossRef]
- Mantyla, E.; Aho, V.; Kann, M.; Vihinen-Ranta, M. Cytoplasmic Parvovirus Capsids Recruit Importin Beta for Nuclear Delivery. J. Virol. 2020, 94, e01532-19. [Google Scholar] [CrossRef]
- Porwal, M.; Cohen, S.; Snoussi, K.; Popa-Wagner, R.; Anderson, F.; Dugot-Senant, N.; Wodrich, H.; Dinsart, C.; Kleinschmidt, J.A.; Pante, N.; et al. Parvoviruses cause nuclear envelope breakdown by activating key enzymes of mitosis. PLoS Pathog. 2013, 9, e1003671. [Google Scholar] [CrossRef] [PubMed]
- Fay, N.; Pante, N. Nuclear entry of DNA viruses. Front. Microbiol. 2015, 6, 467. [Google Scholar] [CrossRef]
- Au, S.; Cohen, S.; Pante, N. Microinjection of Xenopus laevis oocytes as a system for studying nuclear transport of viruses. Methods 2010, 51, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, S.G.; Milich, D.R.; McLachlan, A. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 1991, 65, 575–582. [Google Scholar] [CrossRef]
- Liu, K.; Ludgate, L.; Yuan, Z.; Hu, J. Regulation of multiple stages of hepadnavirus replication by the carboxyl-terminal domain of viral core protein in trans. J. Virol. 2015, 89, 2918–2930. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.; Schwarz, A.; Foss, M.; Zhou, L.; Rabe, B.; Hoellenriegel, J.; Stoeber, M.; Pante, N.; Kann, M. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog. 2010, 6, e1000741. [Google Scholar] [CrossRef]
- Rabe, B.; Vlachou, A.; Pante, N.; Helenius, A.; Kann, M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc. Natl. Acad. Sci. USA 2003, 100, 9849–9854. [Google Scholar] [CrossRef]
- Hu, J.; Tang, L.; Cheng, J.; Zhou, T.; Li, Y.; Chang, J.; Zhao, Q.; Guo, J.T. Hepatitis B virus nucleocapsid uncoating: Biological consequences and regulation by cellular nucleases. Emerg. Microbes Infect. 2021, 10, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ploss, A. Hepatitis B virus cccDNA is formed through distinct repair processes of each strand. Nat. Commun. 2021, 12, 1591. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Georgescu, R.; Yao, N.Y.; Li, H.; O’Donnell, M.E. Cryo-EM structures reveal that RFC recognizes both the 3′- and 5′-DNA ends to load PCNA onto gaps for DNA repair. eLife 2022, 11, e77469. [Google Scholar] [CrossRef] [PubMed]
- Overmeer, R.M.; Gourdin, A.M.; Giglia-Mari, A.; Kool, H.; Houtsmuller, A.B.; Siegal, G.; Fousteri, M.I.; Mullenders, L.H.; Vermeulen, W. Replication factor C recruits DNA polymerase δ to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 2010, 30, 4828–4839. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, K.; Que, L.; Shimadu, M.; Koura, M.; Ishihara, Y.; Wakae, K.; Nakamura, T.; Watashi, K.; Wakita, T.; Muramatsu, M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018, 14, e1007124. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ploss, A. Core components of DNA lagging strand synthesis machinery are essential for hepatitis B virus cccDNA formation. Nat. Microbiol. 2020, 5, 715–726. [Google Scholar] [CrossRef]
- Luo, J.; Luckenbaugh, L.; Hu, H.; Yan, Z.; Gao, L.; Hu, J. Involvement of Host ATR-CHK1 Pathway in Hepatitis B Virus Covalently Closed Circular DNA Formation. mBio 2020, 11, e03423-19. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.L.; Zhang, H.; Kim, E.S.; Yu, X.; Jang, S.; Wang, M.; Guo, H. Proteomic Analysis of Nuclear Hepatitis B Virus Relaxed Circular DNA-Associated Proteins Identifies UV-Damaged DNA Binding Protein as a Host Factor Involved in Covalently Closed Circular DNA Formation. J. Virol. 2022, 96, e0136021. [Google Scholar] [CrossRef] [PubMed]
- Verrier, E.R.; Ligat, G.; Heydmann, L.; Doernbrack, K.; Miller, J.; Maglott-Roth, A.; Juhling, F.; El Saghire, H.; Heuschkel, M.J.; Fujiwara, N.; et al. Cell-based cccDNA reporter assay combined with functional genomics identifies YBX1 as HBV cccDNA host factor and antiviral candidate target. Gut 2022, 72, 1745–1757. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Xu, C.; Zhou, T.; Block, T.M.; Guo, J.T. Characterization of the host factors required for hepadnavirus covalently closed circular (ccc) DNA formation. PLoS ONE 2012, 7, e43270. [Google Scholar] [CrossRef] [PubMed]



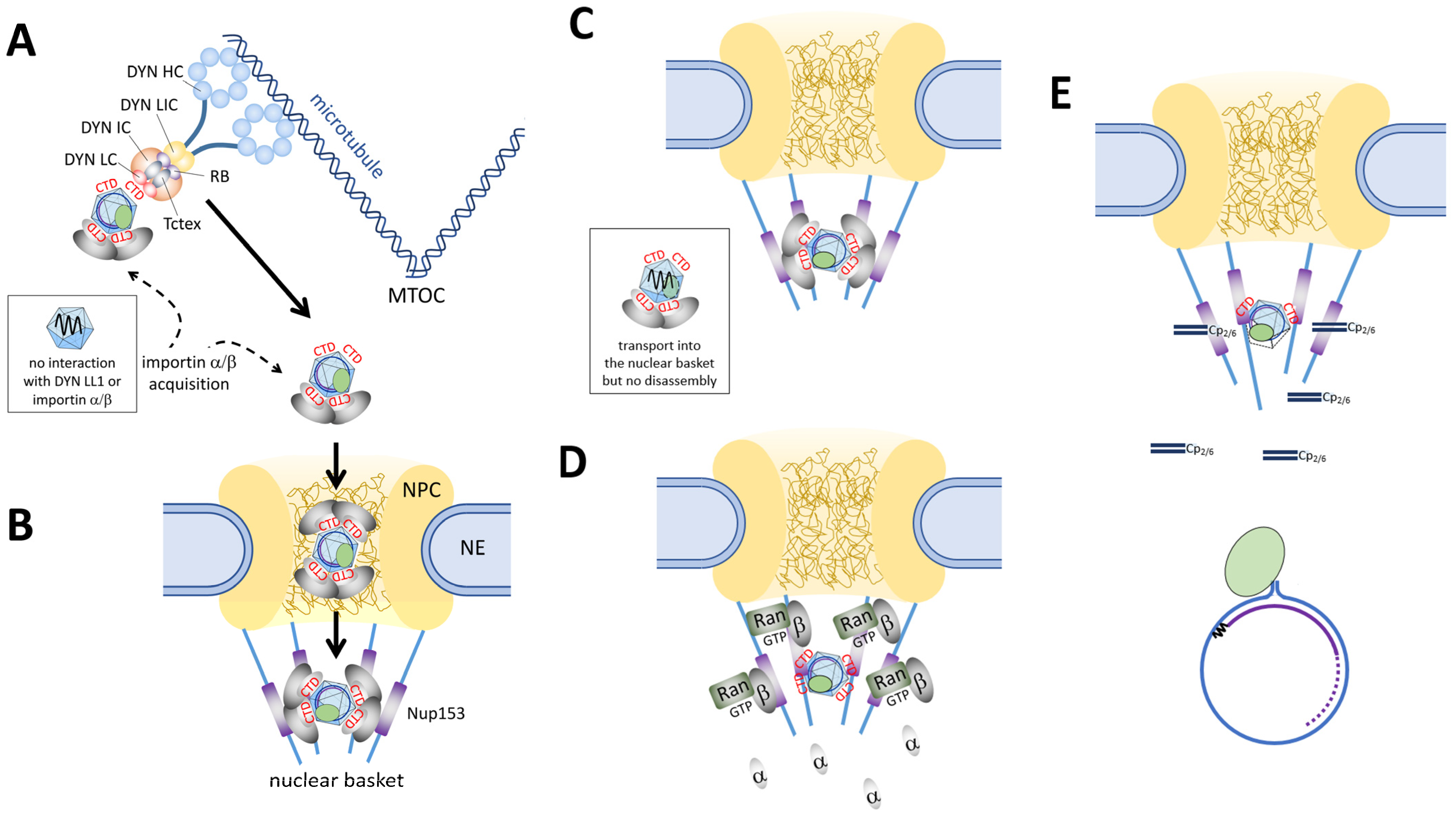
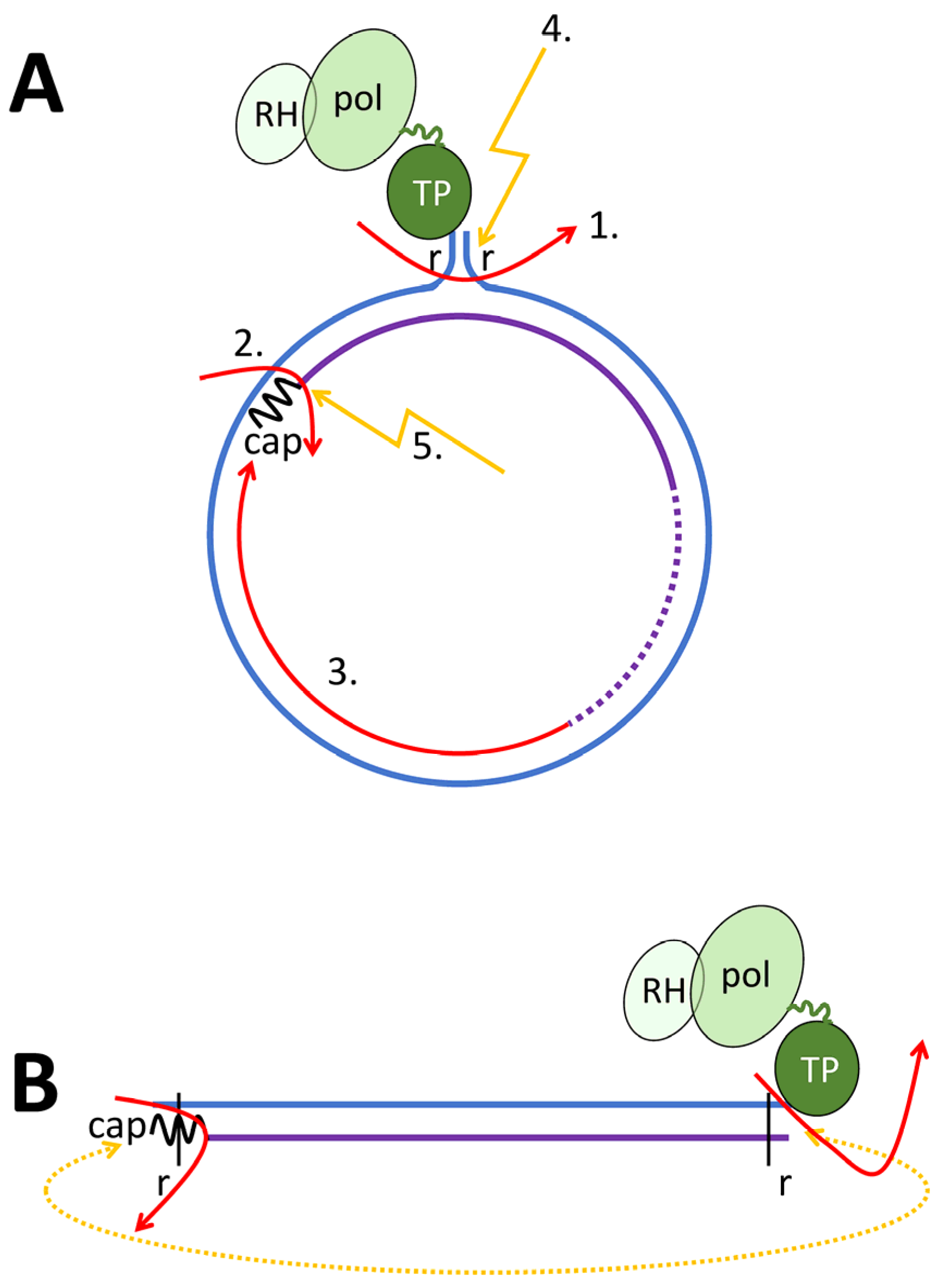
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ringlander, J.; Rydell, G.E.; Kann, M. From the Cytoplasm into the Nucleus—Hepatitis B Virus Travel and Genome Repair. Microorganisms 2025, 13, 157. https://doi.org/10.3390/microorganisms13010157
Ringlander J, Rydell GE, Kann M. From the Cytoplasm into the Nucleus—Hepatitis B Virus Travel and Genome Repair. Microorganisms. 2025; 13(1):157. https://doi.org/10.3390/microorganisms13010157
Chicago/Turabian StyleRinglander, Johan, Gustaf E. Rydell, and Michael Kann. 2025. "From the Cytoplasm into the Nucleus—Hepatitis B Virus Travel and Genome Repair" Microorganisms 13, no. 1: 157. https://doi.org/10.3390/microorganisms13010157
APA StyleRinglander, J., Rydell, G. E., & Kann, M. (2025). From the Cytoplasm into the Nucleus—Hepatitis B Virus Travel and Genome Repair. Microorganisms, 13(1), 157. https://doi.org/10.3390/microorganisms13010157





