Effect of Plateau pika on Soil Microbial Assembly Process and Co-Occurrence Patterns in the Alpine Meadow Ecosystem
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Soil Physicochemical Analysis
2.3. Soil Microbiome Analysis
2.4. Evaluation of Microbe Specialization
2.5. Evaluation of Microbiome Assembly Processes
2.6. Microbial Co-Occurrence Network Construction
2.7. Statistical Analysis
3. Results
3.1. Soil Physiochemistry
3.2. Soil Microbiome Composition and Diversity
3.3. Drivers of Soil Microbiome Diversity
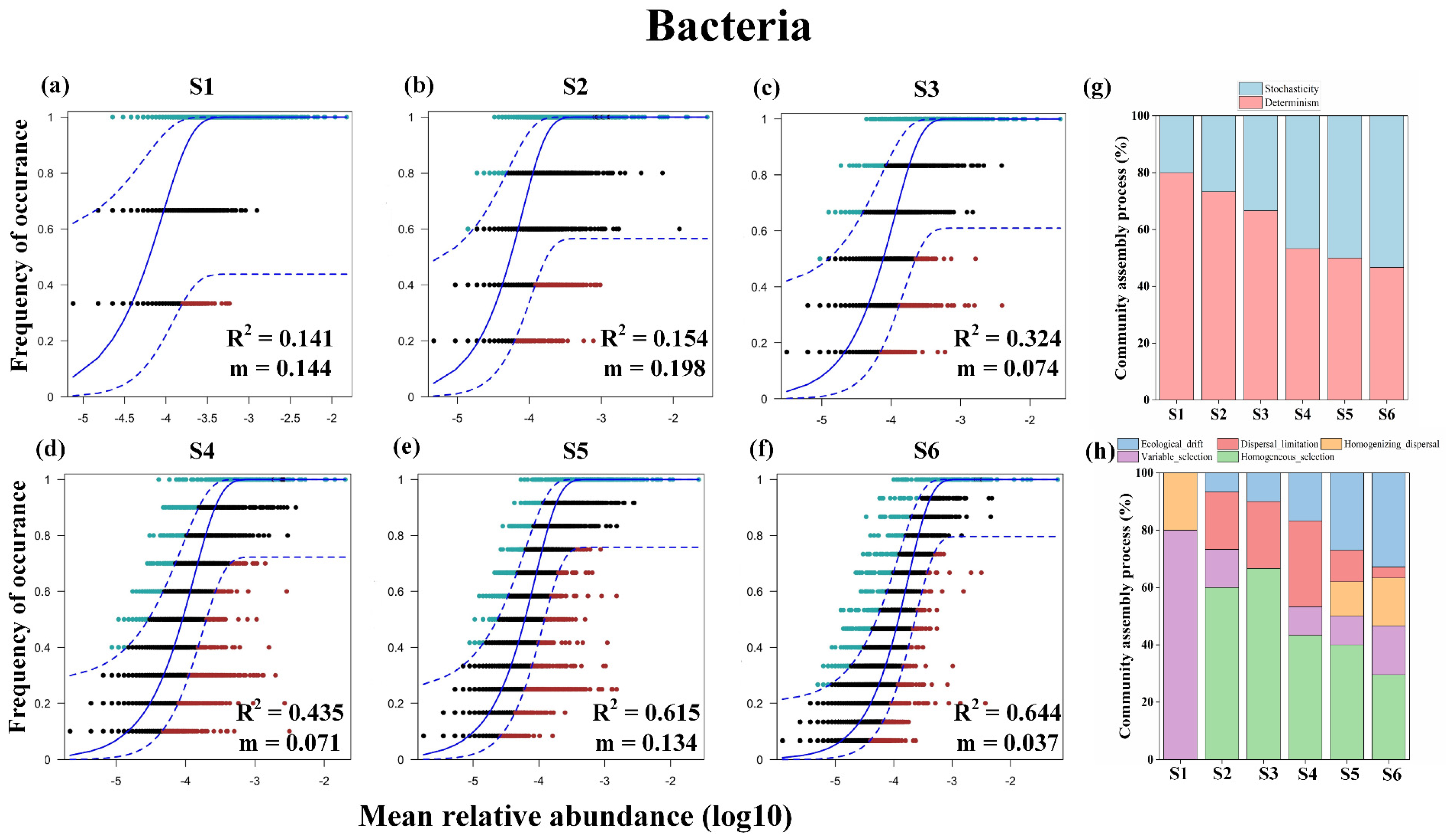
3.4. Microbiome Assembly Processes
3.5. Microbial Co-Occurrence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Brown, L.E.; Hannah, D.M.; Milner, A.M. Vulnerability of alpine stream biodiversity to shrinking glaciers and snowpacks. Glob. Chang. Biol. 2007, 13, 958–966. [Google Scholar] [CrossRef]
- Broadbent, A.A.D.; Snell, H.S.K.; Michas, A.; Pritchard, W.J.; Newbold, L.; Cordero, I.; Goodall, T.; Schallhart, N.; Kaufmann, R.; Griffiths, R.I.; et al. Climate change alters temporal dynamics of alpine soil microbial functioning and biogeochemical cycling via earlier snowmelt. ISME J. 2021, 15, 2264–2275. [Google Scholar] [CrossRef]
- Leifeld, J.; Zimmermann, M.; Fuhrer, J.; Conen, F. Storage and turnover of carbon in grassland soils along an elevation gradient in the Swiss Alps. Glob. Chang. Biol. 2009, 15, 668–679. [Google Scholar] [CrossRef]
- Ims, R.A.; Henden, J.-A.; Strømeng, M.A.; Thingnes, A.V.; Garmo, M.J.; Jepsen, J.U. Arctic greening and bird nest predation risk across tundra ecotones. Nat. Clim. Chang. 2019, 9, 607–610. [Google Scholar] [CrossRef]
- Krug, L.; Erlacher, A.; Markut, K.; Berg, G.; Cernava, T. The microbiome of alpine snow algae shows a specific inter-kingdom connectivity and algae-bacteria interactions with supportive capacities. ISME J. 2020, 14, 2197–2210. [Google Scholar] [CrossRef]
- Chapin, F.S.; Körner, C. Arctic and alpine biodiversity: Patterns, causes and ecosystem consequences. Trends Ecol. Evol. 1994, 9, 45–47. [Google Scholar] [CrossRef]
- Martinsen, V.; Mulder, J.; Austrheim, G.; Hessen, D.O.; Mysterud, A. Effects of Sheep Grazing on Availability and Leaching of Soil Nitrogen in Low-Alpine Grasslands. Arctic Antarct. Alp. Res. 2012, 44, 67–82. [Google Scholar] [CrossRef]
- Su, J.; Ji, W.; Li, H.; Yao, T.; Wang, J.; Nan, Z. Zokor disturbances indicated positive soil microbial responses with carbon cycle and mineral encrustation in alpine grassland. Ecol. Eng. 2020, 144, 105702. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, D.; Meng, B.; Chen, J.; Wang, X.; Jiang, H.; Yu, Y.; Yi, S. Using UAVs to assess the relationship between alpine meadow bare patches and disturbance by pikas in the source region of Yellow River on the Qinghai-Tibetan Plateau. Glob. Ecol. Conserv. 2021, 26, e01517. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, J.; Pang, X.P.; Wang, Q.; Zhou, Y.P.; Guo, Z.G. Soil disturbance and disturbance intensity: Response of soil nutrient concentrations of alpine meadow to Plateau pika bioturbation in the Qinghai-Tibetan Plateau, China. Geoderma 2017, 307, 98–106. [Google Scholar] [CrossRef]
- Rod-Eriksen, L.; Skrutvold, J.; Herfindal, I.; Jensen, H.; Eide, N.E. Highways associated with expansion of boreal scavengers into the alpine tundra of Fennoscandia. J. Appl. Ecol. 2020, 57, 1861–1870. [Google Scholar] [CrossRef]
- Davidson, A.D.; Detling, J.K.; Brown, J.H. Ecological roles and conservation challenges of social, burrowing, herbivorous mammals in the world’s grasslands. Front. Ecol. Environ. 2012, 10, 477–486. [Google Scholar] [CrossRef]
- Che, R.; Wang, Y.; Li, K.; Xu, Z.; Hu, J.; Wang, F.; Rui, Y.; Li, L.; Pang, Z.; Cui, X. Degraded patch formation significantly changed microbial community composition in alpine meadow soils. Soil Tillage Res. 2019, 195, 104426. [Google Scholar] [CrossRef]
- Wang, X.; Bian, Q.; Jiang, Y.; Zhu, L.; Chen, Y.; Liang, Y.; Sun, B. Organic amendments drive shifts in microbial community structure and keystone taxa which increase C mineralization across aggregate size classes. Soil Biol. Biochem. 2021, 153, 108062. [Google Scholar] [CrossRef]
- Eldridge, D.J.; Delgado-Baquerizo, M.; Woodhouse, J.N.; Neilan, B.A. Mammalian engineers drive soil microbial communities and ecosystem functions across a disturbance gradient. J. Anim. Ecol. 2016, 85, 1636–1646. [Google Scholar] [CrossRef]
- Pang, X.P.; Guo, Z.G. Plateau pika disturbances alter plant productivity and soil nutrients in alpine meadows of the Qinghai-Tibetan Plateau, China. Rangel. J. 2017, 39, 133–144. [Google Scholar] [CrossRef]
- Bell, T.; Ager, D.; Song, J.-I.; Newman, J.A.; Thompson, I.P.; Lilley, A.K.; van der Gast, C.J. Larger islands house more bacterial taxa. Science 2005, 308, 1884. [Google Scholar] [CrossRef]
- Li, S.-P.; Wang, P.; Chen, Y.; Wilson, M.C.; Yang, X.; Ma, C.; Lu, J.; Chen, X.-Y.; Wu, J.; Shu, W.-S.; et al. Island biogeography of soil bacteria and fungi: Similar patterns, but different mechanisms. ISME J. 2020, 14, 1886–1896. [Google Scholar] [CrossRef]
- Wu, W.; Lu, H.-P.; Sastri, A.; Yeh, Y.-C.; Gong, G.-C.; Chou, W.-C.; Hsieh, C.-H. Contrasting the relative importance of species sorting and dispersal limitation in shaping marine bacterial versus protist communities. ISME J. 2018, 12, 485–494. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B.H. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Darcy, J.L.; Gendron, E.M.S.; Sommers, P.; Porazinska, D.L.; Schmidt, S.K. Island Biogeography of Cryoconite Hole Bacteria in Antarctica’s Taylor Valley and Around the World. Front. Ecol. Evol. 2018, 6, 413938. [Google Scholar] [CrossRef]
- Van Der Gast, C.J.; Lilley, A.K.; Ager, D.; Thompson, I.P. Island size and bacterial diversity in an archipelago of engineering machines. Environ. Microbiol. 2005, 7, 1220–1226. [Google Scholar] [CrossRef]
- Ge, D.; Lu, L.; Cheng, J.; Xia, L.; Chang, Y.; Wen, Z.; Lv, X.; Du, Y.; Liu, Q.; Yang, Q. An endemic rat species complex is evidence of moderate environmental changes in the terrestrial biodiversity centre of China through the late Quaternary. Sci. Rep. 2017, 7, 46127. [Google Scholar] [CrossRef]
- Pang, X.P.; Yu, C.Q.; Zhang, J.; Wang, Q.; Guo, Z.G.; Tian, Y. Effect of disturbance by Plateau pika on soil nitrogen stocks in alpine meadows. Geoderma 2020, 372, 114392. [Google Scholar] [CrossRef]
- Wilson, M.C.; Smith, A.T. The pika and the watershed: The impact of small mammal poisoning on the ecohydrology of the Qinghai-Tibetan Plateau. Ambio 2015, 44, 16–22. [Google Scholar] [CrossRef]
- Li, W. Is the study subject in ‘Microclimate response of soil to Plateau pika’s disturbance in the northeast Qinghai-Tibet Plateau’ actually plateau zokor? Eur. J. Soil Sci. 2022, 73, e13168. [Google Scholar] [CrossRef]
- Niu, K.; Feng, F.; Xu, Q.; Badingqiuying; Zhang, S. Impoverished soil supports more Plateau pika through lowered diversity of plant functional traits in Tibetan alpine meadows. Agric. Ecosyst. Environ. 2019, 285, 106621. [Google Scholar] [CrossRef]
- Zhao, J.; Tian, L.; Wei, H.; Zhang, T.; Bai, Y.; Li, R.; Tang, Y. Impact of Plateau pika (Ochotona curzoniae) burrowing-induced microtopography on ecosystem respiration of the alpine meadow and steppe on the Tibetan plateau. Plant Soil 2021, 458, 217–230. [Google Scholar] [CrossRef]
- Zheng, Y.; Maitra, P.; Gan, H.Y.; Chen, L.; Li, S.; Tu, T.; Chen, L.; Mi, X.; Gao, C.; Zhang, D.; et al. Soil fungal diversity and community assembly: Affected by island size or type? FEMS Microbiol. Ecol. 2021, 97, fiab062. [Google Scholar] [CrossRef]
- Ye, Z.; Li, J.; Wang, J.; Zhang, C.; Liu, G.; Dong, Q. Diversity and co-occurrence network modularization of bacterial communities determine soil fertility and crop yields in arid fertigation agroecosystems. Biol. Fertil. Soils 2021, 57, 809–824. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Z.-J.; Huang, J.-C.; Sun, S.; Cui, X.; Zhou, W.; He, S. Salinity-driven nitrogen removal and its quantitative molecular mechanisms in artificial tidal wetlands. Water Res. 2021, 202, 117446. [Google Scholar] [CrossRef] [PubMed]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Székely, A.J.; Langenheder, S. The importance of species sorting differs between habitat generalists and specialists in bacterial communities. FEMS Microbiol. Ecol. 2014, 87, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Sloan, W.T.; Lunn, M.; Woodcock, S.; Head, I.M.; Nee, S.; Curtis, T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006, 8, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Yang, Y.; Xu, Y.; Zhang, J.; Lu, Y. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 2020, 14, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef]
- Horner-Devine, M.C.; Lage, M.; Hughes, J.B.; Bohannan, B.J.M. A taxa–area relationship for bacteria. Nature 2004, 432, 750–753. [Google Scholar] [CrossRef]
- Ranjard, L.; Dequiedt, S.; Prévost-Bouré, N.C.; Thioulouse, J.; Saby, N.; Lelievre, M.; Maron, P.A.; Morin, F.; Bispo, A.; Jolivet, C.; et al. Turnover of soil bacterial diversity driven by wide-scale environmental heterogeneity. Nat. Commun. 2013, 4, 1434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, G.; Song, Z.; Wang, J.; Guo, L. Interactions of soil bacteria and fungi with plants during long-term grazing exclusion in semiarid grasslands. Soil Biol. Biochem. 2018, 124, 47–58. [Google Scholar] [CrossRef]
- Wang, J.; Shen, J.; Wu, Y.; Tu, C.; Soininen, J.; Stegen, J.C.; He, J.; Liu, X.; Zhang, L.; Zhang, E. Phylogenetic beta diversity in bacterial assemblages across ecosystems: Deterministic versus stochastic processes. ISME J. 2013, 7, 1310–1321. [Google Scholar] [CrossRef] [PubMed]
- Langenheder, S.; Székely, A.J. Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J. 2011, 5, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.; Martiny, J.B.H.; Allison, S.D. Effects of dispersal and selection on stochastic assembly in microbial communities. ISME J. 2017, 11, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.R.; Stephens, W.Z.; Stagaman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J.M. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016, 10, 655–664. [Google Scholar] [CrossRef]
- Green, J.L.; Holmes, A.J.; Westoby, M.; Oliver, I.; Briscoe, D.; Dangerfield, M.; Gillings, M.; Beattie, A.J. Spatial scaling of microbial eukaryote diversity. Nature 2004, 432, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.K.; Nemergut, D.R.; Darcy, J.L.; Lynch, R. Do bacterial and fungal communities assemble differently during primary succession? Mol. Ecol. 2014, 23, 254–258. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Finlay, B.J. Global dispersal of free-living microbial eukaryote species. Science 2002, 296, 1061–1063. [Google Scholar] [CrossRef]
- Davison, J.; Moora, M.; Öpik, M.; Ainsaar, L.; Ducousso, M.; Hiiesalu, I.; Jairus, T.; Johnson, N.; Jourand, P.; Kalamees, R.; et al. Microbial island biogeography: Isolation shapes the life history characteristics but not diversity of root-symbiotic fungal communities. ISME J. 2018, 12, 2211–2224. [Google Scholar] [CrossRef]
- Egan, C.; Li, D.-W.; Klironomos, J. Detection of arbuscular mycorrhizal fungal spores in the air across different biomes and ecoregions. Fungal Ecol. 2014, 12, 26–31. [Google Scholar] [CrossRef]
- Norros, V.; Penttilä, R.; Suominen, M.; Ovaskainen, O. Dispersal may limit the occurrence of specialist wood decay fungi already at small spatial scales. Oikos 2012, 121, 961–974. [Google Scholar] [CrossRef]
- Zhou, J.; Ning, D. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef]
- I Adams, R.; Miletto, M.; Taylor, J.W.; Bruns, T.D. Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 2013, 7, 1262–1273. [Google Scholar] [CrossRef]
- Xiao, X.; Liang, Y.; Zhou, S.; Zhuang, S.; Sun, B. Fungal community reveals less dispersal limitation and potentially more connected network than that of bacteria in bamboo forest soils. Mol. Ecol. 2018, 27, 550–563. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016, 10, 1891–1901. [Google Scholar] [CrossRef]
- Jiang, Y.; Lei, Y.; Yang, Y.; Korpelainen, H.; Niinemets, Ü.; Li, C. Divergent assemblage patterns and driving forces for bacterial and fungal communities along a glacier forefield chronosequence. Soil Biol. Biochem. 2018, 118, 207–216. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, J.; Liu, Y.; Shi, P.; Wei, G. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem. 2018, 118, 178–186. [Google Scholar] [CrossRef]
- Chen, J.; Nan, J.; Xu, D.; Mo, L.; Zheng, Y.; Chao, L.; Qu, H.; Guo, Y.; Li, F.; Bao, Y. Response differences between soil fungal and bacterial communities under opencast coal mining disturbance conditions. Catena 2020, 194, 104779. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Zheng, Q.; Deng, Y.; Van Nostrand, J.D.; Zhou, J.; Jiao, N. Bacterioplankton community resilience to ocean acidification: Evidence from microbial network analysis. ICES J. Mar. Sci. 2016, 73, 865–875. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Ma, B.; Dai, Z.; Wang, H.; Dsouza, M.; Liu, X.; He, Y.; Wu, J.; Rodrigues, J.L.M.; Gilbert, J.A.; Brookes, P.C.; et al. Distinct Biogeographic Patterns for Archaea, Bacteria, and Fungi along the Vegetation Gradient at the Continental Scale in Eastern China. Msystems 2017, 2, e00174-16. [Google Scholar] [CrossRef]
- Feng, M.; Adams, J.M.; Fan, K.; Shi, Y.; Sun, R.; Wang, D.; Guo, X.; Chu, H. Long-term fertilization influences community assembly processes of soil diazotrophs. Soil Biol. Biochem. 2018, 126, 151–158. [Google Scholar] [CrossRef]
- Morriën, E.; Hannula, S.E.; Snoek, L.B.; Helmsing, N.R.; Zweers, H.; de Hollander, M.; Soto, R.L.; Bouffaud, M.-L.; Buée, M.; Dimmers, W.; et al. Soil networks become more connected and take up more carbon as nature restoration progresses. Nat. Commun. 2017, 8, 14349. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher-Jenull, J.; Ceccherini, M.T.; Pietramellara, G.; Renella, G.; Schloter, M. Beyond microbial diversity for predicting soil functions: A mini review. Pedosphere 2020, 30, 5–17. [Google Scholar] [CrossRef]
- Liao, J.; Cao, X.; Zhao, L.; Wang, J.; Gao, Z.; Wang, M.C.; Huang, Y. The importance of neutral and niche processes for bacterial community assembly differs between habitat generalists and specialists. FEMS Microbiol. Ecol. 2016, 92, fiw174. [Google Scholar] [CrossRef]
- Pandit, S.N.; Kolasa, J.; Cottenie, K. Contrasts between habitat generalists and specialists: An empirical extension to the basic metacommunity framework. Ecology 2009, 90, 2253–2262. [Google Scholar] [CrossRef]
- Sriswasdi, S.; Yang, C.-C.; Iwasaki, W. Generalist species drive microbial dispersion and evolution. Nat. Commun. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Wimp, G.M.; Ries, L.; Lewis, D.; Murphy, S.M. Habitat edge responses of generalist predators are predicted by prey and structural resources. Ecology 2019, 100, e02662. [Google Scholar] [CrossRef]




| TN g/kg | TP g/kg | SOC g/kg | AK mg/kg | AP mg/kg | NO3− mg/kg | NH4+ mg/kg | pH | Moisture% | Heterogeneity | |
|---|---|---|---|---|---|---|---|---|---|---|
| S1 | 3.32 ± 0.09 a | 0.41 ± 0.01 ab | 88.7 ± 3.1 a | 70.5 ± 6.9 bc | 1.25 ± 0.18 ab | 9.9 ± 0.8 b | 72.2 ± 6.4 ab | 5.79 ± 0.06 a | 37.5 ± 2.2 a | 25.0 ± 2.4 |
| S2 | 3.06 ± 0.12 a | 0.42 ± 0.01 ab | 81.7 ± 2.8 ab | 88.6 ± 4.7 a | 1.09 ± 0.09 ab | 19.7 ± 8.9 a | 66.3 ± 6.3 ab | 5.76 ± 0.04 ab | 33.3 ± 1.3 ab | 36.6 ± 4.3 |
| S3 | 2.94 ± 0.15 a | 0.40 ± 0.01 b | 76.4 ± 4.1 b | 74.4 ± 5.1 b | 0.97 ± 0.14 bc | 7.8 ± 1.7 b | 64.4 ± 4.4 ab | 5.78 ± 0.04 a | 32.4 ± 1.0 b | 26.8 ± 2.3 |
| S4 | 2.40 ± 0.17 b | 0.37 ± 0.00 c | 65.2 ± 4.5 c | 56.8 ± 3.1 cd | 0.31 ± 0.04 d | 4.6 ± 0.5 b | 59.4 ± 4.1 b | 5.63 ± 0.05 b | 27.5 ± 1.1 d | 28.3 ± 2.0 |
| S5 | 2.99 ± 0.07 a | 0.35 ± 0.01 d | 77.1 ± 1.2 b | 76.5 ± 4.0 ab | 0.40 ± 0.06 d | 6.6 ± 0.6 b | 75.0 ± 3.5 a | 5.51 ± 0.04 c | 30.5 ± 0.7 bc | 24.0 ± 1.6 |
| S6 | 2.54 ± 0.05 b | 0.43 ± 0.00 a | 65.9 ± 1.1 c | 54.5 ± 1.6 d | 0.77 ± 0.06 c | 6.5 ± 0.7 b | 43.0 ± 2.3 c | 5.81 ± 0.03 a | 29.2 ± 0.8 cd | 16.5 ± 0.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Ye, Z.; Zhang, C.; Wei, X. Effect of Plateau pika on Soil Microbial Assembly Process and Co-Occurrence Patterns in the Alpine Meadow Ecosystem. Microorganisms 2024, 12, 1075. https://doi.org/10.3390/microorganisms12061075
Wang X, Ye Z, Zhang C, Wei X. Effect of Plateau pika on Soil Microbial Assembly Process and Co-Occurrence Patterns in the Alpine Meadow Ecosystem. Microorganisms. 2024; 12(6):1075. https://doi.org/10.3390/microorganisms12061075
Chicago/Turabian StyleWang, Xiangtao, Zhencheng Ye, Chao Zhang, and Xuehong Wei. 2024. "Effect of Plateau pika on Soil Microbial Assembly Process and Co-Occurrence Patterns in the Alpine Meadow Ecosystem" Microorganisms 12, no. 6: 1075. https://doi.org/10.3390/microorganisms12061075
APA StyleWang, X., Ye, Z., Zhang, C., & Wei, X. (2024). Effect of Plateau pika on Soil Microbial Assembly Process and Co-Occurrence Patterns in the Alpine Meadow Ecosystem. Microorganisms, 12(6), 1075. https://doi.org/10.3390/microorganisms12061075





