Abstract
CRESS-DNA encompasses a broad spectrum of viruses documented across diverse organisms such as animals, plants, diatoms, fungi, and marine invertebrates. Despite this prevalence, the full extent of these viruses’ impact on the environment and their respective hosts remains incompletely understood. Furthermore, an increasing number of viruses within this category lack detailed characterization. This investigation focuses on unveiling and characterizing viruses affiliated with the Genomoviridae family identified in liver samples from the bat Molossus molossus. Leveraging viral metagenomics, we identified seven sequences (MmGmV-PA) featuring a circular DNA genome housing two ORFs encoding replication-associated protein (Rep) and capsid protein (Cap). Predictions based on conserved domains typical of the Genomoviridae family were established. Phylogenetic analysis revealed the segregation of these sequences into two clades aligning with the genera Gemycirculavirus (MmGmV-06-PA and MmGmV-07-PA) and Gemykibivirus (MmGmV-01-PA, MmGmV-02-PA, MmGmV-03-PA, MmGmV-05-PA, and MmGmV-09-PA). At the species level, pairwise comparisons based on complete nucleotide sequences indicated the potential existence of three novel species. In summary, our study significantly contributes to an enhanced understanding of the diversity of Genomoviridae within bat samples, shedding light on previously undiscovered viral entities and their potential ecological implications.
1. Introduction
Bats are large natural reservoirs of zoonotic viruses that can cause various viral diseases in mammals as well as in humans [1,2]. Over 20 years, several zoonotic outbreaks with major impacts emerged that led to the deaths of thousands of humans associated with the virome in bats, such as SARS-CoV-2 [3], Nipah [4], Hendra [5], and Herpesvirus [6]. A wide variety of new virus species in bats have been identified and characterized using metagenomics. This technology has helped in several areas, such as precision medicine to diagnose infectious diseases and maintain public health alerts [7,8].
The swift progress of next-generation sequencing (NGS) technology coupled with metagenomics has proven adept at unveiling a spectrum of novel circular ssDNA viruses encoding the Rep protein (Circular Rep-encoding Single-Stranded DNA) or RNA viruses [9]. These advancements extend across various specimen categories, encompassing fecal samples [10], heterogeneous mixtures [11], human subjects [12,13,14], bat populations [15], turtles [16], avian species [17], insects [18,19], plants [20], parasites [21,22], and environmental matrices [23]. This technological synergy facilitates the isolation and comprehensive characterization of viral diversity within diverse ecosystems.
In this framework, the International Committee on Taxonomy of Viruses (ICTV), (accessible at https://ictv.global/taxonomy, accessed on 5 June 2023) assumes a pivotal role. It not only acknowledges but also establishes the standardization of viral species’ nomenclature, employing the binomial system (genus and epithet) for all viruses [24]. Classifying a species within the Genomaviridae family into one of the existing 10 genera mandates a phylogenetic analysis grounded in the Rep protein. To accomplish this, the ICTV imposes a criterion wherein a <78% identity across the entire genome denotes a novel species, and similar thresholds apply to Rep for genre assignment [25]. This systematic approach ensures precision in delineating viral taxonomy, offering a comprehensive framework for understanding and categorizing the evolving landscape of Genomaviridae.
CRESS-DNA viruses infect eukaryotes and are classified in the phylum Cressdnaviricota, in two classes: Arfiviricetese and Repensiviricetes. The Arfiviricetese class contains 12 families (Amesuviridae, Bacilladnaviridae, Circoviridae, Metaxyviridae, Nanoviridae, Naryaviridae, Nenyaviridae, Redondoviridae, Smacoviridae, Vilyaviridae). Repensiviricetes contains only two families: Geminiviridae and Genomoviridae [26,27].
The Genomoviridae family is composed of 10 genera and 237 species distributed among Gemycircularvirus (126 species), Gemyduguivirus (12 species), Gemygorvirus (eight species), Gemykibivirus (50 species), Gemykolovrus (16 species), Gemykrogvirus (13 species), Gemykroznavirus (seven species), Gemytondvirus (one species), Gemytripvirus (one species), and Gemyvongvirus (three species) [25,28]. In general, genomes are small and circular (~1.8–2.4 kb), monopartite, single-stranded ssDNA, and non-enveloped. They contain two open reading frames (ORFs): one encoding the replication-associated protein (Rep) on the complementary strand, and the other encoding the capsid protein (Cap) on the viral strand. Rep is a multifunctional protein containing an endonuclease and a helicase domain. The N-terminal endonuclease domain has three stretches of highly conserved sequences identified as motifs [29]: Motif I (uuTYxQ), Motif II (xHxHx) [30,31], and Motif III (YxxK) [31,32], with the letter (u) indicating bulky hydrophobic residues such as I, L, V, M, F, Y, and W, and the letter (x) representing any residue [33]. The GRS motif is located between motifs II and III [31]. Rep’s helicase activity is measured by conserved motifs such as Walker A (GxxxxGKT), Walker B (uuDDu), and the C-terminal motif (UxxN) [33,34,35].
Initially, the Genomaviridae family housed a solitary genus, Gemycircularvirus, and a lone species, Sclerotinia gemycirculavirus1 (SsHADV-1) [36]. Members of this viral family have been extracted from a wide array of hosts, spanning fungi, animals, plants, diatoms, and insects, with recent characterizations extending to various species of marine invertebrates, particularly crustaceans. Viral metagenomic studies have indicated that CRESS-DNA viruses, such as Geomovirus, are associated with different hosts, such as protozoa of the genera Giardia [21] and Entamoeba [22].
Despite these discoveries, a substantial number of CRESS-DNA viruses remain unclassified, even in protozoa Giardia, belonging to one family (Vilyaviridae) and Entamoeba in two virus families (Naryaviridae and Nenyaviridae [21,22]. It is noteworthy that CRESS viruses do not classify; 71% resemble chimeric Reps (replication-associated proteins encoded by single-stranded DNA viruses (CRESS), which encode proteins associated with circular replication (Rep), since there is the constant exchange of gene fragments that encode the nuclease and helicase domains. Thus, unclassified CRESS viruses present intense chimerism in their Reps [27] and their impact on the environment and their respective hosts remains poorly understood [26].
The identification of novel single-stranded DNA (ssDNA) viruses akin to SsHADV-1, observed across diverse environmental and animal samples, has brought to light a previously unexplored realm of viral biodiversity [13,28,37]. The Gemycircularvirus genus boasts the highest count of viral species within the Genomaviridae family, closely followed by the Gemykibivirus [28]. In this study, we present the comprehensive characterization of seven complete sequences belonging to the Genomoviridae family, stratified into two genera (Gemykibivirus and Gemycircularvirus), along with the identification of three novel species. These findings were derived from bats of the Molossus molossus species in the Northern region of Brazil.
2. Materials and Methods
2.1. Sample Collection
In Caranazal, situated at latitude 2°26′10′′ S and longitude 54°43′49′′ W in the Santarém region of Pará state within the Lower Amazon Mesoregion, we captured 47 bats. These bats were identified as Molossus molossus from the Molossidae family based on external characteristics [28]. The process involved euthanizing individual bats for sample collection, employing xylazine hydrochloride (1 mg/kg) and ketamine hydrochloride (1–2 mg/kg) via intramuscular injection for anesthesia, followed by intracardiac phenobarbital (40 mg/kg) administration once unconsciousness was achieved.
Approval for this research was obtained from the Animal Use Ethics Committee of the Federal University of Western Pará (CEUA/UFOPA) under number 0220220128, and capturing Chiroptera was authorized by the Biodiversity Information and Authorization System. The necropsy procedures were conducted at the Animal Morphology Laboratory of the Federal University of Western Pará, adhering to institutional biosafety norms.
2.2. Processing of Samples
Due to a shortage of reagents, we used pieces of all 47 samples in a single pool. The processing of samples involved a systematic approach to isolate viral particles from liver tissue. Genomoviruses have tropism for liver cells, meaning they are found in greater quantities in liver tissues than in other tissues. When we use tissue in which the virus has lower tropism, we will find a lower concentration of viruses. Therefore, in this tissue, the amount of other microorganisms can interfere with data analysis. The tissue extraction process began with specimen maceration, followed by dilution in 500 µL of Hanks Buffered Saline Solution (HBSS). Subsequent homogenization using a vortex mixer occurred in 2 mL tubes containing lysis matrix C (MP Biomedicals, Santa Ana, CA, USA). After filtration through 0.45 µM filters (Merck Millipore, Billerica, MA, USA) to eliminate eukaryotic and bacterial cell particles, clarified filtrates underwent centrifugation at 32,000 rpm for 1 h. This facilitated the sedimentation of viral particles, which were then carefully resuspended in 250 µL of PBS for nuclease enzyme treatment.
To enhance the purity of the viral isolates, the filtrates underwent treatment with DNase and RNase A at 37 °C for 30 min, followed by Phi29 (Φ29) polymerase enzyme treatment for DNA circular amplification. The entire extraction process, characterized by precise steps and strategic enzymatic treatments, ensured the isolation of high-quality viral particles from liver tissue.
2.3. Nucleic Acid Extraction (DNA/RNA)
For sample preparation, we utilized the QIAamp Viral RNA Mini Kit (GmbH, QIAGEN Strasse 1, 40724 Hilden, Germany) according to the instructions of the manufacturer.
2.4. Preparation of Libraries for the Illumina Platform
Library preparation for the Illumina platform was performed using the Nextera XT DNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA), and sequencing was carried out on the Illumina NovaSeq-6000 platform, producing 250 bp paired reads.
2.5. Bioinformatics Analysis
For data analysis, raw reads underwent pre-processing to remove terminal-matched sequence records, exclude low-quality sequences, and eliminate adapter and primer sequences. The subsequent bioinformatic analysis confirmed the specificity of the analysis, revealing no reads associated with human, plant, fungal, or bacterial sequences. Contigs were compared against the GenBank genetic sequence database using BLASTx and BLASTn.
The predicted gene sequences obtained from BLAST searches were meticulously chosen based on the most favorable outcomes. To confirm and further classify these sequences, reads and/or contigs were aligned against a viral protein database using DIAMOND software version 2.1.8 (double index alignment of next-generation sequencing data). DIAMOND compares translated DNA sequences in pairs of proteins and offers various output formats, including tabular, paired BLAST, and XML. This process aids in refining the taxonomic classification of viruses. It is worth noting that DIAMOND is an open-source algorithm, available as both a desktop application and a command-line tool (CLI). Additionally, it is accessible online, providing highly accurate and rapid results comparable to the sensitivity of the gold standard BLAST tool, with the capability of executing multiple tasks simultaneously at speeds of up to 360 times faster [38]. All sequences generated in this study were deposited in GenBank with the accession numbers: MmGmV_06 PP249602; MmGmV_01 PP249603; MmGmV_03 PP249604; MmGmV_02 PP249605; MmGmV_07 PP249606; MmGmV_09 PP249607; and MmGmV_05 PP249608.
2.6. Genome Annotation
Comparison of DNA virus sequences in studies were obtained from liver samples (pool-B) of M. molossus species and were submitted to an online database DIAMOND and also on the desktop to align the sequences in different taxonomic levels and then obtain the hierarchical structure of the viruses [38].
Furthermore, the comparative analysis of the sequences of the predicted genes was carried out via the online BLASTx program, as it is a protein aligner using a translated DNA sequence. Based on the best results, the sequences were selected to be subsequently aligned [39,40]. All sequences in the analysis were compared with the Genomoviridae family from GenBank and subsequently, complete or almost complete genome sequences were selected and aligned using the MAFFT software, v7 [41]. Conserved RCR motifs were predicted using the online software InterProScan (https://www.ebi.ac.uk/interpro/search/sequence/, accessed on 29 June 2023) and and Motif Finder (https://www.genome.jp/tools/motif/, accessed on 25 June 2023), respectively.
2.7. Genetic Distance
Genetic distance and its standard error were calculated using the maximum likelihood plus gamma correction and bootstrap model with 1000 replications. MEGA software (Version X) was implemented to estimate distance and genetic diversity [42], as well as SDT v1.2 [43] to calculate identity through a combination of pairwise calculations and perform identity score distribution on pairs of sequences. The initial realignment of sequences involved penalizing gaps—a process carried out with the MUSCLE algorithm [44]. Following the calculation of identity scores for each pair of sequences (pairwise scores), the NEIGHBOR component of PHYLIP was employed to construct a tree. This rooted neighbor-joining phylogenetic tree organizes all sequences based on their likely degrees of evolutionary relatedness.
2.8. Phylogenetic Analysis
Phylogenetic trees were constructed using the maximum likelihood approach, using the best evolutionary model (LG + F + R6) inferred with Iq-TREE. Branch support was estimated using a bootstrap test with 1000 replications [45]. The trees were visualized and edited using the Figtree 1.4.2 program (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results
3.1. Identification of Genomoviridae
We identified seven sequences of genomiviruses (MmGmV-01-PA, MmGmV-02-PA, MmGmV-03-PA, MmGmV-05-PA, MmGmV-06-PA, MmGmV-07-PA, and MmGmV-09-PA) in the pool of liver samples from an M. molossus bat. The results of this search are summarized in Table 1. In general, the genomes showed the greatest identity with the CRESS-DNA viral genomes of the Genomaviridae family, with BLASTn identity ranging from 74.03 to 98.71%, and BLASTp, from 56.90 to 98.00%.

Table 1.
Genome identity of genomoviruses identified in liver tissue of the M. molossus.
3.2. Genome Characterization
All sequences exhibit two putative open reading frames (ORFs), responsible for encoding the replication-associated protein (Rep) with a size range of 194 to 266 amino acids, and the capsid protein (Cap) spanning 130 to 329 amino acids, as detailed in Table 2. It is important to highlight that, between the 5’ ends of the two ORFs, there is an intergenic region, called large, and in some viruses, between the 3’ ends there is a second intergenic region, called small. The presence of one or two intergenic regions is used to distinguish the type (I and II) Genomovirus genomes. The type I genome contains two intergenic regions, and the type II genome has only one intergenic region (Figure 1). When there is no presence of the small intergenic region, there is a juxtaposition of the ORFs at the 3’ terminals (type II genome), in which there is an intron in the Rep coding region. Possibly the introns within the Rep ORF undergo a splicing process constituting the functional Rep protein, as an example of the genomes of the viral sequences of MmGmV_01-PA, MmGmV-06-PA, MmGmV-07-PA (Figure 1), like Genomovirus and Geminivirus [16,46,47]. The MmGmV-07-PA genome is larger than all viral sequences and contains the essential ORFs, in addition to a putative V2 protein like the Hypericum japonicum-associated circular DNA virus (HJasCV) [20].

Table 2.
Identity score of Rep and Cap of MmGmV.
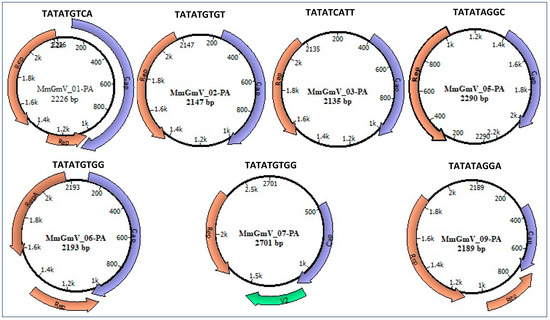
Figure 1.
Genome map of the MmGmV-PA. Circular diagrams depicts the genomes of genomoviruses discovered in Molossus bats. Distinctively colored regions highlight the identified ORFs in MGmV-PA. The nucleotide sequence positioned above each circle signifies the origin of replication for each virus. Within each circle, the virus name and its genome size in base pairs are presented.
We also predicted the origin of replication for the seven genomes. The conserved mononucleotide sequence ‘TATATGTGG’ was identified in the MmGmV-06 and MmGmV-07 viral genomes. Conversely, the mononucleotide sequence ‘TATAT’ exhibits conservation only in positions 1 to 5, displaying variability at its ends. This pattern is consistent among the viruses MmGmV-01-PA, MmGmV-02-PA, MmGmV-03-PA, MmGmV-05-PA, and MmGmV-09-PA. Importantly, these distinctive characteristics align with common features observed in other genera within the Genomoviridae family, as depicted in Figure 1.
The Rep protein of MmGmV-01 exhibits 60.11% identity with the Rep protein of a virus isolated from US wastewater metagenomic samples (QJB18714.1), while the Cap protein shows 43.61% identity with the Red panda feces-associated gemycircularvirus (UBJ26138.1). MmGmV-02-PA and MmGmV-03-PA share 71.11% and 99.48% identity, respectively, with the Rep protein of the Ouratea duparquetiana associated gemykibivirus (QNI80852.1) and canine feces-associated gemycircularvirus (YP_010784661.1). In contrast, the Cap protein exhibits 46.72% and 87.75% identity with metagenomic wastewater (QJB18679.1) and Red panda feces-associated gemycircularvirus (UBJ26188.1). The Rep proteins of MmGmV-05-PA and MmGmV-07-PA share 87.31% and 88.61% identity with the Rep proteins of viruses (QTE03605.1 and QTZ83241.1) isolated Emberiza spodocephala Genomoviridae sp. Additionally, the Cap protein of MmGmV-05 shares 59.63% identity with the putative coat protein of Dragonfly-associated circular virus 3 (YP_009021851.1), while MmGmV-07-PA shares 82.90% identity with the Cap protein Genomoviridae sp. (QXN75602.1).
The Rep protein of MmGmV06-PA is highly similar, with 98% identity, to the RepA of Pteropus-associated Gemycircularvirus5 isolated from fecal samples of Pteropus tonganus bats. Meanwhile, the Cap protein shares 68.25% similarity with the metagenomic animal isolated from haddock tissue (YP_010798116.1). MmGmV09-PA shares 88.50% identity with the Rep protein of the Emberiza spodocephala isolate, and its Cap protein shares 58.97% identity with Genomoviridae sp. (QXN75548.1).
3.3. Analysis of Conserved Motifs
We highlight that the viral genomes of MmGmV-PA belong to circular ssDNA viruses encoding the Rep protein (CRESS-DNA). The Rep protein features crucial motifs for rolling circle replication (RCR), a DNA replication process observed in various single-stranded DNA (ssDNA) viruses [48]. The replication initiator protein (Rep) of geminiviruses is a replicon-specific initiator enzyme and is an essential component of the replisome that carries out viral genome replication its completeness [49].
To gain deeper insights into the sequences, we analyzed conserved motifs within the Rep protein (Figure 2). In the Rep of MmGmV-05-PA, three conserved domains were identified: Geminivirus Rep catalytic domain (Gemini_AL1), putative viral replication protein (Viral_Rep), and Geminivirus Rep protein central domain (Gemini_AL1_M). The Rep of MmGmV-01-PA features Gemini_AL1 and Viral_Rep, while the Rep of MmGmV-02-PA, MmGmV-03-PA, MmGmV-06-PA, and MmGmV-09-PA contains Gemini_AL1 and Gemini_AL1_M. In contrast, the Rep of MmGmV-07-PA exclusively possesses Gemini_AL1. To delve further into the distinctions among the conserved sequences of the Gemini_AL1 geminivirus Rep protein, we scrutinized the motifs present in it. Across all seven MmGmV-PA sequences, the Gemini_AL1 geminivirus Rep catalytic domain, responsible for coding, was consistently identified. The Rep featuring Gemini_AL1 exhibits four conserved regions, denoted as motifs: I, II, GRS domain, and motif III.
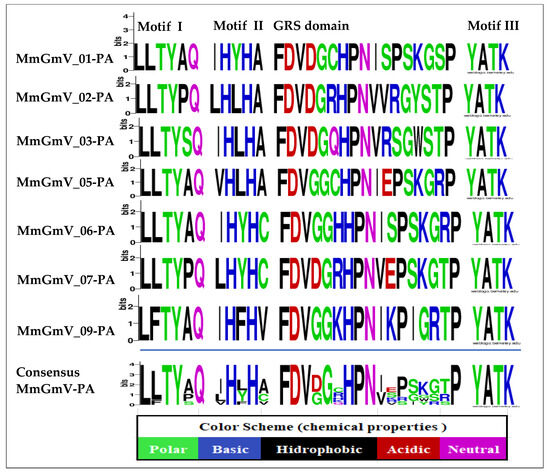
Figure 2.
Amino acid signatures of Rep of MmGmV-PA. Colors represent chemical properties of amino acids. The size of each letter in the consensus sequence represents the frequency of amino acids in the sequences of MmGmV-PA.
In the RCR motif I (six amino acids)—(LLTYxQ), present in the MmGmV Rep, the presence of residue substitutions (“x” for A) in MmGmV (-01, -05, -06 and -09) is identified—(“x” for P) in MmGmV (02 and 07) and MmGmV-03 (“x” for S) in position (5), but we also observed a bulky hydrophobic residue “F” in the sequence of GmV-09 (LFTYAQ), located in second position. RCR motif I is possibly involved in the recognition of iteron sequences associated with the origin of replication and its sequence, and is represented by (uuTYxQ). The letter (u) in the previous representation indicates bulky hydrophobic residues, such as I, L, V, M, F, Y, and W; and the letter (x), any residue [28,30].
Furthermore, the RCR II motif (five amino acids), of MmGmV, which consists of the consensus sequence (xHxHx), presents significant variations within itself and in several species in positions (1, 3 and 5). Histidine is involved in the initial function of coordinating divalent metal ions, Mg2+ or Mn2+, which are important cofactors for endonuclease activity at the origin of replication [28,30].
We observed the RCR motif III (four amino acids—YATK) conserved for all MmGmV sequences, with a lysine residue that is proposed to measure binding and positioning during catalysis. It is worth noting that motif III (YxxK) has only one catalytic tyrosine and may be involved in the cleavage of dsDNA and the covalent attachment of Rep to the catalytic tyrosine residue at the end of the cleaved product [28,30,31,50].
Rep analysis of MmGmV revealed the GRS IV motif (17 amino acids). GRS IV motifs play a role uniquely in geminiviruses (the conserved sequence is found at the N-terminus of the Rep protein) and in genomoviruses (highlights the importance for prevalence and diversity of genomoviruses in nature) [28,31,47]. The Rep contains the motifs RCR I, II, GRS, and III, which are present in several species of the Genomoviridae family, for example three isolates identified in blood samples from patients and blood taken from healthy cattle (HCBI8.215, MSSI2. 225 and HCBI9.212) [13]—the viruses Hypericum japonicum-associated circular DNA virus (HJasCV) [20], the Bemisia-associated genomovirus AdO, [51], the Tadarida brasiliensis gemykibivirus 1 (TbGkyV1), [15], the Drogonfly virus (DfasCV-1, -2, -3) [18], and M. molossus associated Gemykibivirus 1–6 (MAVGs12, 16, 17, 18, 21, 22 and 24) [10].
3.4. Genetic Distances and Phylogenetic Inferences
To infer the phylogenetic tree, we used the complete genome and the Rep protein detected in the MmGmV sequences with other references from the Genomaviridae family (Gemycircularvirus, Gemyduguivirus, Gemygorvirus, Gemykibivirus, Gemykolovrus, Gemykrogvirus, Gemykroznavirus, Gemytondvirus, Gemytripvirus, and Gemyvongvirus) (Figure 3). The tree indicated that the sequences grouped into two clades corresponding to the genera Gemykibivirus (GmV-01, GmV-02, GmV-03, GmV-05 and GmV-09) and Gemycirculavirus (GmV-06 and GmV-07). MmGmV-01 clusters with plant Gemykibivirus species 2 (MK947376/QFR58261), sharing 60.89% pairwise identity based on nucleotide sequence with each other. MmGmV_02 showed the greatest relationship with Gemykibivirus hydro1 (MK483076/QCS35889) and Gemykibivirus draga1 (NC_023872/YP_009021862), sharing 71.41% and 67.91% identity, respectively.
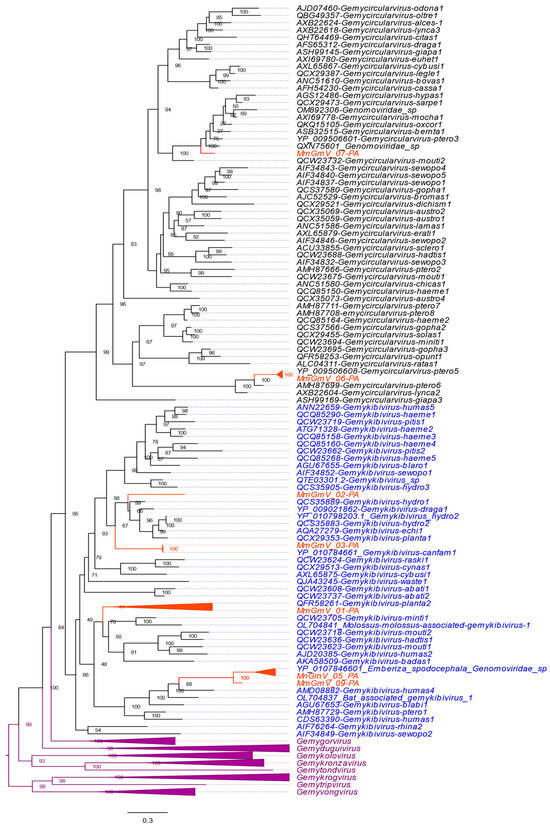
Figure 3.
The phylogenetic tree displays MmGmV-PA Rep protein sequences. Species names in black letters are within the genus Gemycircularvirus, while the genus Gemykibivirus is highlighted in blue letters. The orange triangles represent the grouping of MmGmV-PA viral sequences The remaining families within the Genomoviridae family are represented by a collapsed lilac triangle. The numbers on the branches indicate the support inferred by the bootstrap test with 1000 replications.
MmGmV_03 grouped with the species of Gemykibivirus canfam1 (NC_075339/YP_010784661) isolated in Brazil in 2016. The sequences have 97.70% identity with each other. MmGmV_05-PA grouped with Emberiza spodocephala Genomoviridae sp. discovered in birds in China in 2018 (MW182919/QTE03605) exhibiting 84.12% identity. MmGmV_06-PA has 99% identity with the viruses YP_009506608 (NC_038488/YP_009506608) identified in the guano of Flying foxes (Pteropus tonganus). MmGmV_07-PA clustered in a clade close to an unclassified sequence named Genomoviridae_sp (MW678943/QXN75601) and Gemycircularvirus-ptero3 (NC_038486/YP_009506601) with identity of 76.51% and 64.54%. Meanwhile, MmGmV_09-PA showed clustering close to the sequences MW182919 and MmGmV_05-PA, exhibiting identities of 70.99% and 83.18%, respectively. Because, based on the parameter for classifying the Genomaviridae family < 78% identity for a new species throughout the genome, as well as for Rep to define the genus, in addition to Rep being established based on phylogenetic analysis [16]. MmGmV_01, 02, and 07 are likely to represent new species within the genus Gemykibivirus.
4. Discussion
In this comprehensive investigation, our focus was on the exploration of viral presence within liver samples extracted from molossus bats in the municipality of Santarém, situated in the northern region of Brazil’s state of Pará. The study revealed a noteworthy identification of seven distinct sequences termed MmGmV-PA. Through meticulous sequence analyses, these sequences exhibited a significant level of identity ranging from 74.03% to 98.71%. The genomic organization observed shared commonalities with other members of the Genomoviridae family, encompassing motifs, conserved nonanucleotides in the ori, and two crucial ORFs responsible for encoding Rep and Cap.
The Genomoviridae family, established in 2016, originated with the discovery of the Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 (SsHADV-1) from environmental samples [27]. Since its inception, numerous viruses within this family have been documented across diverse hosts and sample types. Notably, bats of the Tadarida brasiliensis and Molossus molossus species have been studied extensively, with viral species like Tadarida brasiliensis gemykibivirus 1 (TbGkyV1) emerging as the first genomovirus viral sequences from molossus bats in South America [10,15,52,53].
A phylogenetic analysis of the Rep sequences unveiled a classification into two genera: Gemykibivirus (MmGmV-01, MmGmV-02, MmGmV-03, MmGmV-05, and MmGmV-09) and Gemycirculavirus (MmGmV-06 and MmGmV-07). Utilizing the defined criteria for species demarcation (<78% genome-wide pairwise identity), the MmGmV sequences were found to be associated with four known species (MmGmV-03, MmGmV-05, MmGmV-06, and MmGmV-09) and three novel species (MmGmV-01, MmGmV-02, and MmGmV-07).
Notably, the study recognizes the challenges in comparing results from different metagenomics studies due to various factors such as sample types, methodologies, and bioinformatic analyses. Consequently, comprehensive assessments have been made of pathogenic characteristics and epidemiological data related to Genomoviridae in the human sample [12] and a group of CRESS viruses that infect protozoa (Entamoeba and Giardia) named in the eukaryotic family Naryaviridae, Nenyaviridae and Vilyaviridae. They are responsible for many cases of human infectious diseases annually in the world [21,22,54]. Further CRESS-DNA viruses remain elusive [26].
Building upon a prior study’s suggestion of a potential association between CRESS DNA viruses in bats and dietary habits, the current findings underscore the importance of monitoring viruses in bats. Bats, being reservoirs for a diverse array of both known and novel viral species, present a crucial focal point for understanding disease dynamics and cross-species transmission [55,56]. In conclusion, these data significantly contribute to an enhanced comprehension of the diversity of CRESS-DNA viruses within molossus bats, particularly expanding our knowledge with the identification of new species within the Gemykibivirus genus.
5. Conclusions
In conclusion, the investigation contributes to a better understanding of the diversity of CRESS-DNA viruses in bats (M. molossus) and expanded knowledge with the discovery of new species within the genus Gemykibivirus.
Author Contributions
Conceptualization, W.U.A., L.R.R.R., L.F.M., E.L. and A.C.d.C.; methodology, R.d.S.C., W.U.A., L.R.R.R., L.F.M., V.d.S.M., F.V., X.D., E.D., E.L. and A.C.d.C.; investigation, R.d.S.C., W.U.A., L.R.R.R., L.F.M., V.d.S.M., F.V., R.P.P., X.D., E.D., E.L. and A.C.d.C.; data curation, R.d.S.C., W.U.A., L.R.R.R., L.F.M., V.d.S.M., F.V., R.P.P., X.D., E.D., E.L. and A.C.d.C.; writing—original draft preparation, R.d.S.C. and E.L.; writing—review and editing, R.P.P.; supervision, E.L. and A.C.d.C. All authors have read and agreed to the published version of the manuscript.
Funding
R.d.S.C. is supported by a scholarship provided by the Coordination for the Improvement of Higher Education Personnel—Brazil (CAPES). A.C.d.C. is supported by a scholarship from HCFMUSP with funds donated by NUBANK under the #HCCOMVIDA scheme.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We thank Coordenação Geral de Laboratórios de Saúde Pública do Departamento de Articulação Estratégica da Secretaria de Vigilância em Saúde do Ministério da Saúde (CGLAB/DAEVS/SVS-MS), MP Biomedicals do Brasil, Zymo Research Inc. for the donation of reagents for this project. We thank Luciano Monteiro da Silva and Nilton Costa. We thank the Pró-reitoria de pesquisa e pós-graduação of UFPA for supporting the publication costs.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Roes, F.L. On the Evolution of Virulent Zoonotic Viruses in Bats. Biol. Theory 2020, 15, 223–225. [Google Scholar] [CrossRef]
- Wang, L.-F.; Anderson, D.E. Viruses in bats and potential spillover to animals and humans. Curr. Opin. Virol. 2019, 34, 79–89. [Google Scholar] [CrossRef]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef]
- Epstein, J.H.; Anthony, S.J.; Islam, A.; Kilpatrick, A.M.; Khan, S.A.; Balkey, M.D.; Ross, N.; Smith, I.; Zambrana-Torrelio, C.; Tao, Y.; et al. Nipah virus dynamics in bats and implications for spillover to humans. Proc. Natl. Acad. Sci. USA 2020, 117, 29190–29201. [Google Scholar] [CrossRef]
- Khusro, A.; Aarti, C.; Pliego, A.B.; Cipriano-Salazar, M. Hendra Virus Infection in Horses: A Review on Emerging Mystery Paramyxovirus. J. Equine Vet. Sci. 2020, 91, 103149. [Google Scholar] [CrossRef]
- Samies, N.L.; James, S.H. Prevention and treatment of neonatal herpes simplex virus infection. Antivir. Res. 2020, 176, 104721. [Google Scholar] [CrossRef]
- Leguia, M.; Vila-Sanjurjo, A.; Chain, P.S.G.; Berry, I.M.; Jarman, R.G.; Pollett, S. Precision Medicine and Precision Public Health in the Era of Pathogen Next-Generation Sequencing. J. Infect. Dis. 2020, 221, S289–S291. [Google Scholar] [CrossRef]
- Somasekar, S.; Lee, D.; Rule, J.; Naccache, S.N.; Stone, M.; Busch, M.P.; Sanders, C.; Lee, W.M.; Chiu, C.Y. Viral Surveillance in Serum Samples from Patients with Acute Liver Failure By Metagenomic Next-Generation Sequencing. Clin. Infect. Dis. 2017, 65, 1477–1485. [Google Scholar] [CrossRef]
- Zhong, Y.; Xu, F.; Wu, J.; Schubert, J.; Li, M.M. Application of Next Generation Sequencing in Laboratory Medicine. Ann. Lab. Med. 2021, 41, 25–43. [Google Scholar] [CrossRef]
- Bolatti, E.M.; Viarengo, G.; Zorec, T.M.; Cerri, A.; Montani, M.E.; Hosnjak, L.; Casal, P.E.; Bortolotto, E.; Di Domenica, V.; Chouhy, D.; et al. Viral Metagenomic Data Analyses of Five New World Bat Species from Argentina: Identification of 35 Novel DNA Viruses. Microorganisms 2022, 10, 266. [Google Scholar] [CrossRef]
- Han, H.; Wen, H.; Zhao, L.; Liu, J.; Luo, L.; Zhou, C.; Qin, X.; Zhu, Y.; Liu, M.; Qi, R.; et al. Novel coronaviruses, astroviruses, adenoviruses and circoviruses in insectivorous bats from northern China. Zoonoses Public. Health 2017, 64, 636–646. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; He, X.; Ma, J.; Hong, W.; Hu, F.; Zhao, L.; Li, Q.; Zhang, J.; Zhang, C.; et al. Gemykibivirus Genome in Lower Respiratory Tract of Elderly Woman With Unexplained Acute Respiratory Distress Syndrome. Clin. Infect. Dis. 2019, 69, 861–864. [Google Scholar] [CrossRef]
- Lamberto, I.; Gunst, K.; Müller, H.; Zur Hausen, H.; de Villiers, E.-M. Mycovirus-like DNA virus sequences from cattle serum and human brain and serum samples from multiple sclerosis patients. Genome Announc. 2014, 2, e00848-14. [Google Scholar] [CrossRef]
- Bezerra, R.S.; Bitencourt, H.T.; Covas, D.T.; Kashima, S.; Slavov, S.N. Metagenomic identification of human Gemykibivirus-2 (HuGkV-2) in parenterally infected blood donors from the Brazilian Amazon. Int. J. Infect. Dis. 2020, 98, 249–251. [Google Scholar] [CrossRef]
- Bolatti, E.M.; Zorec, T.M.; Montani, M.E.; Hošnjak, L.; Chouhy, D.; Viarengo, G.; Casal, P.E.; Barquez, R.M.; Poljak, M.; Giri, A.A. A Preliminary Study of the Virome of the South American Free-Tailed Bats (Tadarida brasiliensis) and Identification of Two Novel Mammalian Viruses. Viruses 2020, 12, 422. [Google Scholar] [CrossRef]
- Orton, J.P.; Morales, M.; Fontenele, R.S.; Schmidlin, K.; Kraberger, S.; Leavitt, D.J.; Webster, T.H.; Wilson, M.A.; Kusumi, K.; Dolby, G.A.; et al. Virus Discovery in Desert Tortoise Fecal Samples: Novel Circular Single-Stranded DNA Viruses. Viruses 2020, 12, 143. [Google Scholar] [CrossRef]
- Kaszab, E.; Lengyel, G.; Marton, S.; Dán, Á.; Bányai, K.; Fehér, E. Occurrence and genetic diversity of CRESS DNA viruses in wild birds: A Hungarian study. Sci. Rep. 2020, 10, 7036. [Google Scholar] [CrossRef]
- Rosario, K.; Dayaram, A.; Marinov, M.; Ware, J.; Kraberger, S.; Stainton, D.; Breitbart, M.; Varsani, A. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta). J. Gen. Virol. 2012, 93 Pt 12, 2668–2681. [Google Scholar] [CrossRef]
- Dayaram, A.; Potter, K.A.; Pailes, R.; Marinov, M.; Rosenstein, D.D.; Varsani, A. Identification of diverse circular single-stranded DNA viruses in adult dragonflies and damselflies (Insecta: Odonata) of Arizona and Oklahoma, USA. Infect. Genet. Evol. 2015, 30, 278–287. [Google Scholar] [CrossRef]
- Du, Z.; Tang, Y.; Zhang, S.; She, X.; Lan, G.; Varsani, A.; He, Z. Identification and molecular characterization of a single-stranded circular DNA virus with similarities to Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1. Arch. Virol. 2014, 159, 1527–1531. [Google Scholar] [CrossRef]
- Kinsella, C.M.; Bart, A.; Deijs, M.; Broekhuizen, P.; Kaczorowska, J.; Jebbink, M.F.; van Gool, T.; Cotten, M.; van der Hoek, L. Entamoeba and Giardia parasites implicated as hosts of CRESS viruses. Nat. Commun. 2020, 11, 4620. [Google Scholar] [CrossRef]
- Makoa-Meng, M.; Semmar, R.; Antezack, A.; Penant, G.; La Scola, B.; Monnet-Corti, V.; Colson, P. Correlation of Redondovirus and Entamoeba gingivalis Detections in the Human Oral Cavity Suggests That This Amoeba Is Possibly the Redondovirus Host. Int. J. Mol. Sci. 2023, 24, 6303. [Google Scholar] [CrossRef]
- Assis, M.R.d.S.; Vieira, C.B.; Fioretti, J.M.; Rocha, M.S.; de Almeida, P.I.N.; Miagostovich, M.P.; Fumian, T.M. Detection and Molecular Characterization of Gemycircularvirus from Environmental Samples in Brazil. Food Environ. Virol. 2016, 8, 305–309. [Google Scholar] [CrossRef]
- Siddell, S.G.; Walker, P.J.; Lefkowitz, E.J.; Mushegian, A.R.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; Kropinski, A.M.; et al. Binomial nomenclature for virus species: A consultation. Arch. Virol. 2020, 165, 519–525. [Google Scholar] [CrossRef]
- Varsani, A.; Krupovic, M. Family Genomoviridae: 2021 taxonomy update. Arch. Virol. 2021, 166, 2911–2926. [Google Scholar] [CrossRef]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef]
- Kazlauskas, D.; Varsani, A.; Krupovic, M. Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses. Viruses 2018, 10, 187. [Google Scholar] [CrossRef]
- Varsani, A.; Krupovic, M. Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae. Virus Evol. 2017, 3, vew037. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Koonin, E.V.; Wolf, Y.I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990, 262, 145–148. [Google Scholar] [CrossRef]
- Koonin, E.V.; Ilyina, T.V. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J. Gen. Virol. 1992, 73 Pt 10, 2763–2766. [Google Scholar] [CrossRef]
- Nash, T.E.; Dallas, M.B.; Reyes, M.I.; Buhrman, G.K.; Ascencio-Ibañez, J.T.; Hanley-Bowdoin, L. Functional analysis of a novel motif conserved across geminivirus Rep proteins. J. Virol. 2011, 85, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Vega-Rocha, S.; Byeon, I.-J.L.; Gronenborn, B.; Gronenborn, A.M.; Campos-Olivas, R. Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2. J. Mol. Biol. 2007, 367, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Duffy, S.; Breitbart, M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch. Virol. 2012, 157, 1851–1871. [Google Scholar] [CrossRef] [PubMed]
- Deltoro, D.; Ortiz, D.; Ordyan, M.; Sippy, J.; Oh, C.-S.; Keller, N.; Feiss, M.; Catalano, C.E.; Smith, D.E. Walker-A Motif Acts to Coordinate ATP Hydrolysis with Motor Output in Viral DNA Packaging. J. Mol. Biol. 2016, 428, 2709–2729. [Google Scholar] [CrossRef]
- Kanade, M.; Chakraborty, S.; Shelke, S.S.; Gayathri, P. A Distinct Motif in a Prokaryotic Small Ras-Like GTPase Highlights Unifying Features of Walker B Motifs in P-Loop NTPases. J. Mol. Biol. 2020, 432, 5544–5564. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Ghabrial, S.A.; Jiang, D.; Varsani, A. Genomoviridae: A new family of widespread single-stranded DNA viruses. Arch. Virol. 2016, 161, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, S.; Zhang, L.; Qiu, D.; Zhou, X.; Guo, L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci. Adv. 2020, 6, eaay9634. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Xie, C. A poor man’s BLASTX--high-throughput metagenomic protein database search using PAUDA. Bioinformatics 2014, 30, 38–39. [Google Scholar] [CrossRef] [PubMed]
- Pirooznia, M.; Perkins, E.J.; Deng, Y. Batch Blast Extractor: An automated blastx parser application. BMC Genom. 2008, 9 (Suppl. S2), S10. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- de Rezende, R.R.; Mar, T.B.; Páez, L.M.C.; Silva Xavier, A.D.; Xavier, C.A.D.; Navas-Castillo, J.; Zerbini, F.M.; Alfenas-Zerbini, P. Complete genome sequences of two gemycircularviruses associated with non-cultivated plants in Brazil. Arch. Virol. 2018, 163, 3163–3166. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wu, H.; Sun, G.; Yang, S.; Shen, Q.; Wang, X.; Zhang, W. Identification of diverse novel genomoviruses in gut of wild birds. Biosaf. Health 2021, 3, 136–141. [Google Scholar] [CrossRef]
- Krupovic, M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr. Opin. Virol. 2013, 3, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Campos-Olivas, R.; Louis, J.M.; Clerot, D.; Gronenborn, B.; Gronenborn, A.M. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. USA 2002, 99, 10310–10315. [Google Scholar] [CrossRef]
- Steinfeldt, T.; Finsterbusch, T.; Mankertz, A. Demonstration of nicking/joining activity at the origin of DNA replication associated with the rep and rep’ proteins of porcine circovirus type 1. J. Virol. 2006, 80, 6225–6234. [Google Scholar] [CrossRef]
- Nakasu, E.Y.T.; Melo, F.L.; Michereff-Filho, M.; Nagata, T.; Ribeiro, B.M.; Ribeiro, S.G.; Lacorte, C. Discovery of two small circular ssDNA viruses associated with the whitefly Bemisia tabaci. Arch. Virol. 2017, 162, 2835–2838. [Google Scholar] [CrossRef] [PubMed]
- Kemenesi, G.; Kurucz, K.; Zana, B.; Földes, F.; Urbán, P.; Vlaschenko, A.; Kravchenko, K.; Budinski, I.; Szodoray-Parádi, F.; Bücs, S.; et al. Diverse replication-associated protein encoding circular DNA viruses in guano samples of Central-Eastern European bats. Arch. Virol. 2018, 163, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Cibulski, S.P.; Lima, F.E.d.S.; Teixeira, T.F.; Varela, A.P.M.; Scheffer, C.M.; Mayer, F.Q.; Witt, A.A.; Roehe, P.M. Detection of multiple viruses in oropharyngeal samples from Brazilian free-tailed bats (Tadarida brasiliensis) using viral metagenomics. Arch. Virol. 2021, 166, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar]
- Jones, S.; Baizan-Edge, A.; MacFarlane, S.; Torrance, L. Viral Diagnostics in Plants Using Next Generation Sequencing: Computational Analysis in Practice. Front. Plant Sci. 2017, 8, 1770. [Google Scholar] [CrossRef]
- Ghurye, J.S.; Cepeda-Espinoza, V.; Pop, M. Metagenomic Assembly: Overview, Challenges and Applications. Yale J. Biol. Med. 2016, 89, 353–362. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).