Abstract
Since the Lymphogranuloma venereum (LGV) outbreak was first described in Buenos Aires in 2017, the detected strains presented peculiar characteristics. Our goal was to increase the understanding of the strains involved in the LGV outbreak in Argentina. We characterized the ompA gene sequences, using Sanger sequencing, of 88 LGV strains from 239 symptomatic patients in Buenos Aires enrolled between 2017 and 2019, and selected 20 C. trachomatis strains for further characterization using Multilocus Sequence Typing (MLST). Following the ompA gene analysis of the 88 LGV strains, we detected 43% L2b, 31% L1-like, and 26% L2. Among the 38 L2b samples analyzed, there were 7 distinct sequences, 3 of them not previously reported (L2bv12, L2bv13, and L2bv14). Additionally, we detected a strain with a new mutation (AM884176.1:g.59122A>T) found in the position defining L2 or L2b, proposed as L2i. Using MLST, five different sequence types (STs) were detected, including the ST2 (corresponding to the L1-like strains) and a new one (ST60). ST58 was associated with the concomitant presence of another STI and HIV. A high genetic diversity in C. trachomatis LGV strains in Argentina was observed in a short period of time, with a relatively low number of samples from a limited geographical area.
1. Introduction
Chlamydia trachomatis is the most prevalent bacterial sexually transmitted infection (STI) worldwide. In 2020, the World Health Organization (WHO) estimated that approximately 128 million new C. trachomatis sexually transmitted infections occurred globally [1]. In Argentina, there are no general prevalence data, as C. trachomatis detection has not been incorporated into routine diagnostics [2]. A prevalence study in Buenos Aires in 2007 revealed an overall infection rate of 1.9% among adults [3]. Another study conducted in the province of Buenos Aires between 2016 and 2017, focusing on pregnant women, detected C. trachomatis in 18% of them [4]. These isolated studies emphasize the lack of comprehensive nationwide prevalence data for C. trachomatis in Argentina, highlighting the need for broad-scale investigations to grasp its prevalence and impact across the general population.
Since 2003, LGV outbreaks have been reported in men who have sex with men (MSM), most of them infected with the Human Immunodeficiency Virus (HIV), in different countries around the world [5,6,7].
In 2017, the first LGV cases detected in Argentina were similar to those reported in Europe since 2003, mostly HIV-infected male patients who had sex with men. Most cases presented symptomatic manifestations, demonstrating varying degrees of proctitis (mild, moderate, and severe), with associated clinical features such as straining, tenesmus, urgency for bowel movements, and mucous or purulent discharge. In addition to HIV co-infection, 40% of them were diagnosed with at least another associated STI [8,9].
In Argentina, considering the cases reported to the Ministry of Health from the Instituto Nacional de Microbiología Carlos G. Malbran [2] and those contributed by the Laboratorio de Clamidias, there is an average of 60 LGV cases reported per year, which results in an incidence of 20 cases per million inhabitants in the city of Buenos Aires. This incidence value needs to be regarded as the lowest, since, as of today, no other laboratories can identify LGV from non-LGV cases of C. trachomatis and inform them to the Argentinian health surveillance system. Even considering that, this value is 50% higher than the reported LGV rate in Europe, estimated to be 13 cases per million inhabitants [10].
Since the first description of the LGV outbreak in Buenos Aires, the detected strains of C. trachomatis showed unique characteristics. About half of the studied strains belonged to the L2b genotype, 20% to the L2 genotype, and one-third to the L1 genotype, which was absent in the European outbreak [9,11,12,13].
C. trachomatis typing is traditionally based on the sequencing of the highly variable ompA gene [14,15]. Genotyping is important for the identification of LGV strains, since they are more invasive and require extended antibiotic treatment compared to strains with other ompA genotypes [16,17]. Despite this, studying only a single region of the genome is insufficient to comprehend the complexity of genomic diversity in LGV outbreak samples at both global and local levels. To overcome this limitation, Multilocus Sequence Typing (MLST) offers a comprehensive approach. MLST involves PCR amplification and the subsequent DNA sequencing of multiple loci within the genome. Three MLST schemes have been proposed for C. trachomatis typing. Two of them are based on the analysis of constitutively expressed housekeeping genes, providing resolution comparable to complete ompA gene sequencing and making them suitable for evolutionary studies, whilst not as good for outbreak or clinical–epidemiological studies [18,19]. In 2007, Klint et al. introduced a third system, designed for short-term epidemiological studies. The MLST scheme proposed by Klint et al. has undergone slight modifications to optimize processing, resulting in only a one percent reduction in the discriminatory capacity [20]. The current configuration is accessible through the PubMLST//Chlamydiales database (https://pubmlst.org/organisms/chlamydiales-spp, accessed on 24 November 2023). This scheme analyzes five highly variable yet stable genomic loci (hctB, CT058, CT144, CT172, and pbpB), offering up to five times the resolution power of ompA gene sequencing [21].
Considering the scarcity of data regarding the prevalence of C. trachomatis in Argentina and Latin America, and the unique aspect of the 2017 LGV outbreak in Buenos Aires, characterized by the presence of an L1-like strain in one-third of the diagnosed cases, there exists a significant gap in understanding the epidemiology of LGV in the region. Furthermore, unlike in Europe, there is a notable lack of epidemiological and microbiological data on lymphogranuloma venereum in South America. This study aims to address this gap by investigating the epidemiology and its correlations with the clinical presentation of LGV in Argentina. To achieve this, we have applied ompA sequencing and MLST.
2. Materials and Methods
A total of 239 anorectal swabs were obtained from patients with symptoms compatible with proctitis between September 2017 and August 2019. Adult patients were prospectively included from a public hospital (Hospital Juan A. Fernández, Buenos Aires, Argentina) and a private health center (Centro Privado de Cirugía y Coloproctología, Buenos Aires, Argentina) in Buenos Aires, Argentina. Inclusion criteria encompassed individuals aged 18 years and above who provided written informed consent, seeking consultation for anorectal symptoms including rectal tenesmus, anal discharge, bowel urgency, and proctalgia, and who had not taken antimicrobial drugs in the previous month. Exclusion criteria included individuals under 18 years of age, those with a history of pelvic radiotherapy, and those previously diagnosed with inflammatory bowel disease. No patient was referred by another participant in the study. Inflammatory bowel disease and pelvic radiotherapy patients were excluded. Patients were treated according to current STI guidelines [22,23].
A thorough anamnesis was taken, and anal samples were collected and analyzed at the Laboratorio de Clamidias (Facultad de Farmacia y Bioquímica, Universidad de Buenos Aires, Buenos Aires, Argentina). Briefly, total DNA was isolated from clinical samples using Quick-DNA MiniPrep (Zymo Research Corporation, Irvine, CA, USA). C. trachomatis was tested using a real-time PCR targeting the cryptic plasmid (CHLAMYDIA tr. Q-PCR Alert kit; ELITech Molecular Diagnostics, Bothell, WA, USA). All real-time PCR-positive samples were genotyped by the PCR-RFLP of the ompA gene, as previously described. A nested PCR targeting the ompA gene was initially performed. The first PCR round generated a 1.1 kb DNA fragment of the ompA gene [24]. Then, a nested PCR was conducted, yielding a DNA fragment spanning from positions c.55 to c.1067 within the ompA gene of the C. trachomatis DNA sequence L2b:JN795427.1. RFLP analysis of the nested PCR-positive samples followed the protocol described by Sayada et al., with minor adjustments [25]. Briefly, 10 µL of the positive nested PCR product underwent digestion with 2.5 U AluI (Promega Corporation, Madison, WI, USA) following the manufacturer’s guidelines. Subsequently, samples were subjected to 10% polyacrylamide gel electrophoresis, stained with Gel Red (GENBIOTECH SRL, Gel Red 10,000 X in 500 µL water per the manufacturer’s instructions) for 15 min, and photographed under UV light [24]. When necessary, additional analyses were performed using a second enzyme (HinfI, CfoI, or a combination of EcoRI and DdeI). Genotypes were confirmed by sequencing the nested PCR product using capillary electrophoresis sequencing (Sanger sequencing), which covers 86% of the ompA gene (Macrogen, Inc., Seoul, Republic of Korea). These sequences were analyzed against the NIH databases using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 24 November 2023). These sequences have been deposited in GenBank (MN548736.1 to MN548759.1, MN537150.1 to MN537152.1, and MN563608.1 to MN563615.1).
Based on the ompA typing results, 20 C. trachomatis samples were selected for high-resolution typing by MLST according to the Uppsala scheme (https://pubmlst.org/static/organisms/chlamydiales-spp/Protocol_MLST_C_trachomatis_Uppsala.pdf, accessed on 24 November 2023) [20]. Selected samples were subjected to PCR amplification and sequencing of the five target regions (CT144, CT172, CT058, and regions of the pbpB and hctB gene) and the ompA gene. The obtained sequences were analyzed using BioEdit software (version 5.0.9). An allele number was assigned by comparing the sequences at each locus with all corresponding known alleles available in the C. trachomatis PubMLST//Chlamydiales database (https://pubmlst.org/).
Allele profiles based on the five genetic regions generated sequence types (STs). The obtained sequences were deposited in the PubMLST database (Acc. No. 5059–5074). We performed two GrapeTree analyses using the resources provided in the PubMLST//Chlamydiales database (https://pubmlst.org/bigsdb?db=pubmlst_chlamydiales_isolates, accessed on 24 November 2023).
Statistical analysis was performed using InfoStat version 2020 software. The Fisher exact test or the Irwin Fischer test were used for categorical variable analysis. Statistical significance was defined as p < 0.05.
3. Results
3.1. Patient Characteristics and ompA Gene Variability in C. trachomatis Strains
The study included 239 patients aged 18 years and older (18–70, mean 33), comprising 9 cisgender women, 8 transgender women, and 222 cisgender men. All reported engaging in unprotected sexual intercourse. The overall HIV positivity rate among the participants was 63%. C. trachomatis was detected by PCR in 107 (45%) of the 239 patients, 1 cisgender woman, 2 transgender women, and 104 cisgender men, all of them MSM. The mean age of C. trachomatis-infected patients was 34 years (18–59 years), and 78% were HIV-positive and undergoing treatment. PCR-RFLP genotyping revealed that 88 out of 107 (82%) belonged to an LGV genotype. Of the LGV-positive patients, 87 were MSM and 1 was a transgender woman. The mean age was 34 years (21–59 years), and 86% were HIV-positive and receiving treatment.
Successful ompA sequencing was obtained for 104 (97%) positive samples, of which 85% (n = 88) were LGV and 15% (n = 16) were non-LGV genotypes. Regarding LGV genotypes, close to half were identified as the ompA genotype L2b (n = 38, 43%), whereas one-fourth of the samples were the ompA genotype L2 (n = 23, 26%) and one-third were similar to the L1 genotype (n = 27, 31%). The 27 L1 strains identified by PCR-RFLP were identical in the ompA sequence analysis and exhibited up to 10 mutations compared to the L1/440 based on the length of the obtained sequence (DQ064294.1: c.268G>A, c.348T>A, c.462C>T, c.471G>A, c.474A>G, c.477C>T, c.594C>T, c.931A>G, c.1017C>T, and c.1020A>C.). Hence, we named it as L1-like. Moreover, one of these L1-like strains had an additional ompA mutation (DQ064294.1:c.507A>C). The mutation in this L1-like strain (at position 931 in the L1/440 reference sequence DQ064294.1:c.931A>G) has not been previously described.
Among the 23 L2 strains, 22 were identical to the reference sequence AM884176.1, and 1 had a mutation at position AM884176.1: g.59122A>T. Notably, this single mutation in the ompA gene was located precisely at position 485, which is the position that distinguishes L2 from L2b. Consequently, this particular strain cannot be classified according to the convention used for L2/L2b [26]. We have initially designated this variation, which has not been previously documented in the databases, as “L2nv (new variant)” (additional information in the Supplementary Table S1). The distribution of the LGV L2b ompA genotype designations in our sample set is summarized in Table 1. Among the 38 samples analyzed, 29 (76%) were classified as the L2b genotype. Notable variations were observed, with a subset of samples falling into distinct ompA genotype designations, such as L2bv1 (four, 10%), L2bv5 (one, 2.6%), and L2bv11 (one, 2.6%). Three different strains (7.9%) lacked ompA designations in the referenced study by Helena M. B. Seth-Smith et al. (2021) [27] and were not found in the BLAST database. Following the nomenclature used in that study, we named them L2bv12, L2bv13, and L2bv14 [27]. Noteworthy, all the found mutations generated conservative amino acid substitutions, except for one demonstrating semi-conservative alteration. All mutations, except one, were confined within the variable domains of the ompA gene [28,29,30,31,32].

Table 1.
Variability of the ompA gene in Chlamydia trachomatis LGV L2b.
3.2. Novel C. trachomatis MLST Sequence Type and Uncommon Genotype Prevalence
For the MLST analysis, we selected a total of 20 samples, with 10 identified as the L2b genotype, 6 as L1-like, and 4 as L2. Within the L2b subset, we analyzed two L2b types, three L2bv1 types, one L2bv5 type, one L2bv11 type, one L2bv12 type, one L2bv13 type, and one L2bv14 type. Within L1-like samples, we included the one featuring the additional mutation. Among the L2 samples, three showed identical sequences to the reference sequence AM884176.1, while one exhibited a mutation at position AM884176.1: g.59122A>T.
The analysis resulted in high-quality sequencing results for 16 out of the 20 samples (Table 2). Unfortunately, four samples corresponding to the L2b ompA genotype could not be analyzed due to insufficient amplification of the target genes (two L2bv1, one L2bv11, and one L2bv13). We identified the following five distinct sequence types according to the Uppsala scheme: ST2, ST58, ST141, ST143, and a novel ST designated as number 60 (Table 2).

Table 2.
Allelic profiles of sequence types (STs) according to the Uppsala scheme and sample distribution.
Upon closer examination of the five identified STs, three of them corresponded to previously well-known sequence types (ST58, ST141, and ST143). The L1-like samples, identified as genotype ST2, represent the only ST2 sequences with DNA sequences available in the PubMLST database. Additionally, an unprecedented combination of allele loci was observed, generating a novel ST (CT058 13; CT144 17; CT172 13; hctB 18; pbpB 28) designated as ST60, recorded under the accession number 5067 in the PubMLST database.
According to the ompA analysis, the samples classified as belonging to the L2b genotype were identified as either MLST ST58 or ST143, both of which have been previously associated with L2 and L2b strains. Among the four L2 samples, one exhibited ST58, one exhibited ST141 (both of which were formerly associated with L2 genotypes), another one was MLST ST60, and the fourth sample, L2nv, also resulted in ST58 by MLST analysis (Table 3).

Table 3.
MLST-derived STs and ompA genotypes.
To contextualize the relationship of C. trachomatis STs from the Buenos Aires outbreak, we performed two GrapeTree analyses using data from the PubMLST database (Figure 1). Of the 560 STs associated with C. trachomatis Uppsala scheme, we identified 9 STs (ST = 2, 49, 51, 58, 60, 141, 142, 143, and 144) associated with LGV based on sequences and epidemiological information. Our samples were associated with STs related with L2 and L2b (ST = 58, 141, and 143), a newly associated ST2 linked to the L1-like, and a novel ST60 (ompA = L2), centrally positioned in the diagram (Figure 1b), connecting with all other STs.
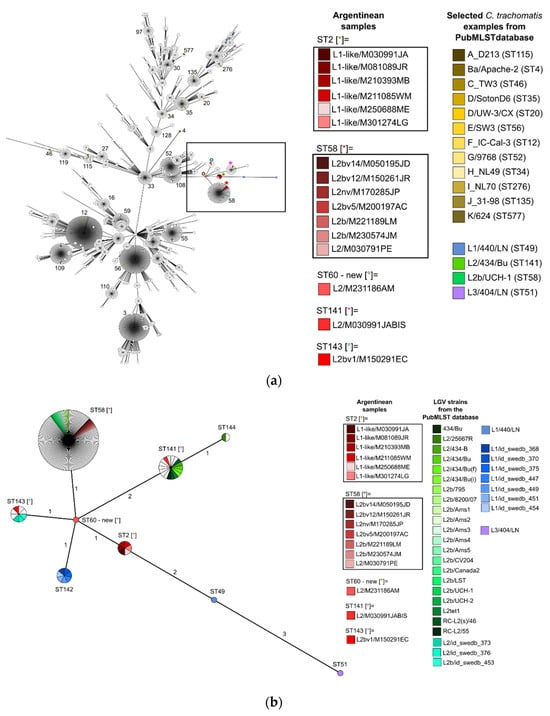
Figure 1.
GrapeTrees showing the relationship of C. trachomatis STs from patients of the Buenos Aires outbreak in the context of all characterized samples published in the Chlamydiales PubMLST database with the Uppsala scheme. For reference: Argentinian samples are highlighted in shades of red, C. trachomatis examples from the PubMLST database are referenced in other colors, and the remaining samples in the database that meet the selection criteria remain white. Different colored asterisks and small circles indicated in square brackets within the figure correspond to the different STs of the Argentinean samples as shown in the legend of the graph. (a) GrapeTree clustering of 560 STs available in the database containing 3402 samples. The black box indicates where Argentinean samples are located. The most common STs are indicated by number; (b) GrapeTree clustering of all (n = 160) available LGV strains and their available STs (https://pubmlst.org/bigsdb?db=pubmlst_chlamydiales_isolates (accessed on 24 November 2023)). Sphere size is proportional to the number of samples of each sequence type. The numbers on the branches represent the number of alleles that varies between STs (single- or double-locus variants).
3.3. Comparative Analysis of Sequence Types by MLST and Clinical Symptoms
An analysis of the patient’s clinical manifestations and STs was performed (Table 4). This analysis focused on the subset of 16 patients for whom MLST results were available, enabling a comparison with clinical symptomatology. Symptoms in this subset of patients were diverse. Notably, fistula formation was consistently absent in all cases. Most cases presented with rectal syndrome (14 patients). ST58 was associated with other STIs besides HIV (p = 0.0291). The frequency of HIV infection was higher in patients with C. trachomatis ST58, and having an inflammatory tumor was more frequent in patients with ST2, but no statistically significant association was reached. Concerning sexual behavior, 14 patients declared to have active and passive anal sexual practices and oral–anal intercourse, 1 patient claimed passive practices, and another passive practice and oral–anal intercourse. All of them were MSM.

Table 4.
Comparison of STs (sequence types by MLST) with patient symptomatology.
4. Discussion
This study provides significant contributions to understanding the genetic diversity of C. trachomatis LGV strains in Argentina, filling a notable gap in the data not only within Argentina but also across South America. It represents the first comprehensive investigation of this issue in the region. In addition to ompA gene sequencing, our study employed Multilocus Sequence Typing (MLST), offering a more comprehensive approach. However, a limitation of this study is the absence of complete genome analysis from the samples, which could have provided additional insights. Our results reveal a substantial variability in the ompA gene sequences of the L2b strains, which, in contrast to the diversity observed in the European outbreak, is notable for occurring within a relatively short timeframe and limited geographical area. It is important to note that the European variability stems from studies conducted over several years, encompassing diverse geographical regions and a large number of samples [12,27,33,34,35]. We identified three novel variants, designated as L2bv12, L2bv13, and L2bv14, and one strain with a mutation AM884176.1: g.59122A>T that defies classification as either L2 or L2b. For this strain, named as L2nv, we propose the designation L2i for this variant according to Helena M. B. Seth-Smith et al. (2021) [26,27]. Given the complexity and lack of consensus surrounding the nomenclature of Chlamydia trachomatis strains, coupled with the current challenge of distinguishing between closely related variants, we advocate for a systematic approach in assigning such designations, based on the work of Helena M. B. Seth-Smith et al., 2021 [27]. As the ompA of L2b was defined while being 1bp different from the ompA of L2, we suggest that strains exhibiting the mutation characterizing L2b in the ompA gene (A485G) be classified as variants of L2b (L2bv12, L2bv13, and L2bv14). Conversely, strains lacking the L2b mutation could be assigned a new letter designation (L2i) to denote their distinct identity.
The sequences of strains initially labeled as L1 exhibited remarkable homogeneity and differed significantly from the standard L1. Therefore, we provisionally designate them as L1-like pending further detailed analysis. These strains were assigned to ST2, which, as of our study, lacked uploaded sequences and associated epidemiological characteristics. It is noteworthy that we have had a high proportion of L1-like strains since 2017, indicating a unique epidemiological trend not previously documented in South America and sporadically globally reported [27]. All our L1-like samples exhibited up to 10 mutations compared to L1/440 in the ompA gene. Nine of these mutations had been previously documented in a strain from a patient in the United States diagnosed in the early 1980s (GQ413956.1) that was published in a 2010 study employing MLST on strains isolated from MSM in Europe and the United States. Furthermore, compared to the strain found in the US patient, our study strains exhibit differences in two of five alleles, as determined by the MLST Uppsala scheme (CT144 and pbpB) [36]. These findings highlight the emergence of novel ompA genotypes within the circulating LGV strains in Buenos Aires, Argentina, with an epidemiological pattern different from Europe and the US [33,37,38,39]. The discovery of the L1-like strain raises questions about its appearance and evolution; thus, a study of complete genomes would provide a better understanding.
In addition, a new sequence type, ST60, was identified within the L2 genotype. Despite the homogeneity of L2 based on ompA sequences, it exhibited three different STs, with each sample analyzed representing a unique ST, highlighting significant variability within the strains denoted as the L2 type. Furthermore, the variant analysis conducted by Seth-Smith et al. focused on the evolution of C. trachomatis lymphogranuloma venereum from the European outbreak, revealing a common reversion to the ompA genotype L2 with the L2b genomic backbone [27,40]. In contrast to our results, all genomes analyzed in that study turned out to be ST58 according to the Uppsala scheme of C. trachomatis. Therefore, there is a possibility that our samples, classified as L2 according to the ompA gene study and MLST ST58, also exhibit an L2b genomic backbone. However, this remains uncertain without the analysis of complete genomes [41,42,43,44].
In previous studies, we have compared LGV and non-LGV genotypes in symptomatic patients in Buenos Aires. Among patients presenting with anorectal symptoms, LGV genotypes were found to predominate over non-LGV genotypes. While clinical manifestations are not pathognomonic of a specific biovar, notable differences were observed between the two groups. Specifically, older age and HIV-positive status were significantly higher in the LGV group. Anal discharge, bleeding, severe proctitis, and anal ulcers were more commonly reported in the LGV group [9]. In this study, we delved deeper into the LGV group and utilized MLST data to conduct a comparative analysis of sequence types (STs) with associated symptoms. Our analysis revealed that, despite the small number of samples studied, it was possible to demonstrate the association of ST58 with other STIs besides HIV. The confirmation of the differences in clinical manifestation for ST2 (the presence of an inflammatory tumor) and the correlation between a greater frequency of HIV and ST58 can be supported by studying a larger set of samples.
The prevalence of LGV was high in our study, probably because it was a population exhibiting symptoms associated with proctitis. At the same time, the modest sample size limits the conclusions drawn from our results. For example, among LGV-infected patients, there was a higher frequency of HIV-infected individuals than the overall HIV prevalence in the study population, or even the HIV prevalence in C. trachomatis-infected patients, but no statistical significance was demonstrated.
In summary, our findings not only reveal the presence of previously unreported genotypes but also emphasize the evolving landscape of LGV strains in Buenos Aires. The absence of L1-like strains in global databases and their emergence in our region points out the importance of continued genomic and epidemiologic surveillance, highlighting the unique dynamics of C. trachomatis genotypes in different geographical contexts [12,13,38,45]. Within the L2b strains, we identified three novel ompA L2b variants. Additionally, a new ompA L2i variant was proposed, adding to the observed variability. The discovery of a novel sequence type (ST60) within the L2 genotype further contributes to the genetic diversity uncovered in our study. Furthermore, despite the relatively small sample size, our study revealed a noteworthy diversity of sequence types within the L2 group. Lastly, ST58 was associated with the concomitant presence of another STI in addition to HIV. All of these findings underscore the importance of further investigation into the genomic intricacies of C. trachomatis strains and their comparison against the epidemiological characteristics of the patients in our region.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12030587/s1, Table S1: Summary of Patient Characteristics, including Gender, Sexual Behavior, Disease Type (HIV+/HIV−), ompA Genotype, ompA Mutations, and MLST Results.
Author Contributions
Conceptualization, M.R.F.; validation, K.A.B. and M.R.F.; formal analysis, K.A.B. and B.H.; investigation, K.A.B., D.L.A., D.C., L.L.R. and L.S.L.; resources, A.C.E., M.L.G.V., D.L.A., D.C., L.L.R. and L.S.L.; data curation, K.A.B., L.L.R. and L.S.L.; writing—original draft preparation, K.A.B.; writing—review and editing, M.R.F., A.C.E., M.L.G.V. and B.H.; visualization, K.A.B.; supervision, M.R.F. and B.H.; project administration, M.R.F.; funding acquisition, M.R.F.; statistical analysis, O.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universidad de Buenos Aires, UBACyT, grant number UBACYT 20020150100223BA and UBACYT 20020190100357BA.
Data Availability Statement
All the data presented are available in PubMLST database (https://pubmlst.org/, Acc. No. 5059-5074) and in GenBank (https://www.ncbi.nlm.nih.gov/genbank/, Acc. No. MN548736.1 to MN548759.1, MN537150.1 to MN537152.1, and MN563608.1 to MN563615.1, accessed on 24 November 2023). The study was approved by an Ethics Committee (“Detección de C. trachomatis en pacientes con rectitis infecciosa: prevalencia y tipificación”, Gobierno de la Ciudad de Buenos Aires, No. 201723) and patients provided written informed consent.
Acknowledgments
We express our sincere gratitude to the dedicated staff of the two hospitals and the Centro Privado de Cirugía y Coloproctología in Buenos Aires. Their invaluable encouragement, generous assistance, and collaborative efforts were essential in the successful completion of this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Health Organization. Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections, 2021. Who 2021, 53, 1689–1699. [Google Scholar]
- Ministerio de Salud, Presidencia de la Nación, Argentina. Boletin No 39: Respuesta Al VIH y Las ITS En La Argentina; Ministerio de Salud, Presidencia de la Nación: Buenos Aires, Argentina, 2022. [Google Scholar]
- Fermepin, M.R.; Entrocassi, A.C.; Sauka, D.H.; Vaulet, M.L.G.; Corominas, A.I. Chlamydia trachomatis Serovars in Buenos Aires, Argentina: Predominance of Serovar E in Ophthalmia neonatorum. Sex. Transm. Dis. 2007, 34, 1041. [Google Scholar] [CrossRef]
- Entrocassi, A.C.; Paisan, L.G.; Costa, M.Q.; Vaulet, M.L.G.; Sosa, D.; Ramos, C.; Junges, J.; D’Errico, M.; Varela, C.; Acosta, E.; et al. P3.12 Frequency and Distribution of Chlamydia trachomatis Infection among Young Pregnant Women in Argentina. Sex. Transm. Infect. 2017, 93 (Suppl. S2), A97. [Google Scholar] [CrossRef]
- Nieuwenhuis, R.F.; Ossewaarde, J.M.; Götz, H.M.; Dees, J.; Thio, H.B.; Thomeer, M.G.J.; Den Hollander, J.C.; Neumann, M.H.A.; Van Der Meijden, W.I. Resurgence of Lymphogranuloma Venereum in Western Europe: An Outbreak of Chlamydia trachomatis Serovar L2 Proctitis in The Netherlands among Men Who Have Sex with Men. Clin. Infect. Dis. 2004, 39, 996–1003. [Google Scholar] [CrossRef]
- Meyer, T.; Arndt, R.; Von Krosigk, A.; Plettenberg, A. Repeated Detection of Lymphogranuloma Venereum Caused by Chlamydia trachomatis L2 in Homosexual Men in Hamburg. Sex. Transm. Infect. 2005, 81, 91. [Google Scholar] [CrossRef]
- Vall Mayans, M.; Sanz Colomo, B.; Ossewaarde, J.M. First Case of LGV Confirmed in Barcelona. Euro Surveill. 2005, 10, 2634. [Google Scholar] [CrossRef]
- Ministerio de Salud, Presidencia de la Nación, Argentina. Casos de Linfogranuloma Venéreo (LGV) En Argentina; Ministerio de Salud, Presidencia de la Nación: Buenos Aires, Argentina, 2018; Available online: https://www.argentina.gob.ar/sites/default/files/alerta_linfogranuloma_venereo_se332018.pdf (accessed on 3 July 2021).
- La Rosa, L.; Svidler López, L.; Entrocassi, A.C.; López Aquino, D.; Caffarena, D.; Büttner, K.A.; Gallo Vaulet, L.; Rodríguez Fermepin, M. Chlamydia trachomatis Anorectal Infections by LGV (L1, L2 and L2b) and Non-LGV Serotypes in Symptomatic Patients in Buenos Aires, Argentina. Int. J. STD AIDS 2021, 32, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Lymphogranuloma venereum. In ECDC. ANNUAL Epidemiological Report for 2019; ECDC: Stockholm, Sweden, 2022. [Google Scholar] [CrossRef]
- van de Laar, M.J.W. The Emergence of LGV in Western Europe: What Do We Know, What Can We Do? Euro Surveill. 2006, 11, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Spaargaren, J.; Fennema, H.S.A.; Morré, S.A.; De Vries, H.J.C.; Coutinho, R.A. New Lymphogranuloma Venereum Chlamydia trachomatis Variant, Amsterdam. Emerg. Infect. Dis. 2005, 11, 1090. [Google Scholar] [CrossRef]
- Harris, S.R.; Clarke, I.N.; Seth-Smith, H.M.B.; Solomon, A.W.; Cutcliffe, L.T.; Marsh, P.; Skilton, R.J.; Holland, M.J.; Mabey, D.; Peeling, R.W.; et al. Whole Genome Analysis of Diverse Chlamydia trachomatis Strains Identifies Phylogenetic Relationships Masked by Current Clinical Typing. Nat. Genet. 2012, 44, 413. [Google Scholar] [CrossRef]
- Sun, G.; Pal, S.; Sarcon, A.K.; Kim, S.; Sugawara, E.; Nikaido, H.; Cocco, M.J.; Peterson, E.M.; De La Maza, L.M. Structural and Functional Analyses of the Major Outer Membrane Protein of Chlamydia Trachomatis. J. Bacteriol. 2007, 189, 6222–6235. [Google Scholar] [CrossRef]
- De Vries, H.J.C.; Schim Van Der Loeff, M.F.; Bruisten, S.M. High-Resolution Typing of Chlamydia Trachomatis: Epidemiological and Clinical Uses. Curr. Opin. Infect. Dis. 2015, 28, 61–71. [Google Scholar] [CrossRef]
- de Vries, H.J.C.; Nori, A.V.; Kiellberg Larsen, H.; Kreuter, A.; Padovese, V.; Pallawela, S.; Vall-Mayans, M.; Ross, J. 2021 European Guideline on the Management of Proctitis, Proctocolitis and Enteritis Caused by Sexually Transmissible Pathogens. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 1434–1443. [Google Scholar] [CrossRef]
- Stoner, B.P.; Cohen, S.E. Lymphogranuloma Venereum 2015: Clinical Presentation, Diagnosis, and Treatment. Clin. Infect. Dis. 2015, 61 (Suppl. S8), S865–S873. [Google Scholar] [CrossRef]
- Dean, D.; Bruno, W.J.; Wan, R.; Gomes, J.P.; Devignot, S.; Mehari, T.; De Vries, H.J.C.; Morré, S.A.; Myers, G.; Read, T.D.; et al. Predicting Phenotype and Emerging Strains among Chlamydia trachomatis Infections. Emerg. Infect. Dis. 2009, 15, 1385–1394. [Google Scholar] [CrossRef]
- Pannekoek, Y.; Morelli, G.; Kusecek, B.; Morré, S.A.; Ossewaarde, J.M.; Langerak, A.A.; Van Der Ende, A. Multi Locus Sequence Typing of Chlamydiales: Clonal Groupings within the Obligate Intracellular Bacteria Chlamydia Trachomatis. BMC Microbiol. 2008, 8, 42. [Google Scholar] [CrossRef]
- Herrmann, B.; Isaksson, J.; Ryberg, M.; Tångrot, J.; Saleh, I.; Versteeg, B.; Gravningen, K.; Bruisten, S. Global Multilocus Sequence Type Analysis of Chlamydia trachomatis Strains from 16 Countries. J. Clin. Microbiol. 2015, 53, 2172–2179. [Google Scholar] [CrossRef]
- Gravningen, K.; Christerson, L.; Furberg, A.S.; Simonsen, G.S.; Ödman, K.; Ståhlsten, A.; Herrmann, B. Multilocus Sequence Typing of Genital Chlamydia trachomatis in Norway Reveals Multiple New Sequence Types and a Large Genetic Diversity. PLoS ONE 2012, 7, e34452. [Google Scholar] [CrossRef] [PubMed]
- Kimberly, A.; Bolan, M.A. Sexually Transmitted Diseases Treatment Guidelines, 2015. MMWR. Recomm. Rep. Morb. Mortal. Wkly. Report. Recomm. Rep. 2015, 64, 1. [Google Scholar]
- de Vries, H.J.C.; de Barbeyrac, B.; de Vrieze, N.H.N.; Viset, J.D.; White, J.A.; Vall-Mayans, M.; Unemo, M. 2019 European Guideline on the Management of Lymphogranuloma Venereum. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Walboomers, J.M.M.; Roosendaal, R.; Van Doornum, G.J.J.; MacLaren, D.M.; Meijer, C.J.L.M.; Van den Brule, A.J.C. Direct Detection and Genotyping of Chlamydia trachomatis in Cervical Scrapes by Using Polymerase Chain Reaction and Restriction Fragment Length Polymorphism Analysis. J. Clin. Microbiol. 1993, 31, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Sayada, C.; Denamur, E.; Orfila, J.; Catalan, F.; Elion, J. Rapid Genotyping of the Chlamydia trachomatis Major Outer Membrane Protein by the Polymerase Chain Reaction. FEMS Microbiol. Lett. 1991, 67, 73–78. [Google Scholar] [CrossRef][Green Version]
- Thomson, N.R.; Holden, M.T.G.; Carder, C.; Lennard, N.; Lockey, S.J.; Marsh, P.; Skipp, P.; O’Connor, C.D.; Goodhead, I.; Norbertzcak, H.; et al. Chlamydia Trachomatis: Genome Sequence Analysis of Lymphogranuloma Venereum Isolates. Genome Res. 2008, 18, 161. [Google Scholar] [CrossRef]
- Seth-Smith, H.M.B.; Bénard, A.; Bruisten, S.M.; Versteeg, B.; Herrmann, B.; Kok, J.; Carter, I.; Peuchant, O.; Bébéar, C.; Lewis, D.A.; et al. Ongoing Evolution of Chlamydia trachomatis Lymphogranuloma Venereum: Exploring the Genomic Diversity of Circulating Strains. Microb. Genom. 2021, 7, 000599. [Google Scholar] [CrossRef] [PubMed]
- Feher, V.A.; Randall, A.; Baldi, P.; Bush, R.M.; de la Maza, L.M.; Amaro, R.E. A3-Dimensional Trimeric β-Barrel Model for Chlamydia MOMP Contains Conserved and Novel Elements of Gram-Negative Bacterial Porins. PLoS ONE 2013, 8, e68934. [Google Scholar] [CrossRef]
- Rodríguez-Marañón, M.J.; Bush, R.M.; Peterson, E.M.; Schirmer, T.; de la Maza, L.M. Prediction of the Membrane-Spanning Beta-Strands of the Major Outer Membrane Protein of Chlamydia. Protein Sci. 2002, 11, 1854–1861. [Google Scholar] [CrossRef]
- Zhong, G.; Brunham, R.C. Antigenic Determinants of the Chlamydial Major Outer Membrane Protein Resolved at a Single Amino Acid Level. Infect. Immun. 1991, 59, 1141. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Harris, S.R.; Seth-Smith, H.M.B.; Parmar, S.; Andersson, P.; Giffard, P.M.; Schachter, J.; Moncada, J.; Ellison, L.; Vaulet, M.L.G.; et al. Comprehensive Global Genome Dynamics of Chlamydia trachomatis Show Ancient Diversification Followed by Contemporary Mixing and Recent Lineage Expansion. Genome Res. 2017, 27, 1220–1229. [Google Scholar] [CrossRef]
- Brunelle, B.W.; Sensabaugh, G.F. Nucleotide and Phylogenetic Analyses of the Chlamydia trachomatis OmpA Gene Indicates It Is a Hotspot for Mutation. BMC Res. Notes 2012, 5, 53. [Google Scholar] [CrossRef][Green Version]
- Marangoni, A.; Foschi, C.; Tartari, F.; Gaspari, V.; Re, M.C. Lymphogranuloma Venereum Genovariants in Men Having Sex with Men in Italy. Sex. Transm. Infect. 2021, 97, 441–445. [Google Scholar] [CrossRef]
- Olivia Peuchant, A.T.; Clément Sperandio, N.H.; Cécile Laurier-Nadalié, C.B.; de Barbeyrac, B. Changing Pattern of Chlamydia trachomatis Strains in Lymphogranuloma Venereum Outbreak, France, 2010–2015. Emerg. Infect. Dis. 2016, 22, 1945–1947. [Google Scholar] [CrossRef]
- Piñeiro, L.; Bernal, S.; Bordes, A.; Palomares, J.C.; Gilarranz, R.; von Wichmann, M.A.; Cilla, G. Minimum Spread of the New Swedish Variant of Chlamydia trachomatis and Distribution of C. Trachomatis OmpA Genotypes in Three Geographically Distant Areas of Spain, 2011–2012. Infection 2014, 42, 905–912. [Google Scholar] [CrossRef]
- Christerson, L.; de Vries, H.J.C.; de Barbeyrac, B.; Gaydos, C.A.; Henrich, B.; Hoffmann, S.; Schachter, J.; Thorvaldsen, J.; Vall-Mayans, M.; Klint, M.; et al. Typing of Lymphogranuloma Venereum Chlamydia trachomatis Strains. Emerg. Infect. Dis. 2010, 16, 1777–1779. [Google Scholar] [CrossRef]
- Rodríguez-Domínguez, M.; Puerta, T.; Menéndez, B.; González-Alba, J.M.; Rodríguez, C.; Hellín, T.; Vera, M.; González-Sainz, F.J.; Clavo, P.; Villa, M.; et al. Clinical and Epidemiological Characterization of a Lymphogranuloma Venereum Outbreak in Madrid, Spain: Co-Circulation of Two Variants. Clin. Microbiol. Infect. 2014, 20, 219–225. [Google Scholar] [CrossRef]
- Kendall, B.A.; Tardif, K.D.; Schlaberg, R. Chlamydia trachomatis L. Serovars and Dominance of Novel L2b OmpA Variants, U.S.A. Sex. Transm. Infect. 2014, 90, 336. [Google Scholar] [CrossRef]
- Gomes, J.P.; Nunes, A.; Florindo, C.; Ferreira, M.A.; Santo, I.; Azevedo, J.; Borrego, M.J. Lymphogranuloma Venereum in Portugal: Unusual Events and New Variants during 2007. Sex. Transm. Dis. 2009, 36, 88–91. [Google Scholar] [CrossRef]
- Touati, A.; Peuchant, O.; Hénin, N.; Bébéar, C.; de Barbeyrac, B. The L2b Real-Time PCR Targeting the PmpH Gene of Chlamydia trachomatis Used for the Diagnosis of Lymphogranuloma Venereum Is Not Specific to L2b Strains. Clin. Microbiol. Infect. 2016, 22, 574-e7. [Google Scholar] [CrossRef][Green Version]
- Borges, V.; Cordeiro, D.; Salas, A.I.; Lodhia, Z.; Correia, C.; Isidro, J.; Fernandes, C.; Rodrigues, A.M.; Azevedo, J.; Alves, J.; et al. Chlamydia Trachomatis: When the Virulence-Associated Genome Backbone Imports a Prevalence-Associated Major Antigen Signature. Microb. Genom. 2019, 5, e000313. [Google Scholar] [CrossRef] [PubMed]
- Somboonna, N.; Wan, R.; Ojcius, D.M.; Pettengill, M.A.; Joseph, S.J.; Chang, A.; Hsu, R.; Read, T.D.; Dean, D. Hypervirulent Chlamydia trachomatis Clinical Strain Is a Recombinant between Lymphogranuloma Venereum (L(2)) and D Lineages. MBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Seth-Smith, H.M.; Galán, J.C.; Goldenberger, D.; Lewis, D.A.; Peuchant, O.; Bébéar, C.; De Barbeyrac, B.; Bénard, A.; Carter, I.; Kok, J.; et al. Concern Regarding the Alleged Spread of Hypervirulent Lymphogranuloma Venereum Chlamydia trachomatis Strain in Europe. Eurosurveillance 2017, 22, 30511. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.J.; Didelot, X.; Rothschild, J.; De Vries, H.J.C.; Morré, S.A.; Read, T.D.; Dean, D. Population Genomics of Chlamydia Trachomatis: Insights on Drift, Selection, Recombination, and Population Structure. Mol. Biol. Evol. 2012, 29, 3933–3946. [Google Scholar] [CrossRef] [PubMed]
- Stary, G.; Meyer, T.; Bangert, C.; Kohrgruber, N.; Gmeinhart, B.; Kirnbauer, R.; Jantschitsch, C.; Rieger, A.; Stary, A.; Geusau, A. New Chlamydia trachomatis L2 Strains Identified in a Recent Outbreak of Lymphogranuloma Venereum in Vienna, Austria. Sex. Transm. Dis. 2008, 35, 377–382. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).