Abstract
Microbial communities play an important role in the fitness of mosquito hosts. However, the factors shaping microbial communities in wild populations, with regard to interactions among microbial species, are still largely unknown. Previous research has demonstrated that two of the most studied mosquito symbionts, the bacteria Wolbachia and Asaia, seem to compete or not compete, depending on the genetic background of the reference mosquito host. The large diversity of Wolbachia–Asaia strain combinations that infect natural populations of mosquitoes may offer a relevant opportunity to select suitable phenotypes for the suppression of pathogen transmission and for the manipulation of host reproduction. We surveyed Wolbachia and Asaia in 44 mosquito populations belonging to 11 different species of the genera Anopheles, Aedes, and Culex using qualitative PCR. Through quantitative PCR, the amounts of both bacteria were assessed in different mosquito organs, and through metagenomics, we determined the microbiota compositions in some selected mosquito populations. We show that variation in microbial community structure is likely associated with the species/strain of mosquito, its geographical position, and tissue localization. Together, our results shed light on the interactions among different bacterial species in the microbial communities of mosquito vectors, and this can aid the development and/or improvement of methods for symbiotic control of insect vectors.
1. Introduction
Microbial interactions within mosquitoes of different species can have significant effects both on the physiology of the host, and more generally on their biology, and on their susceptibility to pathogens. At the same time, these interactions can condition the effectiveness of control methods based on symbionts, the so-called symbiotic control (SC) [1,2]. From this point of view, little is known about how these interactions can interfere with the efficacy of different SC methods.
Much of this is because the existing studies were mainly conducted using laboratory-bred mosquito populations, which, obviously, can also be very significantly different from field mosquitoes due to the composition of their microbiota [3].
Two bacteria have attracted great interest for their potential in the control of mosquito-borne diseases (MBDs): Wolbachia, obligate intracellular bacteria found in many insect species, and Asaia, Gram-negative bacteria also widely distributed in insects [4,5,6].
The potential of Wolbachia in controlling MBDs has been proven for years, having been corroborated by numerous studies. In fact, the “forced” introduction of some strains of Wolbachia in Aedes aegypti has shown great efficacy in reducing dengue virus and its competence for transmission [7,8,9,10]. It has also been consistently demonstrated that Ae. aegypti, being resistant to dengue virus infection, is capable of rapidly displacing natural/susceptible populations. Consequently, dengue control with Wolbachia-based strategies is still ongoing today in various regions of the world [8,9,10,11,12]. Similar approaches have also been proposed for other mosquito species, including some malaria vectors [13,14].
Concerning Asaia, its strict ecological association with many different mosquito species has attracted much interest in the frame of the paratransgenic control of malaria and other MBDs. Indeed, Asaia may infect most of the members of a population if not all, including all developmental stages and several anatomical districts, thus acclaiming itself as one of the best paratransgenic agents. In this frame, at the laboratory level, paratransgenic strains of Asaia that inhibit malaria transmission have been produced [15,16]. Moreover, Asaia can stimulate the basal level of mosquito immunity to naturally reduce the development of malaria parasite oocysts in Anopheles stephensi, thus expanding its potential in SC approaches, not only through paratransgenesis, but also as a potential effector for insect immune priming [17,18]. The establishment of Wolbachia in the host mosquito can be inhibited by the co-presence of other bacteria, which could limit its effectiveness in controlling some MBDs [19]. In some genetic backgrounds, the presence of Wolbachia, and therefore its potential control efficacy, is related to the absence/presence of Asaia strains. A possible explanation for this competition could lie in the fact that Asaia and Wolbachia potentially compete for the same resources, but this still needs to be better clarified. Nevertheless, experimental evidence of a competition between the two symbionts has been reported in mosquitoes of the genus Anopheles and in Ae. aegypti [20,21,22]. In order to be able to evaluate the effectiveness of control methods based on symbionts, and, in this specific case, on Asaia and Wolbachia, a detailed study of the distribution of these two bacteria in different species/strains of mosquitoes coming from regions characterized by different eco-ethological contexts has been performed. Indeed, the large diversity of Wolbachia–Asaia strain combinations able to infect natural populations of mosquitoes may offer a relevant opportunity to select suitable phenotypes for the suppression of pathogen transmission and for the manipulation of host reproduction. Here, we surveyed Wolbachia and Asaia in 44 mosquito populations using molecular tools, showing that variation in microbial community structure is likely associated with several factors like mosquito species/strains, localization within the mosquito tissues, and the geographical localization of the mosquito.
2. Materials and Methods
2.1. Mosquito Collection and Identification
Mosquitoes were collected in four locations in 2022 and 2023: Italy, Cameroon (Africa), Crete (Greece), and Ohio (USA) (Table S1).
2.1.1. European Collections
In Italy, Aedes koreicus and Aedes. japonicus were collected in Feltre, Pedavena, Sospirolo, and Alano di Piave (Veneto region), whereas Aedes albopictus were collected in Sospirolo, Pedavena, Feltre, and Petriolo (Marche region). Mosquito larvae were mainly collected from artificial containers by dipping and delivered to the laboratory in a box fridge. Mosquitoes were identified morphologically according to Montarsi et al., 2013 [23] and molecularly as described in Schneider et al., 2016 [24]. Culex pipiens mosquitoes were collected in Camerino (Marche), Pedavena (Veneto), and Pisa (Toscana). The mosquito genera were evaluated using morphological keys [25], whereas the species were molecularly identified as described in Fotakis et al., 2022 [26].
In Crete, Ae. albopictus specimens were collected in Gazi, Heraklion, Rethymnon, and Hersonissos; Cx. pipiens specimens were collected in Rethymnon, Heraklion, and Hersonissos, using BG-Sentinel 2 traps (Blue line S.r.l, Forlì, Italy). The mosquito genera were evaluated using morphological keys [25], whereas the species were molecularly identified as described in Fotakis et al., 2022 [26].
2.1.2. African Collections
In Cameroon, Anopheles gambiae, Anopheles coluzzii, Anopheles arabiensis, Anopheles pharoensis, Anopheles ziemanni, and Ae. albopictus were collected from 7 locations belonging to 5 different eco-geographical areas (Santchou, Yaoundé, Adamaoua, Douala, Mbandjock, Mbalmayo, and Yangah). Mosquitoes were collected using Center for Disease Control Light Traps, Human Landing Catches, and Prokopack aspirators. Adults were identified morphologically using the identification keys [27,28]. An. gambiae complex specimens were identified using the SYBR green-based assay described by Chabi et al., 2019 [29]. An. phaorensis and An. ziemanni samples were sequenced at cytochrome C oxidase subunit 1 (COI) loci for species confirmation [30].
A cohort of Ae. aegypti and Anopheles funestus collected in Ouagadougou (Burkina Faso) in 2008 were included in the study.
2.1.3. USA Collections
In Ohio, Ae. japonicus mosquitoes were collected in Wooster city using gravid traps baited with yeasts. Mosquitoes were morphologically and molecularly identified as described in Nanfack-Minkeu et al., 2023 [31].
2.2. DNA Extraction
Before the DNA extraction, the insect surface was sterilized in 70% ethanol and rinsed for three times in sterile PBS. Samples were homogenized with sterile 1 mm wide glass beads (Next Advance Inc., New York, NY, USA) for 30 s at 6800 rpm with an automatic tissue homogenizer (Precellys 24, Bertin Instrument, Montigny-le-Bretonneux, France). Genomic DNA was extracted using a JetFlex Genomic DNA Purification kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The concentration and purity of the DNA was determined using a NanoDrop™ spectrophotometer (Thermo Scientific, Waltham, MA, USA). Finally, the DNA was stored at −20 °C prior to analysis.
2.3. Wolbachia and Asaia Detection
A total of 1098 mosquitoes were tested to detect the presence of the bacteria Asaia and Wolbachia using specific oligonucleotides (Table 1). For Wolbachia, a semi-nested PCR targeting the Wolbachia 16S rRNA gene was performed using 50 ng genomic DNA, 1X Buffer, 0.25 mM dNTPs, 0.9U DreamTaq Polymerase (Thermo Scientific, Waltham, MA, USA), 240 nM Wol-For, 160 nM Wol-rev2, and 120 nM Wol-rev3 [21]. The amplification cycle consisted of an initial denaturation at 95 °C for 3 min, followed by 5 cycles consisting of denaturation at 95 °C for 30 s, annealing at 54 °C for 30 s, and extension at 72 °C for 30 s, and 25 cycles consisting of denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 30 s, concluding with a final extension step of 10 min at 72 °C. For Asaia, specific PCR targeting of the 16S rRNA gene was performed using 50 ng genomic DNA, 1X Buffer, 0.25 mM dNTPs, 0.9U DreamTaq Polymerase (Thermo Scientific, Waltham, MA, USA), and 200 nM of AsaiaNewFor and AsaiaNewRev oligonucleotides [5]. The amplification protocol included initial denaturation at 95 °C for 3 min, followed by 30 cycles consisting of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s, concluding with a final extension step of 10 min at 72 °C. The PCR products were electrophoresed on a 1% agarose gel to verify the presence and size of the specific amplicons.

Table 1.
Oligonucleotides used in the study.
2.4. Metagenomics Analysis
For each of the species analyzed, the microbiota profiles of 10 single mosquitoes were measured. Ae. japonicus, collected in Italy and Ohio, and Ae. albopictus and Cx. pipiens, collected in Italy and Crete, were analyzed through NGS analysis using the bacterial target 16S rRNA gene. Blank extractions were analyzed as negative controls to evaluate possible bacterial contaminations occurring during the template preparation. Moreover, 16S rRNA gene profiling was conducted with SYNBIOTEC srl (Camerino, Italy). Library preparation was performed by covering the hypervariable region V3-V4 of the 16S rRNA gene using the oligonucleotides 341F and 785R [33]. The data were pre-processed using the Illumina MiSeq—2 × 250 PE—V2 nano, and the readings were sorted by amplicon in line barcodes. No quantifiable libraries were produced from the negative controls.
Quality control, taxonomic attribution and diversity, and abundance analyses were performed in Qiime2 version 2023.5 [34]. Qiime tools were imported and the “CasavaOneEightSingleLanePerSampleDirFmt” format was used to import the 16S sequences. Readings were merged with the qiime vsearch merge-pair. Quality control and denoising were performed using qiime quality filter q-score and qiime deblur denoise 16S. For the taxonomy identification, Silva138 [35] and qiime feature-classifier classify-sklearn were used. Plots were obtained with qiime taxa barplot and GraphPad (https://www.graphpad.com/) (accessed on 14 February 2024). Rarefaction curves, Shannon alpha diversity, and Bray–Curtis PCoA were obtained with qiime diversity alpha-rarefaction, qiime diversity alpha-group-significance, and qiime emperor plot. The data presented in this study are deposited in the NCBI repository (accession number PRJNA1069599).
2.5. Asaia and Wolbachia Quantification via qPCR
Asaia and Wolbachia density was evaluated in organs (male and female guts, male and female reproductive organs, and female salivary glands) of Ae. albopictus (collected in Petriolo, Marche region), Cx. pipiens (collected in Camerino, Marche region), and An. stephensi (laboratory colony) via qPCR. Eight pools of 10 organs were obtained by dissecting 15-day-old mosquitoes in a drop of sterile 1× PBS using sterile needles under a stereomicroscope (Olympus, Tokyo, Japan). Samples were homogenized and the DNA was extracted as described above. PCR assays were performed using 1XHOT FIREPol® EvaGreen® qPCR Supermix (Tartu, Estonia), 200 nM of oligonucleotides, and 50 ng of genomic DNA. Specific oligonucleotides targeting the 16S DNA were used to quantify Asaia and Wolbachia. Moreover, the genes Ae-rps7, Cx-rps3, and As-rps7 were amplified as housekeeping genes (for Ae. albopictus, Cx. pipiens, and An. stephensi, respectively) [21,32]. All gene sequences are summarized in Table 1. Reactions were run on a CFX thermocycler (Bio-Rad, Hercules, CA, USA) using the following cycling conditions: 1 cycle of 95 °C for 12 min, 40 cycles of 95 °C for 1 min, 60 °C for 1 min, and 74 °C for 30 s. The melting peak for each target was obtained with the following steps: 65 °C to 95 °C for 5 s with an increment of 0.5 °C. The quantity of amplified targets was measured using standard curves obtained by eight serial dilutions of specific plasmids for each amplicon (from 2 to 2 × 10−7 ng). The standard curves used in the experiments had the following parameters (E = efficiency; R2 = correlation coefficient): Asaia: E = 91.8%, R2 = 0.9983; Wolbachia: E = 91.2%, R2 = 0.997; Ae-rps7: E = 99.5, R2 = 0.9996; Cx-rps3: E = 100.6, R2 = 0.9985; and As-rps7: E = 96.2, R2 = 0.999. Amounts of Asaia and Wolbachia were estimated as relative quantities, calculating the number gene copy ratio (number gene copy of 16S rRNA/number gene copy of housekeeping gene). The amounts of Asaia and Wolbachia were estimated using the Bio-Rad CFX Maestro Software 2.3 (version 5.3.022.1030) and the GraphPad software (http://www.graphpad.com) (accessed on 16 February 2024). Data were obtained from the average of eight pools per organ. The value of each pool resulted from the average of two technical replicates that were compared through the Mann–Whitney test.
3. Results
3.1. Asaia–Wolbachia Distribution in Different Mosquito Populations
Specific PCR tests were used to verify the circulation of the two symbionts in different species and populations of mosquitoes belonging to the genera Anopheles, Aedes, and Culex.
Concerning the genus Anopheles, Asaia was detected in all species and populations tested, while Wolbachia was never detected (Table 2). The relative prevalence of Asaia is highly variable and seems to be dependent on the geographical region of Cameroon in which the mosquitoes were collected. Indeed, those collected in Yangah and Amadoua show relative prevalence rates spanning from 2.1 to 29.2%, while those collected in other areas of Cameroon show prevalence rates spanning from about 33 to 100%. In An. funestus collected in Burkina Faso, Asaia was detected in 46.4% of the samples.

Table 2.
Samples analyzed for Asaia and Wolbachia.
Concerning the genus Aedes, Asaia was detected in all of the populations tested (Table 2). In all Ae. albopictus populations, the prevalence of Wolbachia was 100%, and the prevalence of Asaia ranged from 91 to 100%. In Ae. aegypti, Ae. koreicus, and Ae. japonicus, the prevalences of Asaia were 42.9%, 97–100%, and 97.4–100%, respectively, while Wolbachia was never detected.
An exception to these data is represented by the circulation of the two symbionts in the populations collected in Greece, on the island of Crete. The populations of Ae. albopictus analyzed always revealed the presence of Wolbachia in all of the tested insects. As regards Asaia, the percentages fluctuated from 0 to 61.5%.
Moreover, in Ae. japonicus, the circulation of Asaia detected in the Italian populations (from 97.4 to 100%) was much higher than that recorded in the American population (49%). In consideration of this, two comparative metabarcoding analyses were conducted: the first compared the Italian Ae. albopictus with its Greek counterpart; the second compared Ae. japonicus collected in the field in Italy versus in the United States. Both analyses revealed that the composition of the microbiota varied between the compared populations with a more than likely impact on the circulation of Asaia. Nevertheless, while the microbiota of the Italian and Greek populations of Ae. albopictus were dominated by the presence of Wolbachia, and therefore their differences concern a quantitative minority share of the microbiota represented by various bacterial species (Figure 1A), the microbiota of the Italian and North American populations of Ae. japonicus differed markedly. In fact, while Asaia was largely the predominant bacterium in the Italian population, Pseudomonas prevailed in the American population (Figure 1B).
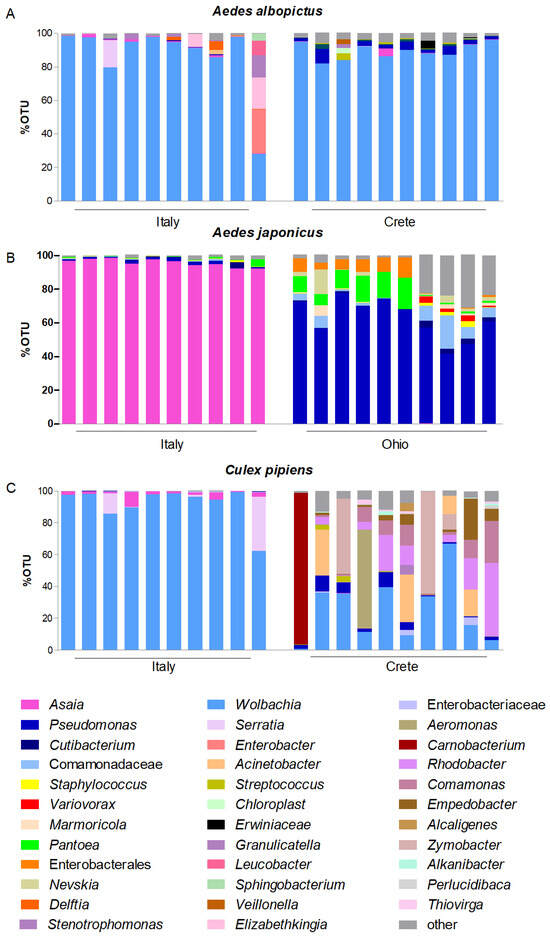
Figure 1.
Stacked bar plots showing the relative abundances of bacterial taxa distributed among mosquito species: (A) Aedes albopictus; (B) Ae. japonicus; (C) Cx. pipiens. X-axis indicates the mosquito samples and Y-axis indicates the relative abundance of bacterial taxa calculated as a percentage of the Operational Taxonomic Units (OTUs). Only OTUs > 2% of the total readings are represented.
Concerning the genus Culex, both Asaia and Wolbachia were detected in all of the Italian and Greek populations of Cx. pipiens tested (Table 2). In these populations, the prevalence of Wolbachia was 100% in all of the populations, while Asaia was found in 100% of the Italian mosquitoes, with a range of 36.4 to 54.5% in the Greek ones.
Similarly to the findings in some Aedes populations, the circulation of Asaia detected in the Italian populations of Cx. pipiens was higher than that recorded in the Greek populations. Thus, comparative metagenomic analyses were conducted, revealing that the microbiota associated with the two populations differed very substantially: while in the Italian population Wolbachia was definitely the predominant bacterium in all of the samples analyzed, confining Asaia to a limited quantity, in the Greek population, the situation was much more varied, with Wolbachia being confined to decidedly more modest proportions and a greater richness of other bacteria such as Pseudomonas and Acinetobacter (Figure 1C).
Regarding the 16S results, the principal coordinates analysis (PCoA) confirmed both the large differences between mosquitoes collected in Italy compared to those collected in Ohio and Crete, and the strong similarity of the microbial composition in Ae. albopictus and Cx. pipiens collected in Italy. Furthermore, the high quantity of Asaia in the Italian population of Ae. japonicus appears to have affected the entire microbial community enough to isolate this population from the others (Figure S1).
3.2. Competition in Different Mosquito Organs
The coexistence of Wolbachia and Asaia was found only in Cx. pipiens and Ae. albopictus; therefore, we verified in which anatomical organs the competition between these two symbionts occurred through quantitative PCR. As shown in Figure 2, the comparative analysis of male and female guts, reproductive organs, and female salivary glands clearly indicated that competition occurs only in the reproductive organs of both sexes. In these organs, in both sexes and in both species, the presence of Wolbachia was much greater than that of Asaia.
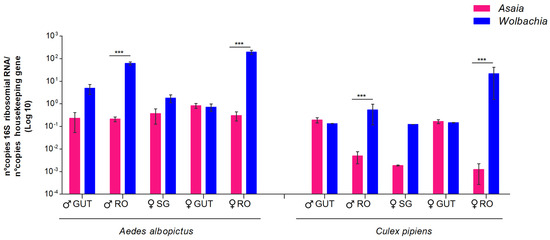
Figure 2.
Quantitative detection of Asaia and Wolbachia in organs of Ae. albopictus and Cx. pipiens mosquito species obtained via qPCR. The relative amount of the bacteria is expressed as a ratio of bacterial 16S rRNA and mosquito rps7 (Ae. albopictus) or rps3 genes (Cx. pipiens) copies in a logarithmic scale. Abundance results from the mean ± SEM of eight pools (10 organs) for each species. Statistically significant differences are represented by asterisks (p < 0.001), as determined through multiple comparisons using the Mann-Whitney test. ♀: female; ♂: male; RO: reproductive organs; SG: salivary glands.
Quantitative PCR was also used to compare the amount of Asaia in the reproductive organs and guts of mosquitoes with Wolbachia (Ae. albopictus) and without Wolbachia (An. stephensi). We recorded a notably greater presence of Asaia in both the guts and reproductive organs of both sexes in An. stephensi (Figure 3).
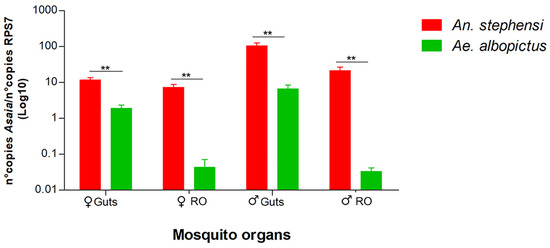
Figure 3.
Quantitative detection of Asaia in organs of An. stephensi and Ae. albopictus mosquito species obtained via qPCR. The relative amount of the bacteria is expressed as a ratio of bacterial 16S rRNA and mosquito rps7 genes (An. stephensi and Ae. albopictus) copies in a logarithmic scale. Abundance results from the mean ± SEM of eight pools (10 organs) for each species. Statistically significant differences are represented by asterisks (p < 0.01), as determined through multiple comparisons using the Mann–Whitney test. ♀: female; ♂: male; RO: reproductive organs.
4. Discussion
We have previously demonstrated a role for Wolbachia in preventing some mosquito species from conducting a stable and successful Asaia infection in the gonads [21]. It has also been proposed that Asaia plays a role in the difficulty of Wolbachia to infect anopheline mosquitoes [20,21].
Therefore, in this study, we analyzed mosquitoes from different geographical contexts to analyze a large diversity of mosquito populations in order to better understand the circulations of the two symbiotic bacteria in different host vectors.
In all of the anopheline populations analyzed, we never found the presence of Wolbachia. This is not surprising, since although Wolbachia has been reported to be present in some wild populations of anophelines, the infection frequencies are very low [36,37,38,39]. On the other hand, we found Asaia in all of these populations, albeit with variable infection frequencies that appeared to be strongly correlated to the geographical location of the mosquito population analyzed. For instance, we analyzed several anopheline populations collected in seven different areas in Cameroon. Interestingly, in the populations collected in two areas located in the north of the country, the circulation of Asaia was much lower than those recorded in populations collected in other areas of the nation. This would seem to suggest a circulation of Asaia somehow conditioned by eco-ethological factors and not only by the host species.
In the analyzed Aedes species, the situation was substantially different and much more varied. In Ae. aegypti, Ae. koreicus, and Ae. japonicus, we did not record the presence of Wolbachia, while Asaia was always present, albeit with significant fluctuations. These data are consistent with the literature; in fact, for Ae. aegypti, Ae. koreicus, and Ae. japonicus, there have only been episodic and very rare reports of Wolbachia infections [40,41,42,43].
On the other hand, the circulation of Asaia in these species has been strongly described [41,44,45,46]. Nonetheless, the comparison of the circulation of Asaia in North American and Italian populations of Ae. japonicus revealed very different frequencies, being significantly higher in the Italian ones.
The comparative metagenomic analysis between an Italian and a North American population highlighted that in the Italian population, Asaia was the dominant bacterium, while in that of Ohio, the large presence of Pseudomonas and Pantoea relegated Asaia in very small amounts, confirming the different flows of microbial competition in these different host populations.
For Ae. albopictus, very high infection rates of both bacteria have been confirmed and reported in many tested populations [47,48,49], and a similar situation was detected in the Culex populations we analyzed in this study.
Nonetheless, even in the Greek populations of Ae. albopictus and Cx. pipiens, the circulation of Asaia was found to be lower than in the Italian populations. The comparative metagenomic analysis between the Italian and Greek populations once again highlighted that in the Italian population, although Wolbachia was the dominant bacterium, Asaia was still present in most of the individuals, while in the Greek populations, the different microbiota composition translated into very small quantities of Asaia circulating in the host, thus confirming different patterns of microbial competition in these different host populations.
Nevertheless, these species offered us the chance to better define the kind of competition that occurs between these two symbionts: dissecting the scale of this competition at the tissue level shows that competition occurs mainly in the reproductive organs, in which the high quantity of Wolbachia seems to strongly limit the circulation of Asaia.
Indeed, the comparison of Asaia circulation between An. stephensi and Ae. albopictus seems to present a further demonstration of the mutualistically exclusive relationship between the two symbionts. In An. stephensi, in which Wolbachia was absent, the amounts of Asaia found were much higher than those found in Ae. albopictus, both in the guts and in the reproductive organs.
5. Conclusions
The circulation of Wolbachia and Asaia in mosquito vectors appears to be conditioned by different factors such as the host species, the geographical location of the vector, and the reference tissue of the mosquito. Although we cannot exclude other factors, such as the genetic characteristics of different strains of the symbionts and the eco-ethological contexts of reference of the mosquito, our study is further evidence of the competitive relationship between Asaia and Wolbachia in many mosquito vectors. Since they represent two of the most used and/or proposed symbionts for symbiotic control methods, our evidence represents a relevant contribution to understanding the Wolbachia–Asaia strain combinations able to infect natural populations of mosquitoes, aiming to select suitable phenotypes for the suppression of pathogen transmission and for the manipulation of host reproduction.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12030545/s1, Table S1: Field-collected mosquitoes included in the study; Figure S1: Principal coordinates analysis (PCoA) plots of samples, colored according to the sample sites. The comparisons among the microbial composition were analyzed using the Bray-Curtis-emperor method.
Author Contributions
Conceptualization, G.F.; methodology, M.I.K.N., A.C., C.D., M.F., M.P.A.M., F.N.-M. and C.C.; software, P.L.C.; validation, A.C., C.D., I.R. and G.F.; formal analysis, A.C.; investigation, M.I.K.N. and A.C.; resources, G.F. and C.D.; data curation, A.C.; writing—original draft preparation, G.F. and A.C.; writing—review and editing, C.D. and I.R.; visualization, A.C. and G.F.; supervision, G.F.; project administration, G.F.; funding acquisition, G.F. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the European Union—NextGenerationEU, with the grant PRIN 2022 PNRR MUR P20225TJWB to G.F. Marie Paul Audrey Mayi was supported by a long-term fellowship from the European Molecular Biology Organization (EMBO) (EMBO ALTF 369-2022). The funding body did not have any role in the experimental design, collection of data, analysis, or interpretation of data, or in the writing of the manuscript.
Data Availability Statement
All of the readings related to the 16S Miseq analysis (Bioproject PRJNA1069599) have been deposited in the EMBL Nucleotide Sequence Database (NCBI).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wang, J.; Gao, L.; Aksoy, S. Microbiota in disease-transmitting vectors. Nat. Rev. Microbiol. 2023, 21, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Riehle, M.A.; Jacobs-Lorena, M. Using bacteria to express and display anti-parasite molecules in mosquitoes: Current and future strategies. Insect Biochem. Mol. Biol. 2005, 35, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Didion, E.M.; Doyle, M.; Benoit, J.B. Bacterial Communities of Lab and Field Northern House Mosquitoes (Diptera: Culicidae) Throughout Diapause. J. Med. Entomol. 2022, 59, 648–658. [Google Scholar] [CrossRef]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef]
- Favia, G.; Ricci, I.; Damiani, C.; Raddadi, N.; Crotti, E.; Marzorati, M.; Rizzi, A.; Urso, R.; Brusetti, L.; Borin, S.; et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl. Acad. Sci. USA 2007, 104, 9047–9051. [Google Scholar] [CrossRef] [PubMed]
- Mercant Osuna, A.; Gidley, A.; Mayi, M.P.A.; Bamou, R.; Dhokiya, V.; Antonio-Nkondjio, C.; Jeffries, C.L.; Walker, T. Diverse novel Wolbachia bacteria strains and genera-specific co-infections with Asaia bacteria in Culicine mosquitoes from ecologically diverse regions of Cameroon. Wellcome Open Res. 2023, 8, 267. [Google Scholar] [CrossRef]
- Frentiu, F.D.; Zakir, T.; Walker, T.; Popovici, J.; Pyke, A.T.; van den Hurk, A.; McGraw, E.A.; O’Neill, S.L. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl. Trop. Dis. 2014, 8, e2688. [Google Scholar] [CrossRef]
- Pinto, S.B.; Riback, T.I.S.; Sylvestre, G.; Costa, G.; Peixoto, J.; Dias, F.B.S.; Tanamas, S.K.; Simmons, C.P.; Dufault, S.M.; Ryan, P.A.; et al. Effectiveness of Wolbachia-infected mosquito deployments in reducing the incidence of dengue and other Aedes-borne diseases in Niterói, Brazil: A quasi-experimental study. PLoS Negl. Trop. Dis. 2021, 15, e0009556. [Google Scholar] [CrossRef]
- Utarini, A.; Indriani, C.; Ahmad, R.A.; Tantowijoyo, W.; Arguni, E.; Ansari, M.R.; Supriyati, E.; Wardana, D.S.; Meitika, Y.; Ernesia, I.; et al. Efficacy of Wolbachia-Infected Mosquito Deployments for the Control of Dengue. N. Engl. J. Med. 2021, 384, 2177–2186. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Ahmad, N.W.; Keong, W.M.; Ling, C.Y.; Ahmad, N.A.; Golding, N.; Tierney, N.; Jelip, J.; Putit, P.W.; Mokhtar, N.; et al. Introduction of Aedes aegypti mosquitoes carrying wAlbB Wolbachia sharply decreases dengue incidence in disease hotspots. iScience 2024, 27, 108942. [Google Scholar] [CrossRef]
- Ahmad, N.A.; Mancini, M.V.; Ant, T.H.; Martinez, J.; Kamarul, G.M.R.; Nazni, W.A.; Hoffmann, A.A.; Sinkins, S.P. Wolbachia strain w AlbB maintains high density and dengue inhibition following introduction into a field population of Aedes aegypti. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 20190809. [Google Scholar] [CrossRef] [PubMed]
- Nazni, W.A.; Hoffmann, A.A.; NoorAfizah, A.; Cheong, Y.L.; Mancini, M.V.; Golding, N.; Kamarul, G.M.R.; Arif, M.A.K.; Thohir, H.; NurSyamimi, H.; et al. Establishment of Wolbachia Strain wAlbB in Malaysian Populations of Aedes aegypti for Dengue Control. Curr. Biol. 2019, 29, 4241–4248.e4245. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Joshi, D.; Dong, Y.; Lu, P.; Zhou, G.; Pan, X.; Xu, Y.; Dimopoulos, G.; Xi, Z. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science 2013, 340, 748–751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xi, Z. Wolbachia Transinfection Via Embryonic Microinjection. Methods Mol. Biol. 2024, 2739, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Shane, J.L.; Grogan, C.L.; Cwalina, C.; Lampe, D.J. Blood meal-induced inhibition of vector-borne disease by transgenic microbiota. Nat. Commun. 2018, 9, 4127. [Google Scholar] [CrossRef] [PubMed]
- Grogan, C.; Bennett, M.; Lampe, D.J. An evaluation of fusion partner proteins for paratransgenesis in Asaia bogorensis. PLoS ONE 2022, 17, e0273568. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, A.; Damiani, C.; Mancini, M.V.; Valzano, M.; Rossi, P.; Serrao, A.; Ricci, I.; Favia, G. Asaia Activates Immune Genes in Mosquito Eliciting an Anti-Plasmodium Response: Implications in Malaria Control. Front. Genet. 2019, 10, 836. [Google Scholar] [CrossRef] [PubMed]
- Gonella, E.; Mandrioli, M.; Tedeschi, R.; Crotti, E.; Pontini, M.; Alma, A. Activation of Immune Genes in Leafhoppers by Phytoplasmas and Symbiotic Bacteria. Front. Physiol. 2019, 10, 795. [Google Scholar] [CrossRef]
- Pascar, J.; Middleton, H.; Dorus, S. Aedes aegypti microbiome composition covaries with the density of Wolbachia infection. Microbiome 2023, 11, 255. [Google Scholar] [CrossRef]
- Hughes, G.L.; Dodson, B.L.; Johnson, R.M.; Murdock, C.C.; Tsujimoto, H.; Suzuki, Y.; Patt, A.A.; Cui, L.; Nossa, C.W.; Barry, R.M.; et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 2014, 111, 12498–12503. [Google Scholar] [CrossRef]
- Rossi, P.; Ricci, I.; Cappelli, A.; Damiani, C.; Ulissi, U.; Mancini, M.V.; Valzano, M.; Capone, A.; Epis, S.; Crotti, E.; et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit. Vectors 2015, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Andreychuk, S.; Yakob, L. Mathematical modelling to assess the feasibility of Wolbachia in malaria vector biocontrol. J. Theor. Biol. 2022, 542, 111110. [Google Scholar] [CrossRef] [PubMed]
- Montarsi, F.; Martini, S.; Dal Pont, M.; Delai, N.; Ferro Milone, N.; Mazzucato, M.; Soppelsa, F.; Cazzola, L.; Cazzin, S.; Ravagnan, S.; et al. Distribution and habitat characterization of the recently introduced invasive mosquito Aedes koreicus [Hulecoeteomyia koreica], a new potential vector and pest in north-eastern Italy. Parasit. Vectors 2013, 6, 292. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Valentini, A.; Dejean, T.; Montarsi, F.; Taberlet, P.; Glaizot, O.; Fumagalli, L. Detection of Invasive Mosquito Vectors Using Environmental DNA (eDNA) from Water Samples. PLoS ONE 2016, 11, e0162493. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.; Petrić, D.; Zgomba, M.; Boase, C.; Madon, M.; Dahl, C.; Kaiser, A. Mosquitoes: Identification, Ecology and Control, 3rd ed.; Springer Nature: Cham, Switzerland, 2020. [Google Scholar]
- Fotakis, E.A.; Mavridis, K.; Kampouraki, A.; Balaska, S.; Tanti, F.; Vlachos, G.; Gewehr, S.; Mourelatos, S.; Papadakis, A.; Kavalou, M.; et al. Mosquito population structure, pathogen surveillance and insecticide resistance monitoring in urban regions of Crete, Greece. PLoS Negl. Trop. Dis. 2022, 16, e0010186. [Google Scholar] [CrossRef] [PubMed]
- Gillies, M.; De Meillon, B. The Anophelinae of Africa South of the Sahara; South African Institute for Medical Research: Johannesburg, South Africa, 1968; p. 343. [Google Scholar]
- Gillies, M.; Coetzee, M. Supplement to the anophelinae of Africa south of the Sahara (afrotropical region). S. Afr. Inst. Med. Res. 1987, 55, 143. [Google Scholar]
- Chabi, J.; Van’t Hof, A.; N’dri, L.K.; Datsomor, A.; Okyere, D.; Njoroge, H.; Pipini, D.; Hadi, M.P.; de Souza, D.K.; Suzuki, T.; et al. Rapid high throughput SYBR green assay for identifying the malaria vectors Anopheles arabiensis, Anopheles coluzzii and Anopheles gambiae s.s. Giles. PLoS ONE 2019, 14, e0215669. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Nanfack-Minkeu, F.; Delong, A.; Luri, M.; Poelstra, J.W. Invasive Aedes japonicus Mosquitoes Dominate the Aedes Fauna Collected with Gravid Traps in Wooster, Northeastern Ohio, USA. Insects 2023, 14, 56. [Google Scholar] [CrossRef]
- Capone, A.; Ricci, I.; Damiani, C.; Mosca, M.; Rossi, P.; Scuppa, P.; Crotti, E.; Epis, S.; Angeletti, M.; Valzano, M.; et al. Interactions between Asaia, Plasmodium and Anopheles: New insights into mosquito symbiosis and implications in malaria symbiotic control. Parasit. Vectors 2013, 6, 182. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Baldini, F.; Rougé, J.; Kreppel, K.; Mkandawile, G.; Mapua, S.A.; Sikulu-Lord, M.; Ferguson, H.M.; Govella, N.; Okumu, F.O. First report of natural Wolbachia infection in the malaria mosquito Anopheles arabiensis in Tanzania. Parasit. Vectors 2018, 11, 635. [Google Scholar] [CrossRef] [PubMed]
- Sawadogo, S.P.; Kabore, D.A.; Tibiri, E.B.; Hughes, A.; Gnankine, O.; Quek, S.; Diabaté, A.; Ranson, H.; Hughes, G.L.; Dabiré, R.K. Lack of robust evidence for a Wolbachia infection in Anopheles gambiae from Burkina Faso. Med. Vet. Entomol. 2022, 36, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Liew, J.W.K.; Wong, W.K.; Pramasivan, S.; Mohamed Hassan, N.; Wan Sulaiman, W.Y.; Jeyaprakasam, N.K.; Leong, C.S.; Low, V.L.; Vythilingam, I. Natural Wolbachia infection in field-collected Anopheles and other mosquito species from Malaysia. Parasit. Vectors 2020, 13, 414. [Google Scholar] [CrossRef] [PubMed]
- Waymire, E.; Duddu, S.; Yared, S.; Getachew, D.; Dengela, D.; Bordenstein, S.R.; Balkew, M.; Zohdy, S.; Irish, S.R.; Carter, T.E. Wolbachia 16S rRNA haplotypes detected in wild Anopheles stephensi in eastern Ethiopia. Parasit. Vectors 2022, 15, 178. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.A.; Robinson, K.L.; Yang, Q.; Callahan, A.G.; Schmidt, T.L.; Axford, J.K.; Coquilleau, M.P.; Staunton, K.M.; Townsend, M.; Ritchie, S.A.; et al. A decade of stability for wMel Wolbachia in natural Aedes aegypti populations. PLoS Pathog. 2022, 18, e1010256. [Google Scholar] [CrossRef]
- Damiani, C.; Cappelli, A.; Comandatore, F.; Montarsi, F.; Serrao, A.; Michelutti, A.; Bertola, M.; Mancini, M.V.; Ricci, I.; Bandi, C.; et al. Wolbachia in Aedes koreicus: Rare Detections and Possible Implications. Insects 2022, 13, 216. [Google Scholar] [CrossRef]
- Muturi, E.J.; Ramirez, J.L.; Rooney, A.P.; Kim, C.H. Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl. Trop. Dis. 2017, 11, e0005377. [Google Scholar] [CrossRef]
- Rosso, F.; Tagliapietra, V.; Albanese, D.; Pindo, M.; Baldacchino, F.; Arnoldi, D.; Donati, C.; Rizzoli, A. Reduced diversity of gut microbiota in two Aedes mosquitoes species in areas of recent invasion. Sci. Rep. 2018, 8, 16091. [Google Scholar] [CrossRef]
- Alfano, N.; Tagliapietra, V.; Rosso, F.; Manica, M.; Arnoldi, D.; Pindo, M.; Rizzoli, A. Changes in Microbiota Across Developmental Stages of. Front. Microbiol. 2019, 10, 2832. [Google Scholar] [CrossRef]
- Crotti, E.; Damiani, C.; Pajoro, M.; Gonella, E.; Rizzi, A.; Ricci, I.; Negri, I.; Scuppa, P.; Rossi, P.; Ballarini, P.; et al. Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ. Microbiol. 2009, 11, 3252–3264. [Google Scholar] [CrossRef] [PubMed]
- Möhlmann, T.W.R.; Vogels, C.B.F.; Göertz, G.P.; Pijlman, G.P.; Ter Braak, C.J.F.; Te Beest, D.E.; Hendriks, M.; Nijhuis, E.H.; Warris, S.; Drolet, B.S.; et al. Impact of Gut Bacteria on the Infection and Transmission of Pathogenic Arboviruses by Biting Midges and Mosquitoes. Microb. Ecol. 2020, 80, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Camargo, C.; Avila, F.W. Characterization of the reproductive tract bacterial microbiota of virgin, mated, and blood-fed Aedes aegypti and Aedes albopictus females. Parasit. Vectors 2021, 14, 592. [Google Scholar] [CrossRef] [PubMed]
- Zouache, K.; Raharimalala, F.N.; Raquin, V.; Tran-Van, V.; Raveloson, L.H.; Ravelonandro, P.; Mavingui, P. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 2011, 75, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Raharimalala, F.N.; Boukraa, S.; Bawin, T.; Boyer, S.; Francis, F. Molecular detection of six (endo-) symbiotic bacteria in Belgian mosquitoes: First step towards the selection of appropriate paratransgenesis candidates. Parasitol. Res. 2016, 115, 1391–1399. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).