Aberrant Bodies: An Alternative Metabolic Homeostasis Allowing Survivability?
Abstract
1. Introduction
2. Historical Overview on the Discovery of Aberrant Bodies
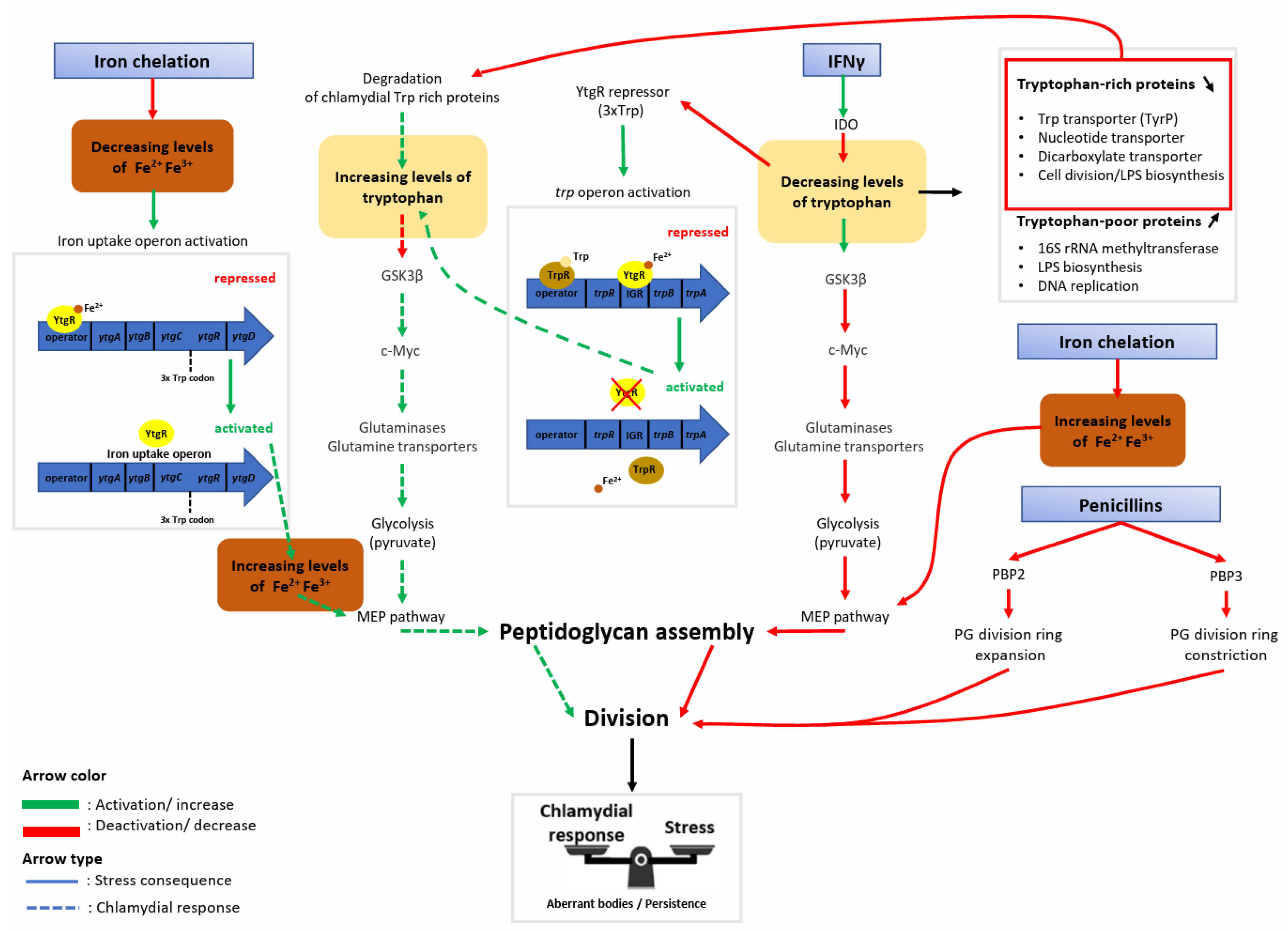
3. The Metabolic Shutdown Induced by Tryptophan Depletion Is Partially Rescued by Chlamydia’s Tryptophan Salvaging Pathway through the Preservation of Host Cell c-Myc
4. The Chlamydial Iron Uptake Operon Is Stimulated upon Iron Starvation and Is Tightly Regulated by Iron and Tryptophan Abundance
5. The Aberrant Body Morphology Induced by Iron Starvation, IFNγ, and β-Lactamase Inhibitors Is Related either to a Defect in Peptidoglycan Ring Assembly or Its Regulation
6. Conclusions/Discussion
Funding
Data Availability Statement
Conflicts of Interest
References
- PAHO/WHO|Chlamydia. Available online: https://www.paho.org/en/topics/chlamydia-infection (accessed on 17 November 2022).
- den Heijer, C.D.J.; Hoebe, C.J.P.A.; Driessen, J.H.M.; Wolffs, P.; van den Broek, I.V.F.; Hoenderboom, B.M.; Williams, R.; de Vries, F.; Dukers-Muijrers, N.H.T.M. Chlamydia trachomatis and the Risk of Pelvic Inflammatory Disease, Ectopic Pregnancy, and Female Infertility: A Retrospective Cohort Study among Primary Care Patients. Clin. Infect. Dis. 2019, 69, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Regan, L.; Greub, G. Emerging role of Chlamydia and Chlamydia-like organisms in adverse pregnancy outcomes. Curr. Opin. Infect. Dis. 2008, 21, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Goy, G.; Jaton, K.; Osterheld, M.-C.; Blumer, S.; Borel, N.; Vial, Y.; Hohlfeld, P.; Pospischil, A.; Greub, G. Role of Chlamydia trachomatis in miscarriage. Emerg. Infect. Dis. 2011, 17, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Clarke, I.N. Evolution of Chlamydia trachomatis. Ann. N. Y. Acad. Sci. 2011, 1230, E11–E18. [Google Scholar] [CrossRef] [PubMed]
- Cunha, B.A. The atypical pneumonias: Clinical diagnosis and importance. Clin. Microbiol. Infect. 2006, 12, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Pichon, N.; Guindre, L.; Laroucau, K.; Cantaloube, M.; Nallatamby, A.; Parreau, S. Chlamydia abortus in Pregnant Woman with Acute Respiratory Distress Syndrome. Emerg. Infect. Dis. 2020, 26, 628. [Google Scholar] [CrossRef] [PubMed]
- Lamoth, F.; Greub, G. Amoebal pathogens as emerging causal agents of pneumonia. FEMS Microbiol. Rev. 2010, 34, 260–280. [Google Scholar] [CrossRef]
- Cheong, H.C.; Lee, C.Y.Q.; Cheok, Y.Y.; Tan, G.M.Y.; Looi, C.Y.; Wong, W.F. Chlamydiaceae: Diseases in Primary Hosts and Zoonosis. Microorganisms 2019, 7, 146. [Google Scholar] [CrossRef]
- Bayramova, F.; Jacquier, N.; Greub, G. Insight in the biology of Chlamydia-related bacteria. Microbes Infect. 2018, 20, 432–440. [Google Scholar] [CrossRef]
- AbdelRahman, Y.M.; Belland, R.J. The chlamydial developmental cycle. FEMS Microbiol. Rev. 2005, 29, 949–959. [Google Scholar] [CrossRef]
- Vouga, M.; Baud, D.; Greub, G. Simkania negevensis may produce long-lasting infections in human pneumocytes and endometrial cells. Pathog. Dis. 2017, 75, ftw115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scherler, A.; Jacquier, N.; Kebbi-Beghdadi, C.; Greub, G. Diverse Stress-Inducing Treatments cause Distinct Aberrant Body Morphologies in the Chlamydia-Related Bacterium, Waddlia chondrophila. Microorganisms 2020, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Kane, C.D.; Byrne, G.I. Differential Effects of Gamma Interferon on Chlamydia trachomatis Growth in Polarized and Nonpolarized Human Epithelial Cells in Culture. Infect. Immun. 1998, 66, 2349–2351. [Google Scholar] [CrossRef] [PubMed]
- Vasilevsky, S.; Gyger, J.; Piersigilli, A.; Pilloux, L.; Greub, G.; Stojanov, M.; Baud, D. Waddlia chondrophila induces systemic infection, organ pathology, and elicits Th1-associated humoral immunity in a murine model of genital infection. Front. Cell. Infect. Microbiol. 2015, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Elwell, C.; Mirrashidi, K.; Engel, J. Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 2016, 14, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Loosli, C.G.; Ritter, M.H. The pathogenesis and histopathology of air-borne pneumonitis virus infection in mice; the effect of penicillin G upon the developing lesion. J. Clin. Investig. 1948, 27, 547. [Google Scholar] [PubMed]
- Weiss, E. The effect of antibiotics on agents of the psittacosis-lymphogranuloma group. I. The effect of penicillin. J. Infect. Dis. 1950, 87, 249–263. [Google Scholar] [CrossRef]
- Hurst, E.W.; Landquist, J.K.; Melvin, P.; Peters, J.M.; Senior, N.; Silk, J.A.; Stacey, G.J. The therapy of experimental psittacosis and lymphogranuloma venereum (inguinale). Br. J. Pharmacol. Chemother. 1953, 8, 297–305. [Google Scholar] [CrossRef]
- Tamura, A.; Manire, G.P. Effect of Penicillin on the Multiplication of Meningopneumonitis Organisms (Chlamydia psittaci). J. Bacteriol. 1968, 96, 875–880. [Google Scholar] [CrossRef]
- Galasso, G.J.; Manire, G.P. Effect of antiserum and antibiotics on persistent infection of HeLa cells with meningopneumonitis virus. J. Immunol. 1961, 86, 382–385. [Google Scholar] [CrossRef]
- Matsumoto, A.; Manire, G.P. Electron Microscopic Observations on the Effects of Penicillin on the Morphology of Chlamydia psittaci. J. Bacteriol. 1970, 101, 278–285. [Google Scholar] [CrossRef]
- Beatty, W.L.; Byrne, G.I.; Morrison, R.P. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 1993, 90, 3998–4002. [Google Scholar] [CrossRef]
- Beatty, W.L.; Morrison, R.P.; Byrne, G.I. Persistent chlamydiae: From cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 1994, 58, 686–699. [Google Scholar] [CrossRef]
- De Barsy, M.; Bottinelli, L.; Greub, G. Antibiotic susceptibility of Estrella lausannensis, a potential emerging pathogen. Microbes Infect. 2014, 16, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Kebbi-Beghdadi, C.; Cisse, O.; Greub, G. Permissivity of Vero cells, human pneumocytes and human endometrial cells to Waddlia chondrophila. Microbes Infect. 2011, 13, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Vouga, M.; Baud, D.; Greub, G. Simkania negevensis, an Example of the Diversity of the Antimicrobial Susceptibility Pattern among Chlamydiales. Antimicrob. Agents Chemother. 2017, 61, e00638-17. [Google Scholar] [CrossRef] [PubMed]
- Sampo, A.; Matsuo, J.; Yamane, C.; Yagita, K.; Nakamura, S.; Shouji, N.; Hayashi, Y.; Yamazaki, T.; Yoshida, M.; Kobayashi, M.; et al. High-temperature adapted primitive Protochlamydia found in Acanthamoeba isolated from a hot spring can grow in immortalized human epithelial HEp-2 cells. Environ. Microbiol. 2014, 16, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Marquis, B.; Ardissone, S.; Greub, G. Temperature affects the host range of Rhabdochlamydia porcellionis and the infectivity of Waddlia chondrophila and Chlamydia trachomatis elementary bodies. bioRxiv 2022. [Google Scholar] [CrossRef]
- Brockett, M.R.; Liechti, G.W. Persistence Alters the Interaction between Chlamydia trachomatis and Its Host Cell. Infect. Immun. 2021, 89, e0068520. [Google Scholar] [CrossRef] [PubMed]
- Bowie, W.R. In vitro activity of clavulanic acid, amoxicillin, and ticarcillin against Chlamydia trachomatis. Antimicrob. Agents Chemother. 1986, 29, 713–715. [Google Scholar] [CrossRef]
- Pokorzynski, N.D.; Hatch, N.D.; Ouellette, S.P.; Carabeo, R.A. The iron-dependent repressor YtgR is a tryptophan-dependent attenuator of the trpRBA operon in Chlamydia trachomatis. Nat. Commun. 2020, 11, 6430. [Google Scholar] [CrossRef]
- Brinkworth, A.J.; Wildung, M.R.; Carabeo, R.A. Genomewide Transcriptional Responses of Iron-Starved Chlamydia trachomatis Reveal Prioritization of Metabolic Precursor Synthesis over Protein Translation. mSystems 2018, 3, e00184-17. [Google Scholar] [CrossRef]
- Thompson, C.C.; Carabeo, R.A. An Optimal Method of Iron Starvation of the Obligate Intracellular Pathogen, Chlamydia Trachomatis. Front. Microbiol. 2011, 2, 20. [Google Scholar] [CrossRef]
- Kahane, S.; Friedman, M.G. Reversibility of heat shock in Chlamydia trachomatis. FEMS Microbiol. Lett. 1992, 97, 25–30. [Google Scholar] [CrossRef][Green Version]
- Huang, Y.; Wurihan, W.; Lu, B.; Zou, Y.; Wang, Y.; Weldon, K.; Fondell, J.D.; Lai, Z.; Wu, X.; Fan, H. Robust Heat Shock Response in Chlamydia Lacking a Typical Heat Shock Sigma Factor. Front. Microbiol. 2022, 12, 812448. [Google Scholar] [CrossRef]
- Deka, S.; Vanover, J.; Sun, J.; Kintner, J.; Whittimore, J.; Schoborg, R.V. An early event in the herpes simplex virus type-2 replication cycle is sufficient to induce Chlamydia trachomatis persistence. Cell. Microbiol. 2007, 9, 725–737. [Google Scholar] [CrossRef]
- Deka, S.; Vanover, J.; Dessus-Babus, S.; Whittimore, J.; Howett, M.K.; Wyrick, P.B.; Schoborg, R.V. Chlamydia trachomatis enters a viable but non-cultivable (persistent) state within herpes simplex virus type 2 (HSV-2) co-infected host cells. Cell. Microbiol. 2006, 8, 149–162. [Google Scholar] [CrossRef]
- Vollmuth, N.; Schlicker, L.; Guo, Y.; Hovhannisyan, P.; Janaki-Raman, S.; Kurmasheva, N.; Schmitz, W.; Schulze, A.; Stelzner, K.; Rajeeve, K.; et al. c-Myc plays a key role in IFN-γ-induced persistence of Chlamydia trachomatis. eLife 2022, 11, e76721. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Campbell, R.; Kintner, J.; Whittimore, J.; Schoborg, R.V. Chlamydia muridarum Enters a Viable but Non-infectious State in Amoxicillin-treated BALB/c Mice. Microbes Infect. Inst. Pasteur 2012, 14, 1177–1185. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Phillips-Campbell, R.; Kintner, J.; Schoborg, R.V. Induction of the Chlamydia muridarum Stress/Persistence Response Increases Azithromycin Treatment Failure in a Murine Model of Infection. Antimicrob. Agents Chemother. 2014, 58, 1782–1784. [Google Scholar] [CrossRef] [PubMed]
- Marti, H.; Borel, N.; Dean, D.; Leonard, C.A. Evaluating the Antibiotic Susceptibility of Chlamydia—New Approaches for in Vitro Assays. Front. Microbiol. 2018, 9, 1414. [Google Scholar] [CrossRef]
- Pospischil, A.; Borel, N.; Chowdhury, E.H.; Guscetti, F. Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet. Microbiol. 2009, 135, 147–156. [Google Scholar] [CrossRef]
- Goellner, S.; Schubert, E.; Liebler-Tenorio, E.; Hotzel, H.; Saluz, H.P.; Sachse, K. Transcriptional Response Patterns of Chlamydophila psittaci in Different In Vitro Models of Persistent Infection. Infect. Immun. 2006, 74, 4801–4808. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, L.; Wang, C.; Xie, Y.; Chen, Z.; Liu, L.; Su, Z.; Wu, Y. Transcriptional Analysis of 10 Selected Genes in a Model of Penicillin G Induced Persistence of Chlamydophila psittaci in HeLa Cells. J. Microbiol. Biotechnol. 2015, 25, 1246–1256. [Google Scholar] [CrossRef]
- Moulder, J.W.; Levy, N.J.; Schulman, L.P. Persistent Infection of Mouse Fibroblasts (L Cells) with Chlamydia psittaci: Evidence for a Cryptic Chlamydial Form. Infect. Immun. 1980, 30, 874–883. [Google Scholar] [CrossRef]
- Jacquier, N.; Frandi, A.; Pillonel, T.; Viollier, P.H.; Greub, G. Cell wall precursors are required to organize the chlamydial division septum. Nat. Commun. 2014, 5, 3578. [Google Scholar] [CrossRef] [PubMed]
- Al-Younes, H.M.; Rudel, T.; Brinkmann, V.; Szczepek, A.J.; Meyer, T.F. Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell. Microbiol. 2001, 3, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Miller, R.D.; Sullivan, E.D.; Theodoropoulos, C.; Mathews, S.A.; Timms, P.; Summersgill, J.T. Protein Expression Profiles of Chlamydia pneumoniae in Models of Persistence versus Those of Heat Shock Stress Response. Infect. Immun. 2006, 74, 3853–3863. [Google Scholar] [CrossRef] [PubMed]
- Mannonen, L.; Kamping, E.; Penttilä, T.; Puolakkainen, M. IFN-γ induced persistent Chlamydia pneumoniae infection in HL and Mono Mac 6 cells: Characterization by real-time quantitative PCR and culture. Microb. Pathog. 2004, 36, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Leonard, C.A.; Dewez, F.; Borel, N. Penicillin G-Induced Chlamydial Stress Response in a Porcine Strain of Chlamydia pecorum. Int. J. Microbiol. 2016, 2016, e3832917. [Google Scholar] [CrossRef] [PubMed]
- Borel, N.; Dumrese, C.; Ziegler, U.; Schifferli, A.; Kaiser, C.; Pospischil, A. Mixed infections with Chlamydia and porcine epidemic diarrhea virus—A new in vitro model of chlamydial persistence. BMC Microbiol. 2010, 10, 201. [Google Scholar] [CrossRef]
- Philips, H.L.; Clarkson, M.J. Spontaneous change from overt to covert infection of Chlamydia pecorum in cycloheximide-treated mouse McCoy cells. Infect. Immun. 1995, 63, 3729–3730. [Google Scholar] [CrossRef]
- Rovero, A.; Kebbi-Beghdadi, C.; Greub, G. Spontaneous Aberrant Bodies Formation in Human Pneumocytes Infected with Estrella lausannensis. Microorganisms 2023, 11, 2368. [Google Scholar] [CrossRef]
- Ardissone, S.; Scherler, A.; Pillonel, T.; Martin, V.; Kebbi-Beghdadi, C.; Greub, G. Transcriptional Landscape of Waddlia chondrophila Aberrant Bodies Induced by Iron Starvation. Microorganisms 2020, 8, 1848. [Google Scholar] [CrossRef]
- Kahane, S.; Fruchter, D.; Dvoskin, B.; Friedman, M.G. Versatility of Simkania negevensis infection in vitro and induction of host cell inflammatory cytokine response. J. Infect. 2007, 55, e13–e21. [Google Scholar] [CrossRef] [PubMed]
- Rottenberg, M.E.; Gigliotti-Rothfuchs, A.; Wigzell, H. The role of IFN-γ in the outcome of chlamydial infection. Curr. Opin. Immunol. 2002, 14, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Pantoja, L.G.; Miller, R.D.; Ramirez, J.A.; Molestina, R.E.; Summersgill, J.T. Characterization of Chlamydia pneumoniae Persistence in HEp-2 Cells Treated with Gamma Interferon. Infect. Immun. 2001, 69, 7927–7932. [Google Scholar] [CrossRef] [PubMed]
- Kak, G.; Raza, M.; Tiwari, B.K. Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol. Concepts 2018, 9, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Ziklo, N.; Vidgen, M.E.; Taing, K.; Huston, W.M.; Timms, P. Dysbiosis of the Vaginal Microbiota and Higher Vaginal Kynurenine/Tryptophan Ratio Reveals an Association with Chlamydia trachomatis Genital Infections. Front. Cell. Infect. Microbiol. 2018, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Fehlner-Gardiner, C.; Roshick, C.; Carlson, J.H.; Hughes, S.; Belland, R.J.; Caldwell, H.D.; McClarty, G. Molecular Basis Defining Human Chlamydia trachomatis Tissue Tropism: A possible role for tryptophan synthase. J. Biol. Chem. 2002, 277, 26893–26903. [Google Scholar] [CrossRef] [PubMed]
- Bonner, C.; Byrne, G.; Jensen, R. Chlamydia exploit the mammalian tryptophan-depletion defense strategy as a counter-defensive cue to trigger a survival state of persistence. Front. Cell. Infect. Microbiol. 2014, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Nelson, D.E. How Chlamydia trachomatis conquered gut microbiome-derived antimicrobial compounds and found a new home in the eye. Proc. Natl. Acad. Sci. USA 2019, 116, 12136–12138. [Google Scholar] [CrossRef]
- Rajeeve, K.; Vollmuth, N.; Janaki-Raman, S.; Wulff, T.F.; Baluapuri, A.; Dejure, F.R.; Huber, C.; Fink, J.; Schmalhofer, M.; Schmitz, W.; et al. Reprogramming of host glutamine metabolism during Chlamydia trachomatis infection and its key role in peptidoglycan synthesis. Nat. Microbiol. 2020, 5, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Albert, T.; Urlbauer, B.; Kohlhuber, F.; Hammersen, B.; Eick, D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt’s lymphoma cell lines. Oncogene 1994, 9, 759–763. [Google Scholar] [PubMed]
- Hentze, M.W.; Muckenthaler, M.U.; Galy, B.; Camaschella, C. Two to tango: Regulation of Mammalian iron metabolism. Cell 2010, 142, 24–38. [Google Scholar] [CrossRef]
- Ouellette, S.; Carabeo, R. A Functional Slow Recycling Pathway of Transferrin is Required for Growth of Chlamydia. Front. Microbiol. 2010, 1, 112. [Google Scholar] [CrossRef]
- Pokorzynski, N.D.; Thompson, C.C.; Carabeo, R.A. Ironing Out the Unconventional Mechanisms of Iron Acquisition and Gene Regulation in Chlamydia. Front. Cell. Infect. Microbiol. 2017, 7, 394. [Google Scholar] [CrossRef]
- Hatch, G.M.; McClarty, G. Phospholipid Composition of Purified Chlamydia trachomatis Mimics That of the Eucaryotic Host Cell. Infect. Immun. 1998, 66, 3727–3735. [Google Scholar] [CrossRef]
- Ouellette, S.P.; Dorsey, F.C.; Moshiach, S.; Cleveland, J.L.; Carabeo, R.A. Chlamydia Species-Dependent Differences in the Growth Requirement for Lysosomes. PLoS ONE 2011, 6, e16783. [Google Scholar] [CrossRef]
- Gough, N.R. Two for One in Iron Homeostasis. Sci. Signal. 2012, 5, ec178. [Google Scholar] [CrossRef]
- Luo, Z.; Neville, S.L.; Campbell, R.; Morey, J.R.; Menon, S.; Thomas, M.; Eijkelkamp, B.A.; Ween, M.P.; Huston, W.M.; Kobe, B.; et al. Structure and Metal Binding Properties of Chlamydia trachomatis YtgA. J. Bacteriol. 2019, 202, e00580-19. [Google Scholar] [CrossRef]
- Pillonel, T.; Tagini, F.; Bertelli, C.; Greub, G. ChlamDB: A comparative genomics database of the phylum Chlamydiae and other members of the Planctomycetes-Verrucomicrobiae-Chlamydiae superphylum. Nucleic Acids Res. 2019, 48, D526–D534. [Google Scholar] [CrossRef]
- Liechti, G.; Kuru, E.; Hall, E.; Kalinda, A.; Brun, Y.V.; VanNieuwenhze, M.; Maurelli, A.T. A new metabolic cell wall labeling method reveals peptidoglycan in Chlamydia trachomatis. Nature 2014, 506, 507–510. [Google Scholar] [CrossRef]
- Moulder, J.W. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan. Infect. Agents Dis. 1993, 2, 87–99. [Google Scholar]
- Jacquier, N.; Viollier, P.H.; Greub, G. The role of peptidoglycan in chlamydial cell division: Towards resolving the chlamydial anomaly. FEMS Microbiol. Rev. 2015, 39, 262–275. [Google Scholar] [CrossRef]
- Packiam, M.; Weinrick, B.; Jacobs, W.R.; Maurelli, A.T. Structural characterization of muropeptides from Chlamydia trachomatis peptidoglycan by mass spectrometry resolves “chlamydial anomaly”. Proc. Natl. Acad. Sci. USA 2015, 112, 11660–11665. [Google Scholar] [CrossRef] [PubMed]
- Frandi, A.; Jacquier, N.; Théraulaz, L.; Greub, G.; Viollier, P.H. FtsZ-independent septal recruitment and function of cell wall remodelling enzymes in chlamydial pathogens. Nat. Commun. 2014, 5, 4200. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, Y.; Ouellette, S.P.; Belland, R.J.; Cox, J.V. Polarized Cell Division of Chlamydia trachomatis. PLoS Pathog. 2016, 12, e1005822. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, N.; Yadav, A.K.; Pillonel, T.; Viollier, P.H.; Cava, F.; Greub, G. A SpoIID Homolog Cleaves Glycan Strands at the Chlamydial Division Septum. mBio 2019, 10, e01128-19. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, N.; Frandi, A.; Viollier, P.H.; Greub, G. Disassembly of a Medial Transenvelope Structure by Antibiotics during Intracellular Division. Chem. Biol. 2015, 22, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Pilhofer, M.; Aistleitner, K.; Biboy, J.; Gray, J.; Kuru, E.; Hall, E.; Brun, Y.V.; VanNieuwenhze, M.S.; Vollmer, W.; Horn, M.; et al. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun. 2013, 4, 2856. [Google Scholar] [CrossRef]
- Liechti, G.W. Localized Peptidoglycan Biosynthesis in Chlamydia trachomatis Conforms to the Polarized Division and Cell Size Reduction Developmental Models. Front. Microbiol. 2021, 12, 733850. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.V.; Abdelrahman, Y.M.; Ouellette, S.P. Penicillin-binding proteins regulate multiple steps in the polarized cell division process of Chlamydia. Sci. Rep. 2020, 10, 12588. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Enciso, G.A.; Boassa, D.; Chander, C.N.; Lou, T.H.; Pairawan, S.S.; Guo, M.C.; Wan, F.Y.M.; Ellisman, M.H.; Sütterlin, C.; et al. Replication-dependent size reduction precedes differentiation in Chlamydia trachomatis. Nat. Commun. 2018, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Liechti, G.; Kuru, E.; Packiam, M.; Hsu, Y.-P.; Tekkam, S.; Hall, E.; Rittichier, J.T.; VanNieuwenhze, M.; Brun, Y.V.; Maurelli, A.T. Pathogenic Chlamydia Lack a Classical Sacculus but Synthesize a Narrow, Mid-cell Peptidoglycan Ring, Regulated by MreB, for Cell Division. PLoS Pathog. 2016, 12, e1005590. [Google Scholar] [CrossRef] [PubMed]
- Testa, C.A.; Brown, M.J. The methylerythritol phosphate pathway and its significance as a novel drug target. Curr. Pharm. Biotechnol. 2003, 4, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Slade, J.A.; Brockett, M.; Singh, R.; Liechti, G.W.; Maurelli, A.T. Fosmidomycin, an inhibitor of isoprenoid synthesis, induces persistence in Chlamydia by inhibiting peptidoglycan assembly. PLoS Pathog. 2019, 15, e1008078. [Google Scholar] [CrossRef]
- Belland, R.J.; Nelson, D.E.; Virok, D.; Crane, D.D.; Hogan, D.; Sturdevant, D.; Beatty, W.L.; Caldwell, H.D. Transcriptome analysis of chlamydial growth during IFN-γ-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA 2003, 100, 15971–15976. [Google Scholar] [CrossRef]
- Ouellette, S.P.; Hatch, T.P.; AbdelRahman, Y.M.; Rose, L.A.; Belland, R.J.; Byrne, G.I. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol. Microbiol. 2006, 62, 1387–1401. [Google Scholar] [CrossRef]
- Sigalova, O.M.; Chaplin, A.V.; Bochkareva, O.O.; Shelyakin, P.V.; Filaretov, V.A.; Akkuratov, E.E.; Burskaia, V.; Gelfand, M.S. Chlamydia Pan-Genomic Analysis Reveals Balance between Host Adaptation and Selective Pressure to Genome Reduction. BMC Genom. 2019, 20, 710. [Google Scholar] [CrossRef]
- Moran, N.A. Microbial minimalism: Genome reduction in bacterial pathogens. Cell 2002, 108, 583–586. [Google Scholar] [CrossRef] [PubMed]

| Peptido-Glycan Synthesis Inhibitors | Glycopeptide Antibiotics | Iron Starvation | Heat Shock | Co-Infection | Protein Synthesis Inhibitors | Immune-Response | Spontaneous | |
|---|---|---|---|---|---|---|---|---|
| Chlamydia trachomatis | ✓[30,31] | ✓[30] | ✓[30,32,33,34] | ✓[35,36] | ✓[37,38] | ✓[30] | ✓[14,23,39] | |
| Chlamydia muridarum | ✓[17,40] | ✓[41] | ||||||
| Chlamydia suis | ✓[42] | ✓[43] | ||||||
| Chlamydia psittaci | ✓[18,22,44] | ✓[44] | ✓[44,45] | ✓[46] | ||||
| Chlamydia pneumoniae | ✓[47] | ✓[48,49] | ✓[49] | ✓[49,50] | ||||
| Chlamydia pecorum | ✓[51] | ✓[52] | ✓[53] | |||||
| Estrella lausannensis | ✓[25] | ✓[54] | ||||||
| Rhabdochlamydia porcellonis | ✓[29] | |||||||
| Waddlia chondrophila | ✓[13,47] | ✓[13] | ✓[13,55] | ✓unpublished | ✓[13] | ✓unpublished | ||
| Simkania negevensis | ✓[27] | ✓[56] | ✓[56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozusnik, T.; Adams, S.E.; Greub, G. Aberrant Bodies: An Alternative Metabolic Homeostasis Allowing Survivability? Microorganisms 2024, 12, 495. https://doi.org/10.3390/microorganisms12030495
Kozusnik T, Adams SE, Greub G. Aberrant Bodies: An Alternative Metabolic Homeostasis Allowing Survivability? Microorganisms. 2024; 12(3):495. https://doi.org/10.3390/microorganisms12030495
Chicago/Turabian StyleKozusnik, Thomas, Simone E. Adams, and Gilbert Greub. 2024. "Aberrant Bodies: An Alternative Metabolic Homeostasis Allowing Survivability?" Microorganisms 12, no. 3: 495. https://doi.org/10.3390/microorganisms12030495
APA StyleKozusnik, T., Adams, S. E., & Greub, G. (2024). Aberrant Bodies: An Alternative Metabolic Homeostasis Allowing Survivability? Microorganisms, 12(3), 495. https://doi.org/10.3390/microorganisms12030495







