Abstract
Traditionally, the role of gut dysbiosis was thought to be limited to pathologies like Clostridioides difficile infection, but studies have shown its role in other intestinal and extraintestinal pathologies. Similarly, recent studies have surfaced showing the strong potential role of the gut microbiome in colorectal cancer, which was traditionally attributed mainly to sporadic or germline mutations. Given that it is the third most common cancer and the second most common cause of cancer-related mortality, 78 grants totaling more than USD 28 million have been granted to improve colon cancer management since 2019. Concerted efforts by several of these studies have identified specific bacterial consortia inducing a proinflammatory environment and promoting genotoxin production, causing the induction or progression of colorectal cancer. In addition, changes in the gut microbiome have also been shown to alter the response to cancer chemotherapy and immunotherapy, thus changing cancer prognosis. Certain bacteria have been identified as biomarkers to predict the efficacy of antineoplastic medications. Given these discoveries, efforts have been made to alter the gut microbiome to promote a favorable diversity to improve cancer progression and the response to therapy. In this review, we expand on the gut microbiome, its association with colorectal cancer, and antineoplastic medications. We also discuss the evolving paradigm of fecal microbiota transplantation in the context of colorectal cancer management.
1. Introduction
The human microbiome is a collection of microbial communities that constantly change as specific communities of microbes occupy anatomical niches within the human body. This colonization process starts soon after birth upon exposure to the vaginal microbiota. Infants continue to be introduced to new flora through routine activities with other humans, including feeding and playing, resulting in the establishment of the microbiome on the skin, gut, and mucosal surfaces. The introduction and reintroduction of flora continue throughout life from our routine interactions.
The microbiome, defined as the number of microorganisms with their genetic material, often significantly impacts human health. It is very different from the microbiota, which refers to the microbial population in different ecosystems in the body [1,2]. These microbiotas carry out many complex biochemical and metabolic functions [3]. The indigenous microbiota can also modify epithelial responses and systemic responses, such as the development and activity of the immune system with alterations in the antitumor responses. Any disturbance to the symbiosis between mammalian hosts and their microbial partners can lead to various diseases, including direct effects on the development and progression of colorectal cancer (CRC). As the third most common cancer in the US, with an estimated burden of 153,020 cases per 100,000 person-years in 2023, and the second most common cause of cancer death, with estimated 52,550 deaths per 100,000 person years, colorectal cancer adds a significant morbidity and healthcare burden to the US [4]. Meanwhile, it has the second highest treatment cost for any cancer, accounting for 12.6% of all cancer treatment costs and equating to a total of USD 24.3 billion annually in 2020 [5]. Hence, there have been ongoing efforts in the research and development of new therapeutic options to treat CRC and, if possible, identify the biomarkers required for its early detection. Manipulating the gut microbiome by means of fecal transplantation is emerging as one of the many novel therapeutic options to treat CRC and improve its response to chemotherapy and immunotherapy. This review focuses on the pivotal connection between the gut microbiome and colorectal cancer, examining its potential for modulating tumorigenesis and presenting treatment strategies [6].
2. Gut Microbiome’s Essential Role in Cancer Prevention
A healthy microbiome is essential in maintaining homeostasis, and certain species have shown their direct effect in preventing cancer. The most extensively studied protective microorganisms are Bifidobacterium and Lactobacillus spp, which have demonstrated anti-cancer properties by employing various mechanisms. These mechanisms encompass their influence on cellular proliferation and apoptosis, their regulation of host immunity, and their neutralization of carcinogenic toxins and xenobiotics [7,8]. However, other colonic bacteria can also exhibit anti-inflammatory and anti-carcinogenic effects. For instance, ingested dietary fibers undergo fermentation by butyrate-producing microbes (Ruminococcus, Clostridium, Eubacterium, and Faecalibacterium), producing short-chain fatty acids (SCFA). SCFA interacts with G protein-coupled receptors, inhibiting histone deacetylase in the colonic epithelium and immune cells. This interaction results in histone hyperacetylation, increasing T-reg numbers and promoting the production of the anti-inflammatory cytokines IL-10 and TGF-beta. Simultaneously, SCFA downregulates the production of the proinflammatory cytokines IL-6 and IL-12 in colonic macrophages, creating an anti-inflammatory microenvironment [9,10]. Moreover, by acting as a histone deacetylase inhibitor, butyrate also promotes the expression of tumor suppressor proteins such as FAS, p21, and p27 [11]. Some bacteria are also known for maintaining the barrier function. Lactobacillus acidophilus, Bifidobacteria bifidum, and Bifidobacteria infantum promote the increased expression of mucin 2 (MUC2), zonula occludens (ZO-1), and occludin, which are an essential part of intestinal epithelial barrier integrity and have shown an association with decreased colorectal tumor incidence and volume [12]. Bifidobacterium longum and Bacillus subtilis have been shown to downregulate proinflammatory cytokines (e.g., IL-17, IL-23, and TNF-α) and upregulate tight junction (TJ) proteins (e.g., claudin-1, occludin, and ZO-1) in colitis mouse models, preserving intestinal barrier function [13,14,15]. One actively investigated mechanism through which the microbiome protects against CRC involves the downregulation of IL-22 by the BL 23 strain of Lactobacillus casei [16]. Even though the species mentioned above are among the many that promote gut homeostasis, other species that tip the balance towards oncogenesis have also been identified, as detailed in the next section.
3. Abnormal Gut Microbiome in Patients with Colorectal Cancer
3.1. Streptococcus Bovis
Streptococcus bovis (S. bovis) is a Gram-positive bacterium, appearing as cocci in chains, with a facultative anaerobic lifestyle, and belongs to the family Streptococcaceae. It has been identified as one of the risk factors for colorectal cancer (CRC) [17,18,19]. S. bovis is typically found in the gastrointestinal tract. The occurrence of S. bovis-induced endocarditis or bacteremia was an early indication of its involvement in colorectal cancer [20]. The link between inflammation and colon carcinogenesis was further confirmed when the pro-inflammatory potential of S. bovis proteins and their carcinogenic properties were observed. It has been discovered that S. bovis actively contributes to the development of CRC, possibly through an inflammation-based sequence of tumor development or propagation involving interleukin (IL)-1, cyclooxygenase-2 (COX-2), and IL-8 [21,22].
3.2. Fusobacteria
Fusobacteria are Gram-negative, non-spore-forming, spindle-shaped obligate anaerobes belonging to the family Fusobacteriaceae, and have recently been at the forefront of discussions regarding the microbiome and tumor-associated pathogens [23]. Studies have shown that they originate from the oral microflora, with enteral transmission being the predominant route for CRC-tissue colonization by F. nucleatum [24,25]. These bacteria produce a unique protein called Fusobacterium adhesin A (FadA), which activates the β-catenin signaling pathway after binding to E-cadherin, a potent oncogenic stimulator [26,27]. Meanwhile, they also produce Fap2, a galactose adhesion hemagglutinin which mediates the colonization and invasion of CRC cells [28]. F. nucleatum promotes oncogenesis via various mechanisms, including promotion of a protumorigenic inflammatory milieu along with the inhibition of anticancer immune responses through genetic and epigenetic alterations [28,29,30,31,32,33,34,35]. In addition, it may also promote chemoresistance in CRC [36,37,38,39]. It has also been shown to be a potential marker for early detection, as well as prognostication and prediction of outcomes in CRC [40,41,42].
3.3. Enterococcus Fecalis
Enterococcus faecalis is a Gram-positive bacterium with a cocci shape from Entercoccaceae family, exhibiting a facultative anaerobic lifestyle. It is a gut commensal bacterium that produces superoxides through the autooxidation of membrane-associated demethylmenaquinone [43]. Infection with E. faecalis leads to superoxide-mediated DNA damage in intestinal epithelial cells. Studies have shown that the abundance of E. faecalis is significantly higher in CRC patients compared with healthy individuals [44,45,46]. Additionally, both in vitro and in vivo experiments have demonstrated that E. faecalis can produce hydroxyl radicals, which are highly mutagenic and can cause DNA breaks, point mutations, and protein–DNA crosslinking. These effects contribute to chromosomal instability and increase the risk of CRC.
3.4. Anaeroplasma
Anaeroplasma is a bacterial genus within the class Mollicutes, and its bacterial characteristics vary based on the specific genus. It survives in anaerobic environments and is typically found in the gastrointestinal and urogenital tract. Studies of its association with patients of CRC are conflicting. It is a fact that chronic colonic inflammation is a risk factor for CRC. In this context, studies have shown a relative higher abundance of Anaeroplasma in colitis mouse models with decrease in levels following healthy gut microbiome transfer [47]. Other studies have shown a higher abundance of Anaeroplasma in mutated APC gene mouse models than controls. The same study showed higher Anaeroplasma in older mice with a higher colonic tumor burden than younger mice with no tumors [48]. Another study showed conflicting results where chronic colitis mouse models treated with AOM (azoxymethane) had relatively lower Anaeroplasma abundance than the controls [49].
3.5. Flavobacteria
Flavobacteria are Gram-negative, rod-shaped bacteria belonging to the family Flavobacteriaceae, with studies showing conflicting evidence. One study showed a decreased concentration of this genus in patients with CRC with an underlying mechanism described as the destruction of mucosa-adherent microbiota in healthy individuals [50]. Another study demonstrated a relative abundance of flavobacteria in patients with colorectal adenomas [51]. Flavonoids are well-known for their anticancer properties, and their biodegradation, secondary to Flavobacteria, could be the primary cause of this pathology [52]. These conflicting results emphasize the possibility that single species do not reflect genus differences as a whole. This might lead to inconsistent results, as emphasized in previous studies although the variation could also reflect differences between adenomas and cancer [53].
3.6. Ruminococcaceae
The Ruminococcaceae family consists of Gram-positive, anaerobic bacteria with varying rod-shaped morphologies and plays an essential role in the fermentation of complex carbohydrates and the metabolism of dietary fibers. Even though these bacteria have shown increased prevalence in probiotics, the promotion of gut barrier integrity and the suppression of colonic carcinogenesis by production of secondary bile acids, studies have shown conflicting evidence [54,55,56]. One study identified the relative abundance of Ruminococcus in the colon of rats with underlying precancerous lesions induced by carcinogen 1,2-dimethyl hydrazine [57]. Another mouse study showed a significant increase in Ruminococcaceae in CRC group, with many other studies showing similar results [58,59,60]. In contrast to this, another study comparing normal, precancerous, and cancerous colonic tissue showed an abundance of Ruminococcus in normal tissues [61]. Another study showed decreased prevalence of Ruminococcus gnavus in mice treated with azoxymethane, which induces inflammation of the colon [49]. The same study further showed a negative correlation with tumor numbers, disease score, and inflammatory T cell subsets and a positive correlation with CD4+, FoxP3+, Tregs, and IL-10-producing T cells [49]. A recent study performing integrated analyses of differentially abundant bacterial groups (ASVs—amplicon sequence variants) of 1056 stool samples to identify biomarkers associated with adenoma and CRC showed Ruminococcaceae UCG-005 as one of the two top-ranking biomarkers between controls and adenoma patients [62]. In the same study, the Ruminococcus gnavus group showed differential abundance between adenoma and cancer compared with healthy controls.
3.7. Acidovarax
Acidovarax are Gram-negative bacteria with variable shapes, exhibiting aerobic or anaerobic characteristics. They consist of acid-degrading bacteria from the family Comamonadaceae, promoting inflammation by metabolizing nitro-aromatic compounds and flagellar proteins that induce local inflammation [63]. This is hypothesized to be the potential reason behind the Acidovarax–adenoma association. Multiple studies analyzing these bacteria at the genus level have shown a relative abundance of Acidovarax in patients with adenoma vs. controls without adenoma [50,64].
3.8. Eubacteria
Eubacteria are Gram-positive anaerobic bacteria with a rod-shaped morphology, belonging to the family Eubacteriaceae. They are identified as bacteria with anti-inflammatory properties, with studies showing decreased anti-inflammatory effects in mice deficient in Faecalibateria and Eubacteria [65]. Another study specifically designed to look at candidate strains with anti-CRC activity showed Eubacterium callanderi to have antiproliferative properties against CRC cells by inducing apoptosis and cell death in a dose-dependent manner [66]. Further, the same study showed higher butyrate concentrations with the peri-tumoral injection of a cell-free supernatant of Eubacteria inhibiting tumor growth, further emphasizing their role in probiotic therapy to prevent CRC [66].
3.9. Bifidobacteria
Bifidobacteria are Gram-positive anaerobic rod-shaped bacteria belonging to the family Bifidobacteriaceae. They have recently garnered a lot of attention because of their beneficial effect on the gut microbiome. They are considered as a health-promoting gut microorganisms, especially beneficial in CRC prevention by means of the downregulation of anti-apoptotic and the upregulation of pro-apoptotic genes including BAD, Bxl-2, caspase-3, caspase-8, caspase-9 and Fas-R [67]. Another recent murine model study emphasized the oncoprotective effect of Bifidobacterium in the colon by means of the downregulation of the urea cycle [68]. Urea cycle activation is linked to microbial dysbiosis and excess urea could enter the macrophages, inhibiting the binding efficiency of p-STAT1 to the SAT1 promoter region, causing accumulation of polyamines and thus skewing macrophages to a pro-tumoral phenotype. These murine models, when treated with urea cycle inhibitors or Bifidobacterium-based probiotics, showed lower expression of Ki67 and CD206, which are markers of increased proliferation and macrophage immunosuppression, respectively. This emphasizes the role of Bifidobacterium-based probiotic supplements in mitigating CRC.
3.10. Others
Besides the abovementioned bacteria, there are many others that have shown associations with CRC. One study showed an increased prevalence of Akkermansia in longstanding colitis models [47]. Another study performed on humans analyzing the oral microbiome showed a decreased prevalence of Prevotella in the CRC vs. healthy group [69]. A few extensive metagenomic analyses of CRC datasets have shown numerous species to be present in the carcinoma-enriched environment, namely Fusobacterium nucleatum, Parvimonas micra, Gemella morbiliorum, Peptostreptococcus spp, Solobacterium moorei, Clostridium symbiosum, Anaerococcus spp, Porphyromonas spp, Prevotella intermedia, Bacteroides fragilis, Streptococcus constellatus, Granuclicatella adiacens, Treponema denticola, Porphyromonas gingivalis and Tannerrella forsthica [24,25,70]. Bacteria that showed increased prevalence in the controls included Roseburia intestinalis, Gordonibacter pamelaeae and Bifidobacterium catenulatum [24].
Expanding on the breadth of data available, studies even show changes in microbiome proportions in the colorectal adenoma vs. carcinoma populations, with various bacteria like Butyricimonas synergistica, Agrobacterium larrymoorei, Bacteroides plebeius, Clostridium scindens, Lachnospiraceae bacterium feline oral taxon 001, Prevotella heparinolytica, Streptococcus mutans, Lachnospiraceae bacterium 19gly4, and Eubacterium hallii showing the best performance in distinguishing colorectal adenoma from CRC populations [53]. Interestingly, other studies have also shown a differential abundance of bacteria in patients with CRC when comparing the right side versus the left side of the colon, with increased prevalence of Haemophilus and Veilonella in right-sided CRC patients vs. increased Roseburia and Akkermansia in left-sided CRC patients [71]. The role of microbiome has also been explored in the context of increasing incidence of young or early onset CRC. It appears that certain bacterial genera such as Akkermansia and Bacteriodes are differentially abundant in young individuals who develop CRC as opposed to several other genera (Bacillus, Staphylococcus, Listeria, Enterococcus, Pseudomonas, Fusobacterium, and Escherichia/Shigella) that are more abundant in CRC arising at the usual age [72]. Approaches such as plasma metabolomics analysis and machine learning are being used to further define the relationship between the altered microbiome in young individuals and CRC to identify microbiome-derived signatures for screening and therapy [73,74].
4. Biochemistry and Microbiome of Patients with Colorectal Cancer
Presently, two hypotheses have been formulated to describe the diverse mechanisms through which the gut microbiota can impact carcinogenesis in CRC.
4.1. Alpha Bug Model
The Alpha Bug Model hypothesis was initially formulated by Cynthia L. Sears and Drew M. Pardoll in 2011 [75]. It states that specific “Alpha-bugs” possess virulence factors capable of not only directly or indirectly initiating carcinogenesis but also displacing protective bacterial species, thereby reshaping the surrounding bacterial community [75]. The foundation for this hypothesis was formed by studying enterotoxigenic Bacteroides fragilis (ETBF), a bacterium that generates Bacteroides fragilis toxin (BFT). Research has demonstrated that the presence of ETBF in the intestines of the multiple intestinal neoplasia (Min) mice (a commonly employed murine model of CRC) led to the development of microscopic adenomas merely one-week post-colonization [76]. Since then, multiple studies have demonstrated the existence of numerous other gut bacteria (alpha-bugs) that can potentially induce a similar chain of events leading to cancer, including superoxide-producing S. bovis, Enterococcus faecalis, and Escherichia coli [77,78,79].
4.2. Driver–Passenger Hypothesis
The novel hypothesis based on the driver–passenger model was introduced by Harold Tjalsma, PhD, in 2012 [80]. This is an extended version of the alpha-bug model. According to this hypothesis, drivers (alpha-bugs) initiate CRC and cause changes in the tumor’s microenvironment, creating conditions suitable for colonization by opportunistic bacteria, known as ‘passengers.’ Passenger bacteria contribute to the progression of the disease and, throughout the illness, may even displace the driver pathogens. Passengers exert no influence on the initiation of tumorigenesis, but they may participate in the progression of the disease. Examples of passengers include S. Gallolyticus, Fusobacterium, and Veillonella spp. Furthermore, certain bacteria can fulfill dual roles as both passengers and drivers. For instance, F. nucleatum can trigger tumorigenesis and is abundant in patients with advanced CRC without being displaced. Supporting evidence for the driver–passenger model was provided through several additional empirical investigations [81]. Nevertheless, a clear differentiation categorizing a specific species as either a “driver” or a “passenger” in CRC continues to elude the definition.
Regardless of the framework considered, the pathophysiology of colorectal cancer concerning the microbiome can be described through the three mechanisms described below: chronic inflammation, metabolism of dietary components, and generation of genotoxins.
4.3. Inflammation
Chronic inflammation and tumorigenesis are intricately interconnected, particularly in the context of CRC. Consequently, individuals with inflammatory bowel disease (IBD) exhibit a significantly heightened likelihood of developing CRC compared with the general population [82]. Moreover, the microbiota can also trigger inflammation, thereby contributing to the oncogenic process. For instance, as mentioned earlier, the Bacteroides fragilis toxin (BFT) produced by B. fragilis impacts local T-cell immunity by activating STAT3 and elevating Th17 and IL-17 levels. STAT3 is an oncogenic transcription factor, and when activated, it is translocated to the cell nucleus and increases the expression of such genes as Bcl-xL, Cyclin D1, and IL-6, thereby inhibiting apoptosis, promoting cellular proliferation and increasing the inflammatory response, therefore promoting abnormal cell line growths, [83]. On the other hand, an increased IL-17 level serves as a chemoattractant for myeloid-derived suppressor cells (MDSC). These cells secrete IL-1, IL-6, and TNF, further fueling chronic inflammation [76,84]. Moreover, IL-6 itself can activate STAT3, leading to a feed-forward loop where STAT3 further increases the production of IL-6 and other inflammatory mediators via the NF-kB-IL-6-Stat3 cascade [83]. Another example of chronic inflammatory changes induced by bacteria can be seen for F. nucleatum. This organism has been shown to increase nuclear factor-kappa B (NF-κB) expression [85,86]. NF-κB is a central immune response regulator and inflammation regulator that upregulates many chemokines (CXCL1, CXCL2, CXCL3) and cytokines (TNFα, IL-1β, IL-6, and IL-8). Both CXCL1 and CXCL2 recruit MDSCs to the tumor microenvironment, which again, in turn, creates a pro-inflammatory and carcinogenic environment. F. nucleatum also activates Wnt signaling pathways while simultaneously suppressing CD3+ T cell-mediated adaptive immunity and fostering inflammatory alterations [85,86]. Moreover, F. nucleatum, Enterococcus faecalis, and Peptostreptococcus anaerobius can disrupt the gut barrier, increasing its permeability, thus exposing Toll-like receptors (TLR) on the surface of immune cells in the colon to these bacteria. This process facilitates bacterial invasion, intensifying the inflammatory response. The resultant inflammatory environment produces reactive oxygen species (ROS), leading to DNA damage. This damage may lead to somatic mutations, for example, in one encoding APC, contributing to the development of carcinomas [87].
4.4. Metabolism of Dietary Components
Bacterial metabolic byproducts can exert direct carcinogenic effects and contribute to inflammatory changes. Although the mechanisms described in the previous sections influencing the tumorigenesis process are primarily linked to specific bacteria, particularly in food product metabolism, the collective impact of the entire microbial community plays a predominant role [88]. During the fermentation of proteins or amino acids within the colon, bacteria can generate diverse substances, including N-nitroso compounds, polyamines, hydrogen sulfide, and ammonia. These substances give rise to ROS, which promote inflammation or can directly affect DNA, promoting mutations and fostering carcinogenesis [89,90,91]. Another family of compounds whose metabolism by gut bacteria can incite carcinogenesis is bile acids. Upon interaction with intestinal bacteria, the primary bile acids transform into secondary bile acids. The latter induces the generation of ROS and reactive nitrogen species (RNS), subsequently leading to DNA damage. Although only about 5% of primary bile acids typically reach the colon, the excessive consumption of fatty food has been shown to increase that amount [92].
4.5. Production of Genotoxins
Genotoxins constitute molecules released by bacteria that can directly damage DNA or alter the activity of tumor suppressor genes. Aside from provoking inflammatory alterations, the BFT secreted by B. fragilis serves as a zinc metalloproteinase that dismantles e-cadherin molecules [27]. E-cadherin, an anti-oncogene product, plays a pivotal role in curbing excessive growth signals and facilitating cell–cell adhesion, thereby contributing to the cohesion of epithelial cells. In its absence, epithelial cells become more susceptible to cancer development [27]. Furthermore, B. fragilis, as well as Enterococcus faecalis, can induce ROS formation that causes direct DNA damage [46,93]. Recent investigations indicate that Escherichia coli expressing polyketide synthases (pks+) can synthesize toxins referred to as cyclomodulins, including cytolethal distending toxins (CDT), cytotoxic necrotizing factor (CNF), cycle-inhibiting factor, and colibactin [94]. These toxins exhibit genotoxic properties or interfere with the normal progression of the cell cycle. In addition, enteropathogenic Escherichia coli secretes an effector protein called EspF, which promotes carcinogenesis by depleting mismatch repair proteins via posttranscriptional mechanisms and depends on EspF mitochondrial targeting [95].
Figure 1 summarizes the above mechanisms of microbiome alterations impacting colorectal carcinogenesis.
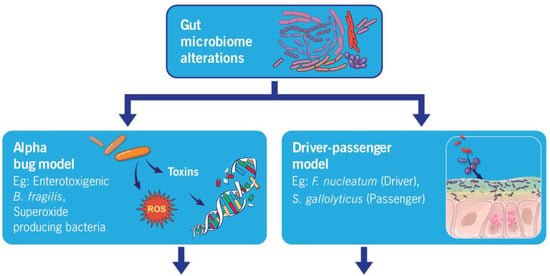
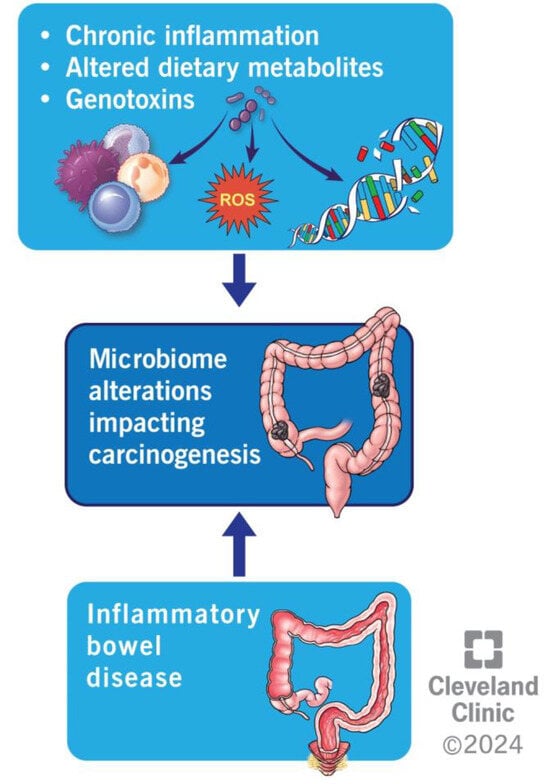
Figure 1.
Summarizing different mechanisms of microbiome alterations impacting colorectal carcinogenesis.
5. Colorectal Cancer and Anti-Neoplastic Medication
The gut microbiome’s influence on antitumor drug therapy primarily occurs via five mechanisms: bacterial translocation, immune regulation, metabolic regulation, enzymatic degradation, and diversity reduction [96]. In this section, we will try to expand on how the response to different systemic therapies for CRC changes with the gut microbiota composition of the host.
5-Fluoro uracil (5-FU) is an antimetabolite primarily used to treat various cancers, especially gastrointestinal cancers, including CRC. It has been shown that 5-FU is associated with a reduction in commensal bacteria such as the genera Streptococcus and Bacteroides and a concomitant enhancement of Gram-negative bacteria such as Clostridium hathewayi and Lachnospiraceae bacterium [97]. In their work on mouse models, Atarashi et al. showed that 17 bacterial strains play a crucial role in enhancing Treg cell abundance and inducing anti-inflammatory cytokines, including IL-10. They found that these 17 strains belong to clusters IV, XIVa, and XVIII of Clostridia and that oral administration of these strains to adult mice improved disease in models of colitis [98]. Another study showed that Lactobacillus plantarum supernatant (LP SN) amplified the efficacy of 5-FU for CRC and reversed the development of chemoresistance by decreasing cancer stem-like cells [99]. It was shown that the LPSN selectively inhibits the expression of specific markers of CSCs, including CD44, CD133, CD166, and ALDH1. The combination therapy of 5-FU and LP SN leads to increased apoptotic activity by inducing caspase-3 activity, inhibiting the growth of CRCs [100]. Additionally, combination therapy was observed to induce an antitumor mechanism by inactivating the Wnt/Beta-catenin signaling of chemo-resistant cells.
Oxaliplatin, a platinum derivative commonly used in colorectal cancer treatment, also induces immunologic cell death that drives antitumor T-cell immunity [101]. A study completed to understand gut microbiome-related changes due to oxaliplatin showed an increasing Gram-negative bacterial population, with sub-analyses showing an increase in Prevotella 2 and Odoribacter bacteria with a significant reduction in Prevotella 1 and Parabacteroides at the genus level in the oxaliplatin treatment group [102]. In another study performed to evaluate impact of microbiota on the efficacy of oxaliplatin and immunotherapy, mice were inoculated with different tumors, namely EL4 lymphoma, MC38 colon cancer, and B16 melanoma [103]. These mice received antibiotics before tumor inoculation and further throughout the study. The results showed a significantly lower tumor regression and survival in antibiotic-treated mice with EL4 tumors when treated with either oxaliplatin or immunotherapy. There was a downregulation of genes associated with phagocytosis and adaptive immune response along with an upregulation of genes related to tissue development and cancer in antibiotic-treated mice. Additionally, this study reported that antibiotics prevented oxaliplatin-induced DNA damage and apoptotic activity by decreasing ROS and the induction of DNA damage response gene ATR and p53 downstream genes (BAX, FAS, CDKN1A, and RB1) [103].
Irinotecan, also known as CPT-11, is a DNA topoisomerase I inhibitor used to treat CRC. Irinotecan is hydrolyzed by carboxylesterase to form an active metabolite, SN-38. UDP-glucuronyl transferases inactivate SN-38 to its SN-38G form. However, once SN-38G enters the small intestine, it is reactivated by bacterial ꞵ-glucuronidase, leading to several adverse reactions, including diarrhea. Studies have evaluated potent bacterial ꞵ-glucuronidase inhibitors to mitigate CPT-11-induced toxicity, but with mixed results [104,105]
Bevacizumab is a monoclonal antibody against vascular endothelial growth factor A (anti-VEGF A) used in metastatic colorectal cancer and other cancers. In a recent case–control study, a difference in gut microbiome composition was observed between patients with a malignant glioma who received a combination of bevacizumab and temozolomide (group 1) vs. temozolomide monotherapy (group 2). They further found that group 1 patients had a higher abundance of Firmicutes, Bacteroides, and Actinobacteria and a lower abundance of Bacteroidetes and Cyanobacteria in their fecal microbiota than group 2 patients. Investigations into the potential role of these microbes in modifying treatment response are needed to assess the possibility for fecal microbiota transplants [106].
To conclude, the gut microbiome’s role in influencing chemotherapy effects is increasingly being understood. The supplementation of probiotics and antibiotic treatment to improve the adverse effect profile and prevent chemoresistance has also increasingly been considered, pending further research [96].
Immunotherapy and its association with gut microbiome is another active area of research. Experimental studies in non-CRC cancers, primarily melanoma, have demonstrated differential effects of the gut microbiome on immunotherapy outcomes [107,108]. It was shown that Bifidobacterium-treated mice displayed significantly improved tumor control when treated with anti-PD1 therapy compared with their non-Bifidobacterium-treated counterparts [107]. Based on a gnotobiotic study involving oral administration of B. fragilis to germ-free mice, it was demonstrated that CTLA blockade resistance of colorectal tumors could be overcome [109]. The findings suggest promise for investigating microbiome modulation, such as the adoptive transfer of bacteria-specific T cells or immunization with bacterial products to boost immunotherapy activity [109]. The microbiome could also help to predict immunotherapy-related toxicity based on findings correlating bacterial species and the development of checkpoint-blockade-induced colitis [108].
6. Fecal Microbiota Transplantation in Colorectal Cancer
Investigations into the potential relationship between fecal microbiota transplantation (FMT) and colorectal cancer have sparked a new frontier in gastrointestinal research. Emerging studies point to the pivotal role of gut microbiota in influencing the risk and progression of colorectal malignancies. Delving into this intricate connection offers a promising avenue for innovative approaches to combating colorectal cancer. Dysbiosis is associated with cancer and poor outcomes in certain forms of cancer therapy, including allogenic stem cell transplantation [110,111,112,113]. FMT is an intervention by which dysbiosis can be reversed by replacing pathogenic gut microbiomes with microbial species that are more abundant in normal, healthy individuals. Even though FMT is most frequently used in treating Clostridioides difficile infection, recent studies show their use in treating other gastrointestinal pathologies like inflammatory bowel disease, hepatic encephalopathy, and colorectal cancer. Therapeutic methods of gut microbiota modification, including FMT and symbiotics, are in the early stages of investigation. Recent research on mouse models has demonstrated a reversal in the microbiome associated with colonic dysplasia post-FMT, with several studies showing near reversal of colonic dysplasia and a significant reduction in pro-oncogenic inflammatory cytokines [114,115,116,117]. These models also show the modulation of immunotherapy efficacy post-FMT. Similar beneficial effects have also been seen in solid tumors other than colorectal cancer [118,119,120]. Regardless, our understanding of this field remains limited, with no similar studies being translated to humans.
For the FMT studies, pathogenic mouse models are created by gavaging azoxymethane (AOM), which is a chemical carcinogen that promotes tumorigenesis, and dextran sodium sulfate (DSS) to disrupt the intestinal barrier and promote colonic inflammation and induces long-lasting colitis. Controls are usually gavaged with isotonic or phosphate-buffered saline (PBS). These mouse models are further gavaged with FMT, either from healthy individuals or from patients with established colorectal cancer. Multiple studies have been conducted with this basic design, showing several unique findings in mice receiving FMT from individuals without CRC compared with pathogenic mice not receiving FMT or receiving FMT from patients with CRC. These findings include the following: (1) A healthier microbiome is associated with a higher percentage of Firmicutes than Bacteroides [117]. (2) Higher alpha diversity is associated with FMT from control groups without CRC [115,117]. (3) Lesser dysplasia, tumor counts, improved mice weight, and longer intestinal lengths are observed in mice receiving FMT from healthy controls [114,115,117]. (4) Increased inflammatory markers such as IFN-gamma, TNF-alpha, Th1, and Th17 cells, increased Ki-67, PCNA immunostaining, beta-catenin, decreased apoptotic cells are observed in mice receiving a pathogenic gut microbiome [114,115,116]. (5) A decrease in the expression of genes responsible for the intestinal barrier function and colonic immunological barrier can be observed (Table 1) [116]. (6) The inheritance of a higher protection from carcinogens (AOM-DSS) is demonstrated by the offspring of mice receiving FMT from wild population exposed to infections, toxins, and mutagens compared with FMT from lab mice [121]. The last finding emphasizes the importance of considering lineages while selecting the correct donor for interventional studies. The findings are outlined in Figure 2.

Table 1.
Review of the literature on mouse model studies showing the efficacy of fecal microbiota transplantation in mice with colorectal cancer and other epithelial tumors.
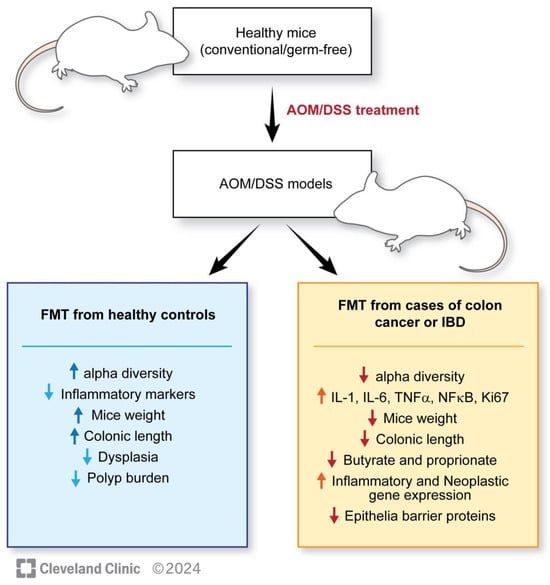
Figure 2.
Outlining the results of mice studies related to FMT and colorectal cancer.
Recent studies have hypothesized the role of a healthy microbiome in improving the response to anti-tumor therapy. It has been demonstrated that mice lavaged with feces from patients who responded to immunotherapy as compared with feces from non-responders showed a change in response to immunotherapy post-lavage, offering a potential role of microbiome manipulation in impacting immunotherapy outcomes [118,119,120]. The most widely studied form of immunotherapy involves immune checkpoint inhibiotors (ICI). They have been found to be effective in multiple tumor types [122]. Many studies have shown decreased antitumor effects of ICIs in dysbiotic mouse models treated with antibiotics, rendering them non-responders. Antibiotic-treated mouse models showed a restoration of therapeutic response to ICIs post-fecal microbiota transplantation from responders [118,119,120]. Further results are summarized in Table 1. Surprisingly, these results go beyond chemotherapy and immunotherapy, with one study showing that germ-free mice undergoing total body irradiation combined with FMT before bone marrow transplantation displayed improved survival compared with ones not receiving FMT, suggesting the role of microbiome in preventing radiation-induced toxicity [74].
The overall impact of gut microbiome alterations on colorectal cancer is summarized in Figure 3.
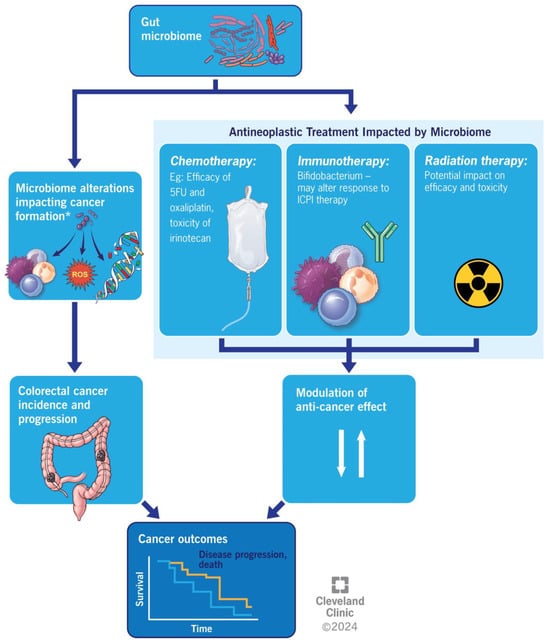
Figure 3.
Illustrating the overall impact of gut microbiome alterations on colorectal cancer outcomes.
7. Conclusions
In CRC patients, certain gut bacteria increase while others decrease, with studies suggesting a potential causative relationship. Even though multiple models have been proposed to understand the microbiome’s oncogenic and cancer-protective properties, the eventual mechanism of action boils down to either the bacterium producing genotoxins or an inflammatory state, either via direct cytokine production or inflammatory metabolite byproducts. Our understanding is constantly improving. Oncotherapy and the gut microbiome show a bidirectional/reciprocal approach rather than a cause–effect relationship. Modalities like chemotherapy, radiotherapy, and immunotherapy alter the gut microbiome drastically. Corollary is also true with changes in the gut microbiome shown to alter the therapeutic response. In such scenarios, FMT emerges as an evolving therapy, showing promise beyond its conventional use in infectious or inflammatory colitis. Even though the mouse studies on FMT and colorectal cancer so far have been promising, the lack of robust human studies remains a notable gap in research.
Funding
The request of English revisions for this research was funded by the Office of Graduate Medical Education at Sinai Hospital of Baltimore.
Acknowledgments
Illustration by Amanda Mendelsohn and Ken Kula. Reprinted with the permission of the Cleveland Clinic Enterprise Creative Services.
Conflicts of Interest
The authors disclose no relevant conflicts of interest.
References
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.-H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef]
- Cancer Stat Facts: Colorectal Cancer. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 2 January 2024).
- Financial Burden of Cancer Care. 2023. Available online: https://progressreport.cancer.gov/after/economic_burden (accessed on 2 January 2024).
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome in Colorectal Cancer Progression. Front. Immunol. 2021, 12, 807648. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, D.; Kim, D.; Cho, J.; Yang, J.; Chung, M.; Kim, K.; Ha, N. Inhibition of proliferation in colon cancer cell lines and harmful enzyme activity of colon bacteria by Bifidobacterium adolescentis SPM0212. Arch. Pharm. Res. 2008, 31, 468–473. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Wang, L.; Zhou, Y.; Hassan, J.S.; Li, M. Distribution and gene mutation of enteric flora carrying β-glucuronidase among patients with colorectal cancer. Int. J. Clin. Exp. Med. 2015, 8, 5310–5316. [Google Scholar]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Kuugbee, E.D.; Shang, X.; Gamallat, Y.; Bamba, D.; Awadasseid, A.; Suliman, M.A.; Zang, S.; Ma, Y.; Chiwala, G.; Xin, Y.; et al. Structural Change in Microbiota by a Probiotic Cocktail Enhances the Gut Barrier and Reduces Cancer via TLR2 Signaling in a Rat Model of Colon Cancer. Dig. Dis. Sci. 2016, 61, 2908–2920. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Li, H.; Li, Y. Effects of Bacillus subtilis on Epithelial Tight Junctions of Mice with Inflammatory Bowel Disease. J. Interferon Cytokine Res. 2016, 36, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Fu, Y.; Wang, L.; Qian, W.; Zheng, F.; Hou, X. Bifidobacterium longum and VSL#3® amelioration of TNBS-induced colitis associated with reduced HMGB1 and epithelial barrier impairment. Dev. Comp. Immunol. 2019, 92, 77–86. [Google Scholar] [PubMed]
- Isidro, R.A.; Lopez, A.; Cruz, M.L.; Gonzalez Torres, M.I.; Chompre, G.; Isidro, A.A.; Appleyard, C.B. The Probiotic VSL#3 Modulates Colonic Macrophages, Inflammation, and Microflora in Acute Trinitrobenzene Sulfonic Acid Colitis. J. Histochem. Cytochem. 2017, 65, 445–461. [Google Scholar] [PubMed]
- Jacouton, E.; Chain, F.; Sokol, H.; Langella, P.; Bermúdez-Humarán, L.G. Probiotic Strain Lactobacillus casei BL23 Prevents Colitis-Associated Colorectal Cancer. Front. Immunol. 2017, 8, 1553. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Madani, R.; Mukhtar, H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis. 2010, 12, 164–171. [Google Scholar] [CrossRef]
- Srivastava, A.; Walter, N.; Atkinson, P. Streptococcus bovis infection of total hip arthroplasty in association with carcinoma of colon. J. Surg. Orthop. Adv. 2010, 19, 125–128. [Google Scholar]
- Boleij, A.; van Gelder, M.M.; Swinkels, D.W.; Tjalsma, H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: Systematic review and meta-analysis. Clin. Infect. Dis. 2011, 53, 870–878. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Abu Bakar, F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: The nature and the underlying mechanisms of its etiological role. J. Exp. Clin. Cancer Res. 2011, 30, 11. [Google Scholar] [CrossRef]
- Biarc, J.; Nguyen, I.S.; Pini, A.; Gossé, F.; Richert, S.; Thiersé, D.; Van Dorsselaer, A.; Leize-Wagner, E.; Raul, F.; Klein, J.-P.; et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis). Carcinogenesis 2004, 25, 1477–1484. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Bakar, F.A. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: Inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer 2010, 9, 249. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wang, R.; Bhattacharya, R.; Boulbes, D.R.; Fan, F.; Xia, L.; Adoni, H.; Ajami, N.J.; Wong, M.C.; Smith, D.P.; et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev. Res. 2017, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Abed, J.; Emgård Johanna, E.M.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Casasanta, M.A.; Yoo, C.C.; Udayasuryan, B.; Sanders, B.E.; Umaña, A.; Zhang, Y.; Peng, H.; Duncan, A.J.; Wang, Y.; Li, L.; et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 2020, 13, eaba9157. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K.; et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef]
- Hamada, T.; Zhang, X.; Mima, K.; Bullman, S.; Sukawa, Y.; Nowak, J.A.; Kosumi, K.; Masugi, Y.; Twombly, T.S.; Cao, Y.; et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol. Res. 2018, 6, 1327–1336. [Google Scholar] [CrossRef]
- Serna, G.; Ruiz-Pace, F.; Hernando, J.; Alonso, L.; Fasani, R.; Landolfi, S.; Comas, R.; Jimenez, J.; Elez, E.; Bullman, S.; et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020, 31, 1366–1375. [Google Scholar] [CrossRef]
- Sayed, I.M.; Chakraborty, A.; Abd El-Hafeez, A.A.; Sharma, A.; Sahan, A.Z.; Huang, W.J.M.; Sahoo, D.; Ghosh, P.; Hazra, T.K.; Das, S. The DNA Glycosylase NEIL2 Suppresses Fusobacterium-Infection-Induced Inflammation and DNA Damage in Colonic Epithelial Cells. Cells 2020, 9, 1980. [Google Scholar] [CrossRef]
- Guo, P.; Tian, Z.; Kong, X.; Yang, L.; Shan, X.; Dong, B.; Ding, X.; Jing, X.; Jiang, C.; Jiang, N.; et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J. Exp. Clin. Cancer Res. 2020, 39, 202. [Google Scholar] [CrossRef]
- Saito, K.; Koido, S.; Odamaki, T.; Kajihara, M.; Kato, K.; Horiuchi, S.; Adachi, S.; Arakawa, H.; Yoshida, S.; Akasu, T.; et al. Metagenomic analyses of the gut microbiota associated with colorectal adenoma. PLoS ONE 2019, 14, e0212406. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Onozawa, H.; Saito, M.; Saito, K.; Kanke, Y.; Watanabe, Y.; Hayase, S.; Sakamoto, W.; Ishigame, T.; Momma, T.; Ohki, S.; et al. Annexin A1 is involved in resistance to 5-FU in colon cancer cells. Oncol. Rep. 2017, 37, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.X.; Chan, W.S.; Tang, K.S.; Zheng, S.Y. (Eds.) Restart based Collective Information Powered Differential Evolution for Solving the 100-Digit Challenge on Single Objective Numerical Optimization. In Proceedings of the 2019 IEEE Congress on Evolutionary Computation (CEC), Wellington, New Zealand, 10–13 June 2019. [Google Scholar]
- Lu, P.; Xu, M.; Xiong, Z.; Zhou, F.; Wang, L. Fusobacterium nucleatum prevents apoptosis in colorectal cancer cells via the ANO1 pathway. Cancer Manag. Res. 2019, 11, 9057–9066. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, A.C.; de Mattos Pereira, L.; Datorre, J.G.; Dos Santos, W.; Berardinelli, G.N.; Matsushita, M.M.; Oliveira, M.A.; Durães, R.O.; Guimarães, D.P.; Reis, R.M. Microbiota Profile and Impact of Fusobacterium nucleatum in Colorectal Cancer Patients of Barretos Cancer Hospital. Front. Oncol. 2019, 9, 813. [Google Scholar] [CrossRef]
- Ito, M.; Kanno, S.; Nosho, K.; Sukawa, Y.; Mitsuhashi, K.; Kurihara, H.; Igarashi, H.; Takahashi, T.; Tachibana, M.; Takahashi, H.; et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 2015, 137, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Kinugasa, H.; Hirai, M.; Terasawa, H.; Yasutomi, E.; Oka, S.; Ohmori, M.; Yamasaki, Y.; Inokuchi, T.; Harada, K.; et al. Heterogeneous distribution of Fusobacterium nucleatum in the progression of colorectal cancer. J. Gastroenterol. Hepatol. 2021, 36, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Winters, M.D.; Schlinke, T.L.; Joyce, W.A.; Glore, S.R.; Huycke, M.M. Prospective case-cohort study of intestinal colonization with enterococci that produce extracellular superoxide and the risk for colorectal adenomas or cancer. Am. J. Gastroenterol. 1998, 93, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef]
- Wang, X.; Huycke, M.M. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 2007, 132, 551–561. [Google Scholar] [CrossRef]
- Gates, T.J.; Yuan, C.; Shetty, M.; Kaiser, T.; Nelson, A.C.; Chauhan, A.; Starr, T.K.; Staley, C.; Subramanian, S. Fecal Microbiota Restoration Modulates the Microbiome in Inflammation-Driven Colorectal Cancer. Cancers 2023, 15, 2260. [Google Scholar] [CrossRef]
- Vitali, F.; Tortora, K.; Di Paola, M.; Bartolucci, G.; Menicatti, M.; De Filippo, C.; Caderni, G. Intestinal microbiota profiles in a genetic model of colon tumorigenesis correlates with colon cancer biomarkers. Sci. Rep. 2022, 12, 1432. [Google Scholar] [CrossRef] [PubMed]
- Alrafas, H.R.; Busbee, P.B.; Chitrala, K.N.; Nagarkatti, M.; Nagarkatti, P. Alterations in the Gut Microbiome and Suppression of Histone Deacetylases by Resveratrol Are Associated with Attenuation of Colonic Inflammation and Protection against Colorectal Cancer. J. Clin. Med. 2020, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Iadsee, N.; Chuaypen, N.; Techawiwattanaboon, T.; Jinato, T.; Patcharatrakul, T.; Malakorn, S.; Petchlorlian, A.; Praditpornsilpa, K.; Patarakul, K. Identification of a novel gut microbiota signature associated with colorectal cancer in Thai population. Sci. Rep. 2023, 13, 6702. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, T.; Chen, G.Y. Flavonoids and Colorectal Cancer Prevention. Antioxidants 2018, 7, 187. [Google Scholar] [CrossRef]
- Hua, H.; Sun, Y.; He, X.; Chen, Y.; Teng, L.; Lu, C. Intestinal Microbiota in Colorectal Adenoma-Carcinoma Sequence. Front. Med. 2022, 9, 888340. [Google Scholar] [CrossRef] [PubMed]
- Centuori, S.M.; Martinez, J.D. Differential Regulation of EGFR–MAPK Signaling by Deoxycholic Acid (DCA) and Ursodeoxycholic Acid (UDCA) in Colon Cancer. Dig. Dis. Sci. 2014, 59, 2367–2380. [Google Scholar] [CrossRef]
- Lee, J.Y.; Arai, H.; Nakamura, Y.; Fukiya, S.; Wada, M.; Yokota, A. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 2013, 54, 3062–3069. [Google Scholar] [CrossRef]
- Pandey, H.; Tang, D.W.T.; Wong, S.H.; Lal, D. Gut Microbiota in Colorectal Cancer: Biological Role and Therapeutic Opportunities. Cancers 2023, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Dong, L.; Wang, T.; Zhang, M.; Hua, W.; Zhang, C.; Pang, X.; Chen, M.; Su, M.; Qiu, Y.; et al. Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiol. Ecol. 2010, 73, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; McDowell, A.; Kim, E.K.; Seo, H.; Lee, W.H.; Moon, C.-M.; Kym, S.-M.; Lee, D.H.; Park, Y.S.; Jee, Y.-K.; et al. Development of a colorectal cancer diagnostic model and dietary risk assessment through gut microbiome analysis. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef]
- Richard, M.L.; Liguori, G.; Lamas, B.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018, 9, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Sarhadi, V.; Lahti, L.; Saberi, F.; Youssef, O.; Kokkola, A.; Karla, T.; Tikkanen, M.; Rautelin, H.; Puolakkainen, P.; Salehi, R.; et al. Gut Microbiota and Host Gene Mutations in Colorectal Cancer Patients and Controls of Iranian and Finnish Origin. Anticancer Res. 2020, 40, 1325–1334. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, D.; Wu, D.; Gao, X.; Shao, F.; Zhao, M.; Wang, J.; Ma, J.; Wang, W.; Qin, X.; et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe 2023, 31, 418–432.e8. [Google Scholar] [CrossRef]
- Wu, Y.; Jiao, N.; Zhu, R.; Zhang, Y.; Wu, D.; Wang, A.-J.; Fang, S.; Tao, L.; Li, Y.; Cheng, S.; et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat. Commun. 2021, 12, 3063. [Google Scholar] [CrossRef]
- Malkan, A.D.; Strollo, W.; Scholand, S.J.; Dudrick, S.J. Implanted-port-catheter-related sepsis caused by Acidovorax avenae and methicillin-sensitive Staphylococcus aureus. J. Clin. Microbiol. 2009, 47, 3358–3361. [Google Scholar] [CrossRef][Green Version]
- Sanapareddy, N.; Legge, R.M.; Jovov, B.; McCoy, A.; Burcal, L.; Araujo-Perez, F.; Randall, T.A.; Galanko, J.; Benson, A.; Sandler, R.S.; et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012, 6, 1858–1868. [Google Scholar] [CrossRef]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; De’angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef]
- Ryu, S.W.; Kim, J.S.; Oh, B.S.; Choi, W.J.; Yu, S.Y.; Bak, J.E.; Park, S.-H.; Kang, S.W.; Lee, J.; Jung, W.Y.; et al. Gut Microbiota Eubacterium callanderi Exerts Anti-Colorectal Cancer Activity. Microbiol. Spectr. 2022, 10, e0253122. [Google Scholar] [CrossRef]
- Faghfoori, Z.; Faghfoori, M.H.; Saber, A.; Izadi, A.; Yari Khosroushahi, A. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021, 21, 258. [Google Scholar] [CrossRef]
- Chen, H.; Tong, T.; Lu, S.Y.; Ji, L.; Xuan, B.; Zhao, G.; Yan, Y.; Song, L.; Zhao, L.; Xie, Y.; et al. Urea cycle activation triggered by host-microbiota maladaptation driving colorectal tumorigenesis. Cell Metab. 2023, 35, 651–666.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jin, K.; Xiong, K.; Jing, W.; Pang, Z.; Feng, M.; Cheng, X. Disease-associated gut microbiome and critical metabolomic alterations in patients with colorectal cancer. Cancer Med. 2023, 12, 15720–15735. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.V.; Visnovska, M.; Biggs, P.J.; Schmeier, S.; Frizelle, F.A. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017, 7, 11590. [Google Scholar] [CrossRef] [PubMed]
- Kneis, B.; Wirtz, S.; Weber, K.; Denz, A.; Gittler, M.; Geppert, C.; Brunner, M.; Krautz, C.; Siebenhüner, A.R.; Schierwagen, R.; et al. Colon Cancer Microbiome Landscaping: Differences in Right- and Left-Sided Colon Cancer and a Tumor Microbiome-Ileal Microbiome Association. Int. J. Mol. Sci. 2023, 24, 3265. [Google Scholar] [CrossRef] [PubMed]
- Barot, S.V.; Sangwan, N.; Nair, K.G.; Schmit, S.L.; Xiang, S.; Kamath, S.; Liska, D.; A Khorana, A. Distinct intratumoral microbiome of young-onset and average-onset colorectal cancer. EBioMedicine 2024, 100, 104980. [Google Scholar] [CrossRef] [PubMed]
- Jayakrishnan, T.; Farha, N.; Mariam, A.; Rotroff, D.M.; Aucejo, F.; Barot, S.V.; Conces, M.; Nair, K.G.; Krishnamurthi, S.S.; Schmit, S.; et al. Metabolomic differences in young-onset versus average-onset colorectal adenocarcinoma. J. Clin. Oncol. 2023, 41 (Suppl. S4), 174. [Google Scholar] [CrossRef]
- Jayakrishnan, T.; Sangwan, N.; Barot, S.; Farha, N.; Mariam, A.; Xiang, S.; Aucejo, F.; Conces, M.; Nair, K.; Krishnamurthi, S.; et al. P-38 Using a machine learning approach to study host-microbiome interactions in early-onset colorectal adenocarcinoma. Ann. Oncol. 2023, 34, S27. [Google Scholar] [CrossRef]
- Sears, C.L.; Pardoll, D.M. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 2011, 203, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Rhee, K.J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.M.; Abrams, V.; Moore, D.R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002, 23, 529–536. [Google Scholar] [CrossRef]
- Swidsinski, A.; Khilkin, M.; Kerjaschki, D.; Schreiber, S.; Ortner, M.; Weber, J.; Lochs, H. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 1998, 115, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Herrera, P.; Kwon, Y.M.; Ricke, S.C. Ecology and pathogenicity of gastrointestinal Streptococcus bovis. Anaerobe 2009, 15, 44–54. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Geng, J.; Song, Q.; Tang, X.; Liang, X.; Fan, H.; Peng, H.; Guo, Q.; Zhang, Z. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Stidham, R.W.; Higgins, P.D.R. Colorectal Cancer in Inflammatory Bowel Disease. Clin. Colon. Rectal Surg. 2018, 31, 168–178. [Google Scholar]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C.; et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Lasry, A.; Zinger, A.; Ben-Neriah, Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016, 17, 230–240. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Windey, K.; De Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Toxicity of Polyamines and Their Metabolic Products. Chem. Res. Toxicol. 2013, 26, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009, 15, 3329–3340. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Singh, A.P.; Sharma, S.; Pagarware, K.; Siraji, R.A.; Ansari, I.; Mandal, A.; Walling, P.; Aijaz, S. Enteropathogenic E. coli effectors EspF and Map independently disrupt tight junctions through distinct mechanisms involving transcriptional and post-transcriptional regulation. Sci. Rep. 2018, 8, 3719. [Google Scholar] [CrossRef]
- Fu, C.; Yang, Z.; Yu, J.; Wei, M. The interaction between gut microbiome and anti-tumor drug therapy. Am. J. Cancer Res. 2021, 11, 5812–5832. [Google Scholar]
- Wan, L.; Li, H.; Sun, G.; Zhang, L.; Xu, H.; Su, F.; He, S.; Xiao, F. Mutational Pattern Induced by 5-Fluorouracil and Oxaliplatin in the Gut Microbiome. Front. Microbiol. 2022, 13, 841458. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Sauter, K.A.; Wood, L.J.; Wong, J.; Iordanov, M.; Magun, B.E. Doxorubicin and daunorubicin induce processing and release of interleukin-1β through activation of the NLRP3 inflammasome. Cancer Biol. Ther. 2011, 11, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Ha, E.M. Combination Therapy of Lactobacillus plantarum Supernatant and 5-Fluouracil Increases Chemosensitivity in Colorectal Cancer Cells. J. Microbiol. Biotechnol. 2016, 26, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Siddik, Z.H. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene 2003, 22, 7265–7279. [Google Scholar] [CrossRef] [PubMed]
- Stojanovska, V.; McQuade, R.M.; Fraser, S.; Prakash, M.; Gondalia, S.; Stavely, R.; Palombo, E.; Apostolopoulos, V.; Sakkal, S.; Nurgali, K. Oxaliplatin-induced changes in microbiota, TLR4+ cells and enhanced HMGB1 expression in the murine colon. PLoS ONE 2018, 13, e0198359. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.-A.; Mani, S.; et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef]
- Yang, W.; Wei, B.; Yan, R. Amoxapine Demonstrates Incomplete Inhibition of β-Glucuronidase Activity from Human Gut Microbiota. SLAS Discov. 2018, 23, 76–83. [Google Scholar] [CrossRef]
- Zhu, J.; Su, J. Alterations of the Gut Microbiome in Recurrent Malignant Gliomas Patients Received Bevacizumab and Temozolomide Combination Treatment and Temozolomide Monotherapy. Indian J. Microbiol. 2022, 62, 23–31. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Segre, J.A. Microbial growth dynamics and human disease. Science 2015, 349, 1058–1059. [Google Scholar] [CrossRef] [PubMed]
- Drewes, J.L.; Housseau, F.; Sears, C.L. Sporadic colorectal cancer: Microbial contributors to disease prevention, development and therapy. Br. J. Cancer 2016, 115, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Jenq, R.R.; Perales, M.A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Zhao, L.; Zhang, X.; Nakatsu, G.; Han, J.; Xu, W.; Xiao, X.; Kwong, T.N.Y.; Tsoi, H.; Wu, W.K.K.; et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017, 153, 1621–1633.e6. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zuo, L.; Guan, B.; Wu, H.; He, Y.; Xu, Z.; Shen, M.; Hu, J.; Qian, J. Microbiota from patients with ulcerative colitis promote colorectal carcinogenesis in mice. Nutrition 2022, 102, 111712. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Zhong, W.; Yang, M.; Xu, M.; Sun, Y.; Ma, J.; Liu, T.; Song, X.; Dong, W.; et al. Gut microbiota from colorectal cancer patients enhances the progression of intestinal adenoma in Apc(min/+) mice. EBioMedicine 2019, 48, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hua, W.; Li, C.; Chang, H.; Liu, R.; Ni, Y.; Sun, H.; Li, Y.; Wang, X.; Hou, M.; et al. Protective Role of Fecal Microbiota Transplantation on Colitis and Colitis-Associated Colon Cancer in Mice Is Associated With Treg Cells. Front. Microbiol. 2019, 10, 2498. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Rosshart, S.P.; Vassallo, B.G.; Angeletti, D.; Hutchinson, D.S.; Morgan, A.P.; Takeda, K.; Hickman, H.D.; McCulloch, J.A.; Badger, J.H.; Ajami, N.J.; et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 2017, 171, 1015–1028.e13. [Google Scholar] [CrossRef]
- Pardoll, D. Cancer and the Immune System: Basic Concepts and Targets for Intervention. Semin. Oncol. 2015, 42, 523–538. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).