Gut Microbiome–Colorectal Cancer Relationship
Abstract
1. Introduction
2. Gut Microbiome’s Essential Role in Cancer Prevention
3. Abnormal Gut Microbiome in Patients with Colorectal Cancer
3.1. Streptococcus Bovis
3.2. Fusobacteria
3.3. Enterococcus Fecalis
3.4. Anaeroplasma
3.5. Flavobacteria
3.6. Ruminococcaceae
3.7. Acidovarax
3.8. Eubacteria
3.9. Bifidobacteria
3.10. Others
4. Biochemistry and Microbiome of Patients with Colorectal Cancer
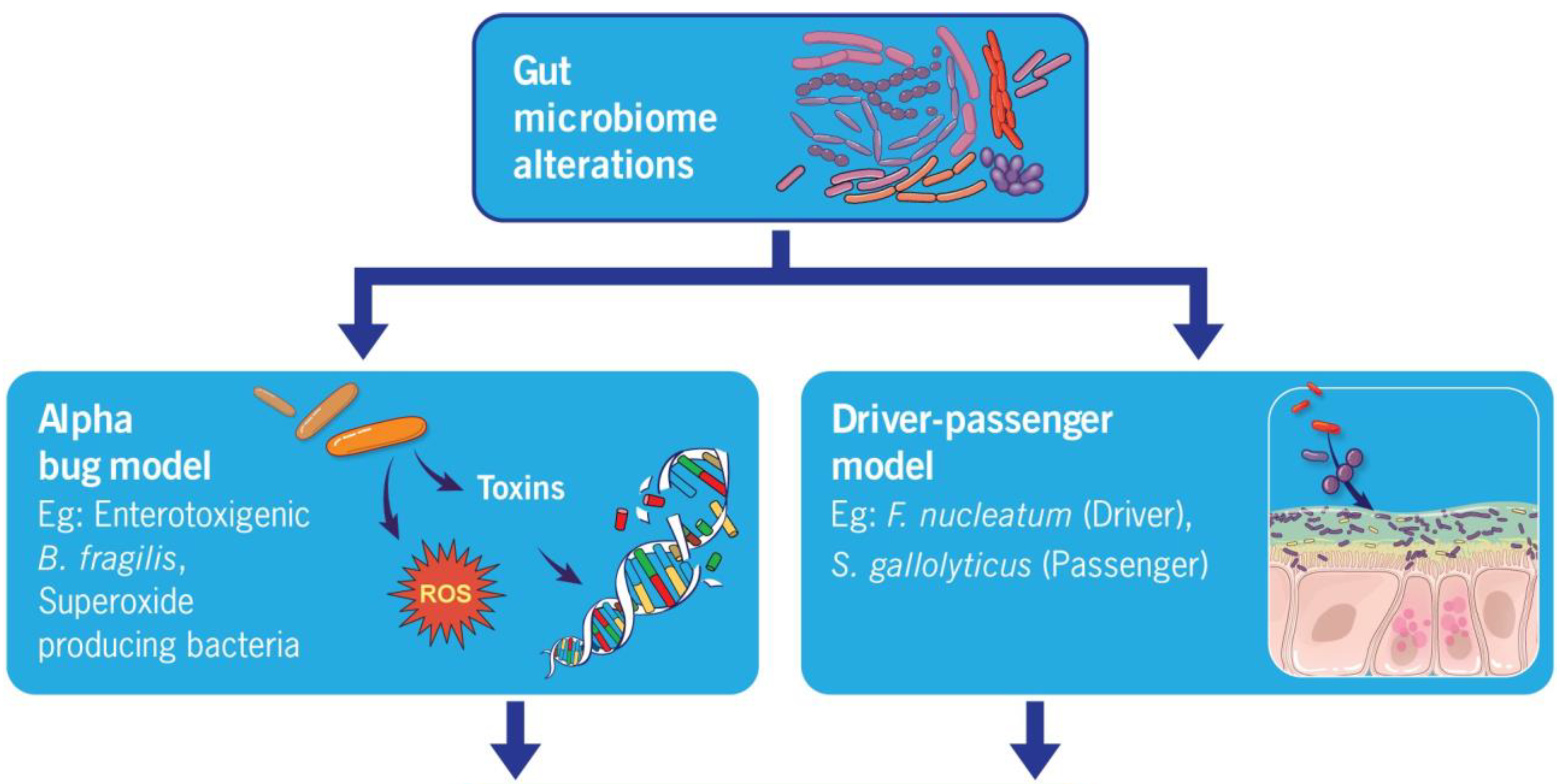
4.1. Alpha Bug Model
4.2. Driver–Passenger Hypothesis
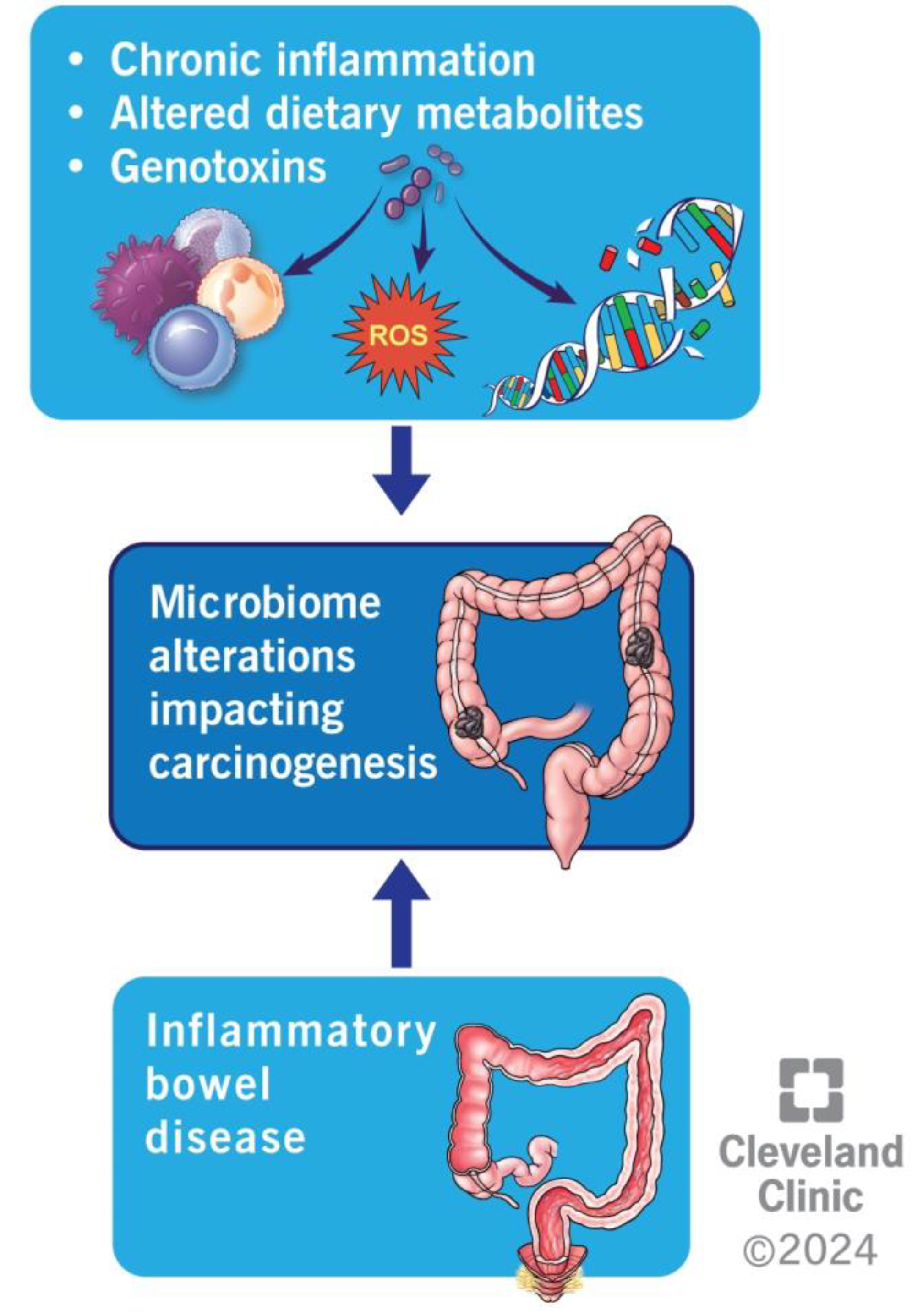
4.3. Inflammation
4.4. Metabolism of Dietary Components
4.5. Production of Genotoxins
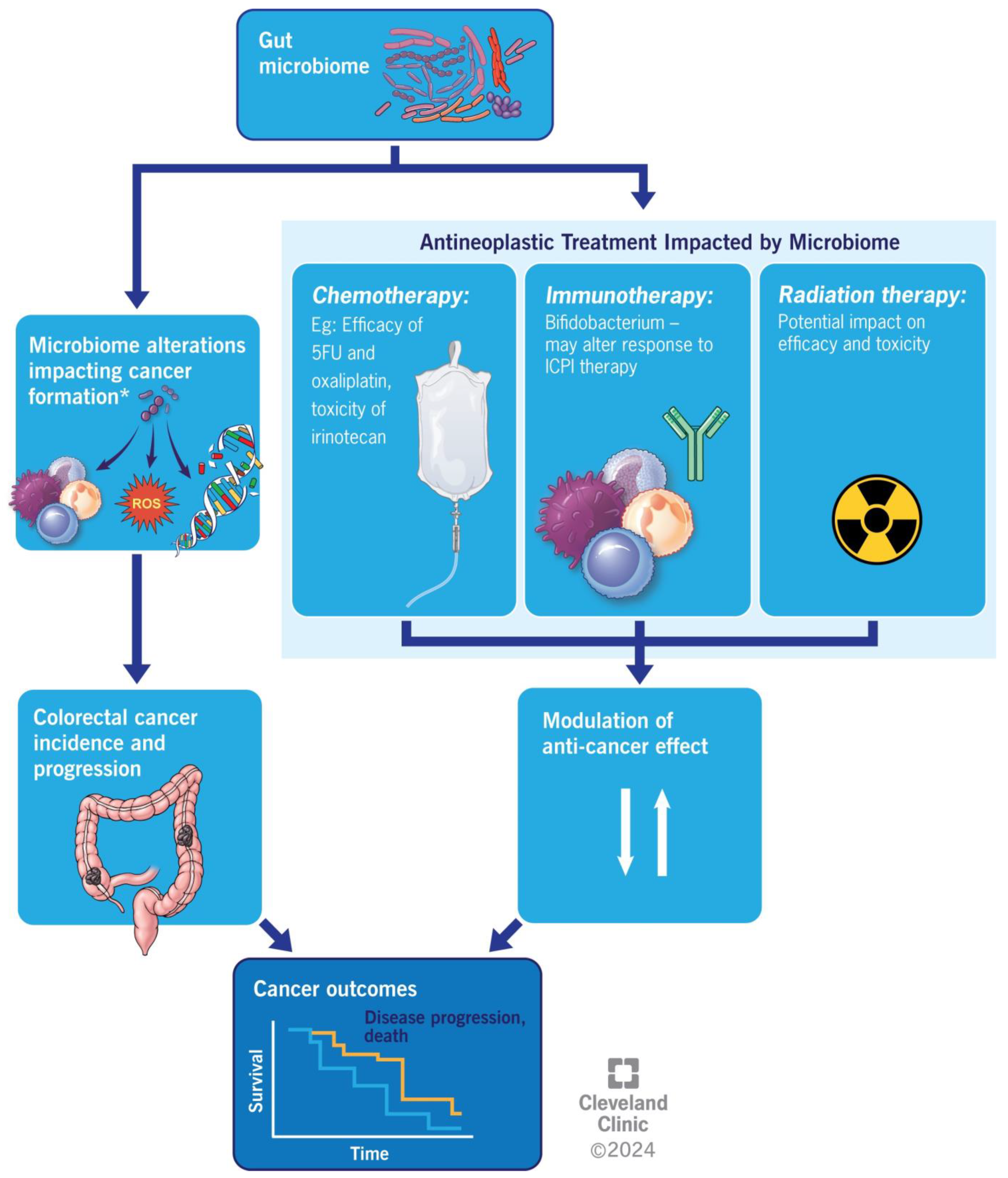
5. Colorectal Cancer and Anti-Neoplastic Medication
6. Fecal Microbiota Transplantation in Colorectal Cancer
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 2020, 8045646. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.-H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef]
- Cancer Stat Facts: Colorectal Cancer. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 2 January 2024).
- Financial Burden of Cancer Care. 2023. Available online: https://progressreport.cancer.gov/after/economic_burden (accessed on 2 January 2024).
- Kim, J.; Lee, H.K. Potential Role of the Gut Microbiome in Colorectal Cancer Progression. Front. Immunol. 2021, 12, 807648. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, D.; Kim, D.; Cho, J.; Yang, J.; Chung, M.; Kim, K.; Ha, N. Inhibition of proliferation in colon cancer cell lines and harmful enzyme activity of colon bacteria by Bifidobacterium adolescentis SPM0212. Arch. Pharm. Res. 2008, 31, 468–473. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Wang, L.; Zhou, Y.; Hassan, J.S.; Li, M. Distribution and gene mutation of enteric flora carrying β-glucuronidase among patients with colorectal cancer. Int. J. Clin. Exp. Med. 2015, 8, 5310–5316. [Google Scholar]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Kuugbee, E.D.; Shang, X.; Gamallat, Y.; Bamba, D.; Awadasseid, A.; Suliman, M.A.; Zang, S.; Ma, Y.; Chiwala, G.; Xin, Y.; et al. Structural Change in Microbiota by a Probiotic Cocktail Enhances the Gut Barrier and Reduces Cancer via TLR2 Signaling in a Rat Model of Colon Cancer. Dig. Dis. Sci. 2016, 61, 2908–2920. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Li, H.; Li, Y. Effects of Bacillus subtilis on Epithelial Tight Junctions of Mice with Inflammatory Bowel Disease. J. Interferon Cytokine Res. 2016, 36, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Fu, Y.; Wang, L.; Qian, W.; Zheng, F.; Hou, X. Bifidobacterium longum and VSL#3® amelioration of TNBS-induced colitis associated with reduced HMGB1 and epithelial barrier impairment. Dev. Comp. Immunol. 2019, 92, 77–86. [Google Scholar] [PubMed]
- Isidro, R.A.; Lopez, A.; Cruz, M.L.; Gonzalez Torres, M.I.; Chompre, G.; Isidro, A.A.; Appleyard, C.B. The Probiotic VSL#3 Modulates Colonic Macrophages, Inflammation, and Microflora in Acute Trinitrobenzene Sulfonic Acid Colitis. J. Histochem. Cytochem. 2017, 65, 445–461. [Google Scholar] [PubMed]
- Jacouton, E.; Chain, F.; Sokol, H.; Langella, P.; Bermúdez-Humarán, L.G. Probiotic Strain Lactobacillus casei BL23 Prevents Colitis-Associated Colorectal Cancer. Front. Immunol. 2017, 8, 1553. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Madani, R.; Mukhtar, H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis. 2010, 12, 164–171. [Google Scholar] [CrossRef]
- Srivastava, A.; Walter, N.; Atkinson, P. Streptococcus bovis infection of total hip arthroplasty in association with carcinoma of colon. J. Surg. Orthop. Adv. 2010, 19, 125–128. [Google Scholar]
- Boleij, A.; van Gelder, M.M.; Swinkels, D.W.; Tjalsma, H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: Systematic review and meta-analysis. Clin. Infect. Dis. 2011, 53, 870–878. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Abu Bakar, F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: The nature and the underlying mechanisms of its etiological role. J. Exp. Clin. Cancer Res. 2011, 30, 11. [Google Scholar] [CrossRef]
- Biarc, J.; Nguyen, I.S.; Pini, A.; Gossé, F.; Richert, S.; Thiersé, D.; Van Dorsselaer, A.; Leize-Wagner, E.; Raul, F.; Klein, J.-P.; et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis). Carcinogenesis 2004, 25, 1477–1484. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Bakar, F.A. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: Inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer 2010, 9, 249. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Wang, R.; Bhattacharya, R.; Boulbes, D.R.; Fan, F.; Xia, L.; Adoni, H.; Ajami, N.J.; Wong, M.C.; Smith, D.P.; et al. Fusobacterium Nucleatum Subspecies Animalis Influences Proinflammatory Cytokine Expression and Monocyte Activation in Human Colorectal Tumors. Cancer Prev. Res. 2017, 10, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Abed, J.; Emgård Johanna, E.M.; Zamir, G.; Faroja, M.; Almogy, G.; Grenov, A.; Sol, A.; Naor, R.; Pikarsky, E.; Atlan, K.A.; et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 2016, 20, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Casasanta, M.A.; Yoo, C.C.; Udayasuryan, B.; Sanders, B.E.; Umaña, A.; Zhang, Y.; Peng, H.; Duncan, A.J.; Wang, Y.; Li, L.; et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 2020, 13, eaba9157. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K.; et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef]
- Hamada, T.; Zhang, X.; Mima, K.; Bullman, S.; Sukawa, Y.; Nowak, J.A.; Kosumi, K.; Masugi, Y.; Twombly, T.S.; Cao, Y.; et al. Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol. Res. 2018, 6, 1327–1336. [Google Scholar] [CrossRef]
- Serna, G.; Ruiz-Pace, F.; Hernando, J.; Alonso, L.; Fasani, R.; Landolfi, S.; Comas, R.; Jimenez, J.; Elez, E.; Bullman, S.; et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020, 31, 1366–1375. [Google Scholar] [CrossRef]
- Sayed, I.M.; Chakraborty, A.; Abd El-Hafeez, A.A.; Sharma, A.; Sahan, A.Z.; Huang, W.J.M.; Sahoo, D.; Ghosh, P.; Hazra, T.K.; Das, S. The DNA Glycosylase NEIL2 Suppresses Fusobacterium-Infection-Induced Inflammation and DNA Damage in Colonic Epithelial Cells. Cells 2020, 9, 1980. [Google Scholar] [CrossRef]
- Guo, P.; Tian, Z.; Kong, X.; Yang, L.; Shan, X.; Dong, B.; Ding, X.; Jing, X.; Jiang, C.; Jiang, N.; et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J. Exp. Clin. Cancer Res. 2020, 39, 202. [Google Scholar] [CrossRef]
- Saito, K.; Koido, S.; Odamaki, T.; Kajihara, M.; Kato, K.; Horiuchi, S.; Adachi, S.; Arakawa, H.; Yoshida, S.; Akasu, T.; et al. Metagenomic analyses of the gut microbiota associated with colorectal adenoma. PLoS ONE 2019, 14, e0212406. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Onozawa, H.; Saito, M.; Saito, K.; Kanke, Y.; Watanabe, Y.; Hayase, S.; Sakamoto, W.; Ishigame, T.; Momma, T.; Ohki, S.; et al. Annexin A1 is involved in resistance to 5-FU in colon cancer cells. Oncol. Rep. 2017, 37, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.X.; Chan, W.S.; Tang, K.S.; Zheng, S.Y. (Eds.) Restart based Collective Information Powered Differential Evolution for Solving the 100-Digit Challenge on Single Objective Numerical Optimization. In Proceedings of the 2019 IEEE Congress on Evolutionary Computation (CEC), Wellington, New Zealand, 10–13 June 2019. [Google Scholar]
- Lu, P.; Xu, M.; Xiong, Z.; Zhou, F.; Wang, L. Fusobacterium nucleatum prevents apoptosis in colorectal cancer cells via the ANO1 pathway. Cancer Manag. Res. 2019, 11, 9057–9066. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, A.C.; de Mattos Pereira, L.; Datorre, J.G.; Dos Santos, W.; Berardinelli, G.N.; Matsushita, M.M.; Oliveira, M.A.; Durães, R.O.; Guimarães, D.P.; Reis, R.M. Microbiota Profile and Impact of Fusobacterium nucleatum in Colorectal Cancer Patients of Barretos Cancer Hospital. Front. Oncol. 2019, 9, 813. [Google Scholar] [CrossRef]
- Ito, M.; Kanno, S.; Nosho, K.; Sukawa, Y.; Mitsuhashi, K.; Kurihara, H.; Igarashi, H.; Takahashi, T.; Tachibana, M.; Takahashi, H.; et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int. J. Cancer 2015, 137, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Kinugasa, H.; Hirai, M.; Terasawa, H.; Yasutomi, E.; Oka, S.; Ohmori, M.; Yamasaki, Y.; Inokuchi, T.; Harada, K.; et al. Heterogeneous distribution of Fusobacterium nucleatum in the progression of colorectal cancer. J. Gastroenterol. Hepatol. 2021, 36, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Winters, M.D.; Schlinke, T.L.; Joyce, W.A.; Glore, S.R.; Huycke, M.M. Prospective case-cohort study of intestinal colonization with enterococci that produce extracellular superoxide and the risk for colorectal adenomas or cancer. Am. J. Gastroenterol. 1998, 93, 2491–2500. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef]
- Wang, X.; Huycke, M.M. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 2007, 132, 551–561. [Google Scholar] [CrossRef]
- Gates, T.J.; Yuan, C.; Shetty, M.; Kaiser, T.; Nelson, A.C.; Chauhan, A.; Starr, T.K.; Staley, C.; Subramanian, S. Fecal Microbiota Restoration Modulates the Microbiome in Inflammation-Driven Colorectal Cancer. Cancers 2023, 15, 2260. [Google Scholar] [CrossRef]
- Vitali, F.; Tortora, K.; Di Paola, M.; Bartolucci, G.; Menicatti, M.; De Filippo, C.; Caderni, G. Intestinal microbiota profiles in a genetic model of colon tumorigenesis correlates with colon cancer biomarkers. Sci. Rep. 2022, 12, 1432. [Google Scholar] [CrossRef] [PubMed]
- Alrafas, H.R.; Busbee, P.B.; Chitrala, K.N.; Nagarkatti, M.; Nagarkatti, P. Alterations in the Gut Microbiome and Suppression of Histone Deacetylases by Resveratrol Are Associated with Attenuation of Colonic Inflammation and Protection against Colorectal Cancer. J. Clin. Med. 2020, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Iadsee, N.; Chuaypen, N.; Techawiwattanaboon, T.; Jinato, T.; Patcharatrakul, T.; Malakorn, S.; Petchlorlian, A.; Praditpornsilpa, K.; Patarakul, K. Identification of a novel gut microbiota signature associated with colorectal cancer in Thai population. Sci. Rep. 2023, 13, 6702. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, T.; Chen, G.Y. Flavonoids and Colorectal Cancer Prevention. Antioxidants 2018, 7, 187. [Google Scholar] [CrossRef]
- Hua, H.; Sun, Y.; He, X.; Chen, Y.; Teng, L.; Lu, C. Intestinal Microbiota in Colorectal Adenoma-Carcinoma Sequence. Front. Med. 2022, 9, 888340. [Google Scholar] [CrossRef] [PubMed]
- Centuori, S.M.; Martinez, J.D. Differential Regulation of EGFR–MAPK Signaling by Deoxycholic Acid (DCA) and Ursodeoxycholic Acid (UDCA) in Colon Cancer. Dig. Dis. Sci. 2014, 59, 2367–2380. [Google Scholar] [CrossRef]
- Lee, J.Y.; Arai, H.; Nakamura, Y.; Fukiya, S.; Wada, M.; Yokota, A. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 2013, 54, 3062–3069. [Google Scholar] [CrossRef]
- Pandey, H.; Tang, D.W.T.; Wong, S.H.; Lal, D. Gut Microbiota in Colorectal Cancer: Biological Role and Therapeutic Opportunities. Cancers 2023, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Dong, L.; Wang, T.; Zhang, M.; Hua, W.; Zhang, C.; Pang, X.; Chen, M.; Su, M.; Qiu, Y.; et al. Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiol. Ecol. 2010, 73, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; McDowell, A.; Kim, E.K.; Seo, H.; Lee, W.H.; Moon, C.-M.; Kym, S.-M.; Lee, D.H.; Park, Y.S.; Jee, Y.-K.; et al. Development of a colorectal cancer diagnostic model and dietary risk assessment through gut microbiome analysis. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef]
- Richard, M.L.; Liguori, G.; Lamas, B.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018, 9, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Sarhadi, V.; Lahti, L.; Saberi, F.; Youssef, O.; Kokkola, A.; Karla, T.; Tikkanen, M.; Rautelin, H.; Puolakkainen, P.; Salehi, R.; et al. Gut Microbiota and Host Gene Mutations in Colorectal Cancer Patients and Controls of Iranian and Finnish Origin. Anticancer Res. 2020, 40, 1325–1334. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, D.; Wu, D.; Gao, X.; Shao, F.; Zhao, M.; Wang, J.; Ma, J.; Wang, W.; Qin, X.; et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe 2023, 31, 418–432.e8. [Google Scholar] [CrossRef]
- Wu, Y.; Jiao, N.; Zhu, R.; Zhang, Y.; Wu, D.; Wang, A.-J.; Fang, S.; Tao, L.; Li, Y.; Cheng, S.; et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat. Commun. 2021, 12, 3063. [Google Scholar] [CrossRef]
- Malkan, A.D.; Strollo, W.; Scholand, S.J.; Dudrick, S.J. Implanted-port-catheter-related sepsis caused by Acidovorax avenae and methicillin-sensitive Staphylococcus aureus. J. Clin. Microbiol. 2009, 47, 3358–3361. [Google Scholar] [CrossRef][Green Version]
- Sanapareddy, N.; Legge, R.M.; Jovov, B.; McCoy, A.; Burcal, L.; Araujo-Perez, F.; Randall, T.A.; Galanko, J.; Benson, A.; Sandler, R.S.; et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012, 6, 1858–1868. [Google Scholar] [CrossRef]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; De’angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef]
- Ryu, S.W.; Kim, J.S.; Oh, B.S.; Choi, W.J.; Yu, S.Y.; Bak, J.E.; Park, S.-H.; Kang, S.W.; Lee, J.; Jung, W.Y.; et al. Gut Microbiota Eubacterium callanderi Exerts Anti-Colorectal Cancer Activity. Microbiol. Spectr. 2022, 10, e0253122. [Google Scholar] [CrossRef]
- Faghfoori, Z.; Faghfoori, M.H.; Saber, A.; Izadi, A.; Yari Khosroushahi, A. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021, 21, 258. [Google Scholar] [CrossRef]
- Chen, H.; Tong, T.; Lu, S.Y.; Ji, L.; Xuan, B.; Zhao, G.; Yan, Y.; Song, L.; Zhao, L.; Xie, Y.; et al. Urea cycle activation triggered by host-microbiota maladaptation driving colorectal tumorigenesis. Cell Metab. 2023, 35, 651–666.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jin, K.; Xiong, K.; Jing, W.; Pang, Z.; Feng, M.; Cheng, X. Disease-associated gut microbiome and critical metabolomic alterations in patients with colorectal cancer. Cancer Med. 2023, 12, 15720–15735. [Google Scholar] [CrossRef] [PubMed]
- Purcell, R.V.; Visnovska, M.; Biggs, P.J.; Schmeier, S.; Frizelle, F.A. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci. Rep. 2017, 7, 11590. [Google Scholar] [CrossRef] [PubMed]
- Kneis, B.; Wirtz, S.; Weber, K.; Denz, A.; Gittler, M.; Geppert, C.; Brunner, M.; Krautz, C.; Siebenhüner, A.R.; Schierwagen, R.; et al. Colon Cancer Microbiome Landscaping: Differences in Right- and Left-Sided Colon Cancer and a Tumor Microbiome-Ileal Microbiome Association. Int. J. Mol. Sci. 2023, 24, 3265. [Google Scholar] [CrossRef] [PubMed]
- Barot, S.V.; Sangwan, N.; Nair, K.G.; Schmit, S.L.; Xiang, S.; Kamath, S.; Liska, D.; A Khorana, A. Distinct intratumoral microbiome of young-onset and average-onset colorectal cancer. EBioMedicine 2024, 100, 104980. [Google Scholar] [CrossRef] [PubMed]
- Jayakrishnan, T.; Farha, N.; Mariam, A.; Rotroff, D.M.; Aucejo, F.; Barot, S.V.; Conces, M.; Nair, K.G.; Krishnamurthi, S.S.; Schmit, S.; et al. Metabolomic differences in young-onset versus average-onset colorectal adenocarcinoma. J. Clin. Oncol. 2023, 41 (Suppl. S4), 174. [Google Scholar] [CrossRef]
- Jayakrishnan, T.; Sangwan, N.; Barot, S.; Farha, N.; Mariam, A.; Xiang, S.; Aucejo, F.; Conces, M.; Nair, K.; Krishnamurthi, S.; et al. P-38 Using a machine learning approach to study host-microbiome interactions in early-onset colorectal adenocarcinoma. Ann. Oncol. 2023, 34, S27. [Google Scholar] [CrossRef]
- Sears, C.L.; Pardoll, D.M. Perspective: Alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 2011, 203, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Rhee, K.J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.M.; Abrams, V.; Moore, D.R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002, 23, 529–536. [Google Scholar] [CrossRef]
- Swidsinski, A.; Khilkin, M.; Kerjaschki, D.; Schreiber, S.; Ortner, M.; Weber, J.; Lochs, H. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 1998, 115, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Herrera, P.; Kwon, Y.M.; Ricke, S.C. Ecology and pathogenicity of gastrointestinal Streptococcus bovis. Anaerobe 2009, 15, 44–54. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Geng, J.; Song, Q.; Tang, X.; Liang, X.; Fan, H.; Peng, H.; Guo, Q.; Zhang, Z. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Stidham, R.W.; Higgins, P.D.R. Colorectal Cancer in Inflammatory Bowel Disease. Clin. Colon. Rectal Surg. 2018, 31, 168–178. [Google Scholar]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C.; et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Lasry, A.; Zinger, A.; Ben-Neriah, Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016, 17, 230–240. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Windey, K.; De Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Toxicity of Polyamines and Their Metabolic Products. Chem. Res. Toxicol. 2013, 26, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009, 15, 3329–3340. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Singh, A.P.; Sharma, S.; Pagarware, K.; Siraji, R.A.; Ansari, I.; Mandal, A.; Walling, P.; Aijaz, S. Enteropathogenic E. coli effectors EspF and Map independently disrupt tight junctions through distinct mechanisms involving transcriptional and post-transcriptional regulation. Sci. Rep. 2018, 8, 3719. [Google Scholar] [CrossRef]
- Fu, C.; Yang, Z.; Yu, J.; Wei, M. The interaction between gut microbiome and anti-tumor drug therapy. Am. J. Cancer Res. 2021, 11, 5812–5832. [Google Scholar]
- Wan, L.; Li, H.; Sun, G.; Zhang, L.; Xu, H.; Su, F.; He, S.; Xiao, F. Mutational Pattern Induced by 5-Fluorouracil and Oxaliplatin in the Gut Microbiome. Front. Microbiol. 2022, 13, 841458. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Sauter, K.A.; Wood, L.J.; Wong, J.; Iordanov, M.; Magun, B.E. Doxorubicin and daunorubicin induce processing and release of interleukin-1β through activation of the NLRP3 inflammasome. Cancer Biol. Ther. 2011, 11, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Ha, E.M. Combination Therapy of Lactobacillus plantarum Supernatant and 5-Fluouracil Increases Chemosensitivity in Colorectal Cancer Cells. J. Microbiol. Biotechnol. 2016, 26, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Siddik, Z.H. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene 2003, 22, 7265–7279. [Google Scholar] [CrossRef] [PubMed]
- Stojanovska, V.; McQuade, R.M.; Fraser, S.; Prakash, M.; Gondalia, S.; Stavely, R.; Palombo, E.; Apostolopoulos, V.; Sakkal, S.; Nurgali, K. Oxaliplatin-induced changes in microbiota, TLR4+ cells and enhanced HMGB1 expression in the murine colon. PLoS ONE 2018, 13, e0198359. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.-A.; Mani, S.; et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef]
- Yang, W.; Wei, B.; Yan, R. Amoxapine Demonstrates Incomplete Inhibition of β-Glucuronidase Activity from Human Gut Microbiota. SLAS Discov. 2018, 23, 76–83. [Google Scholar] [CrossRef]
- Zhu, J.; Su, J. Alterations of the Gut Microbiome in Recurrent Malignant Gliomas Patients Received Bevacizumab and Temozolomide Combination Treatment and Temozolomide Monotherapy. Indian J. Microbiol. 2022, 62, 23–31. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C.; et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Segre, J.A. Microbial growth dynamics and human disease. Science 2015, 349, 1058–1059. [Google Scholar] [CrossRef] [PubMed]
- Drewes, J.L.; Housseau, F.; Sears, C.L. Sporadic colorectal cancer: Microbial contributors to disease prevention, development and therapy. Br. J. Cancer 2016, 115, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Jenq, R.R.; Perales, M.A.; Littmann, E.R.; Morjaria, S.; Ling, L.; No, D.; Gobourne, A.; Viale, A.; Dahi, P.B.; et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014, 124, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Zhao, L.; Zhang, X.; Nakatsu, G.; Han, J.; Xu, W.; Xiao, X.; Kwong, T.N.Y.; Tsoi, H.; Wu, W.K.K.; et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017, 153, 1621–1633.e6. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zuo, L.; Guan, B.; Wu, H.; He, Y.; Xu, Z.; Shen, M.; Hu, J.; Qian, J. Microbiota from patients with ulcerative colitis promote colorectal carcinogenesis in mice. Nutrition 2022, 102, 111712. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Zhong, W.; Yang, M.; Xu, M.; Sun, Y.; Ma, J.; Liu, T.; Song, X.; Dong, W.; et al. Gut microbiota from colorectal cancer patients enhances the progression of intestinal adenoma in Apc(min/+) mice. EBioMedicine 2019, 48, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hua, W.; Li, C.; Chang, H.; Liu, R.; Ni, Y.; Sun, H.; Li, Y.; Wang, X.; Hou, M.; et al. Protective Role of Fecal Microbiota Transplantation on Colitis and Colitis-Associated Colon Cancer in Mice Is Associated With Treg Cells. Front. Microbiol. 2019, 10, 2498. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Rosshart, S.P.; Vassallo, B.G.; Angeletti, D.; Hutchinson, D.S.; Morgan, A.P.; Takeda, K.; Hickman, H.D.; McCulloch, J.A.; Badger, J.H.; Ajami, N.J.; et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 2017, 171, 1015–1028.e13. [Google Scholar] [CrossRef]
- Pardoll, D. Cancer and the Immune System: Basic Concepts and Targets for Intervention. Semin. Oncol. 2015, 42, 523–538. [Google Scholar] [CrossRef]
| Study | Design | Results |
|---|---|---|
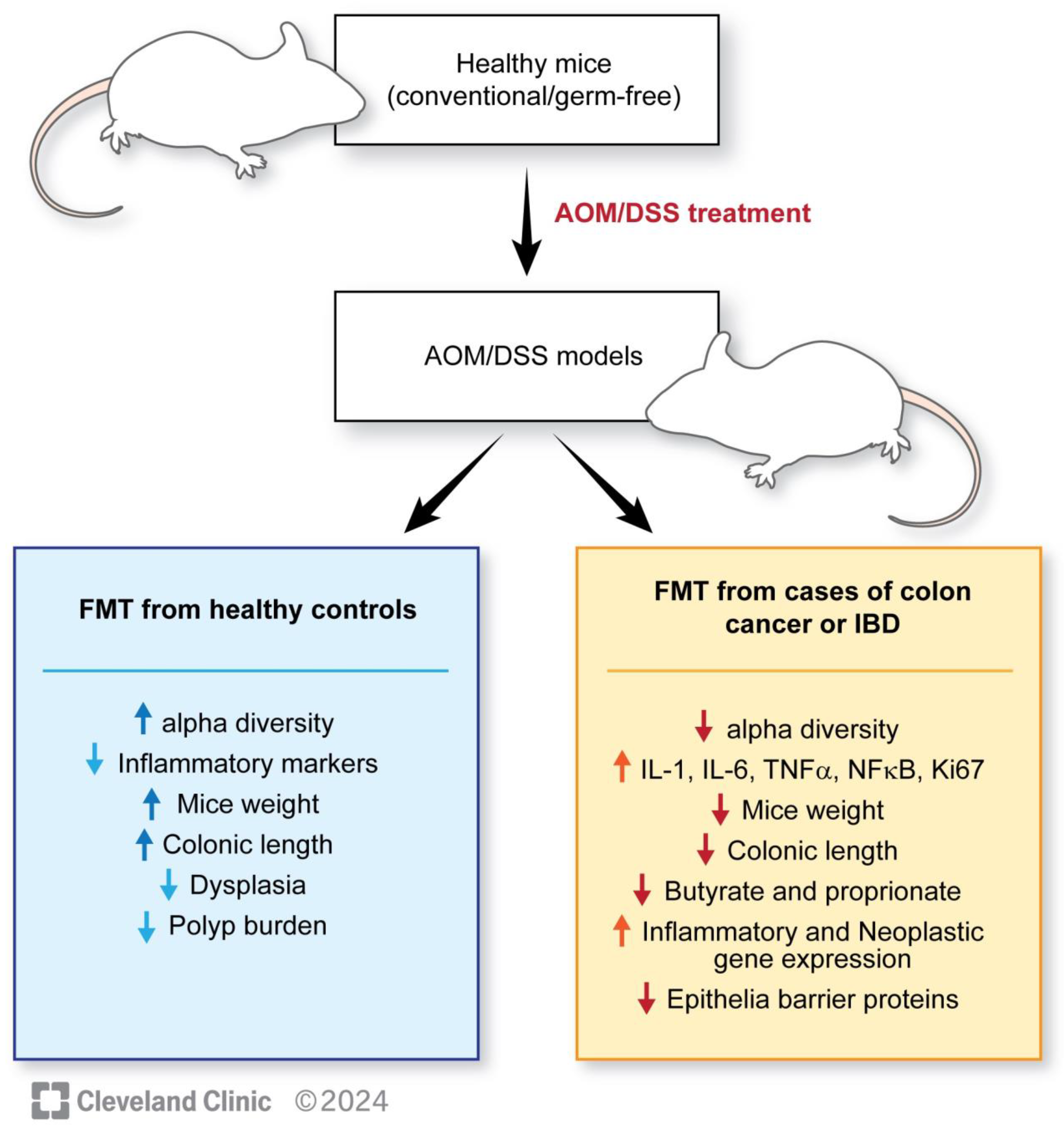
| Wang et al., 2019 [117] | Three mouse groups: CAC+FMT: mice receiving FMT post-AOM-DSS CAC: mice receiving AOM-DSS Control: mice receiving isotonic saline | The CAC group showed an increased percentage of Bacteroides as compared with Firmicutes when compared with CAC-FMT and the control group. Shannon Index, PD Whole Tree Index and Chao1 Index (measures of alpha diversity) showed a significant increase in the CAC + FMT group. PCA for evaluation of beta diversity in CAC mice showed clustering post-FMT. Decrease in IL-1b, IL-6, TNF-alpha, NF-kB, ki-67 and phospho-p65 levels, increase in IL-10, TGF-beta in CAC + FMT levels at 70 days post-FMT. |
| Tian et al., 2022 [115] | AOM-treated mice were divided into three groups: FS-HC: AOM mice gavaged with healthy feces FS-UC: AOM mice gavaged with feces from patients with ulcerative colitis (UC) PBs: AOM mice gavaged with phosphate-buffered saline | FS-UC significantly increased the disease activity index, leading to a lower body weight, shorter colon length, a higher number of polyps, more severe dysplasia, a higher Ki-67-positive burden, increased IFN-y, TNF-alpha, Th1 and Th17 expression, and decreased butyrate and propionate concentrations as compared with FS-HC group. |
| Wong et al., 2017 [114] | The conventional (AOM-treated) and germ-free mouse populations were divided into: CRC-A: conventional mice gavaged with feces of a patient with CRC HC-A: conventional mice gavaged with feces of healthy patients NC-A: conventional mice gavaged with PBS CRC-G: germ-free mice gavaged with feces of patients with CRC HC-G: germ-free mice gavaged with feces of healthy patients | CRC-A group had a higher number of polyps with higher compositive scores, indicative of severe dysplasia and lower bacterial richness. CRC-G group showed increased epithelial proliferation, more Ki-67-positive cells (difference not statistically significant), increased proliferating cell nuclear antigen staining, higher beta-catenin expression, and lower Shannon–Weaver diversity indexes. Both CRC-A and CRC-G groups showed an increase in 33 out of 84 genes associated with inflammation, including Cxcr1, Cxcr2, IL17a, IL22, IL23a, and IFNy-encoding gene. The gene for Tlr-5 was significantly downregulated. Overall, 37 out of the 84 genes involved in cancer pathways also showed upregulation, including Ki-67, Mcm2, Aurka, Cd20, and Bmi1. |
| Li et al., 2019 [116] | 20 C57BL/6 mice and 30 APC gene knockout mice (APC min/+) were used C57BL/6 mice were divided into 2 groups: FMT-CC: gavaged with feces of CRC patients FMT-CH: gavaged with healthy control feces APC min/+ mice were divided into three groups: FMT-AC: gavaged with feces of CRC patients FMT-AH: gavaged with healthy control feces PBS: ones gavaged with PBS | There were no significant changes in mouse, liver, and spleen weight at 8 weeks post-FMT in FMT-AC vs. FMT-AH groups. Overall, 30% of mice in the FMT-AC group showed high-grade dysplasia compared with 10% in the FMT-AH group. Ki-67-positive cells increased in the FMT-AC group. Decrease in ZO-1, occludin, claudin-3, Muc2, cryptdin and Reg3gamma expression and increase in NLRP3, IL-1beta, TNG-alpha and sIgA expression in the small intestine of the FMT-AC group. |
| Rosshart et al., 2017 [121] | Three categories of mice were selected: Lab: offspring of C57BL/6 mice that did not receive any gavaging. LabR: offspring of germ-free mice that received the frozen gut microbiome from SPF C57BL/6 WildR: offspring of mice receiving the microbiome from wild mice | Results post-AOM-DSS induction in all the categories showed that WildR mice had significantly decreased inflammation, AOM-DSS-induced weight loss, and a lower number and surface area of colorectal tumors with significantly low invasiveness scores compared with Lab and LabR mice. |
| Routy et al., 2018 [120] | Mice with established MCA-205 sarcoma and RET melanoma were divided into 2 groups. ATB: mice treated with ampicillin + colistin + streptomycin Control: untreated mice Second part of the study: ATB mice inoculated with MCA-205 tumor cells were divided into 2 groups: ATB-R: Receiving FMT from feces of NSCLC responders ATB-NR: Receiving FMT from feces of NSCLC nonresponders | Significantly compromised antitumor effects and survival of ATB mice with PD-1 mAb or in combination with CTLA-4 mAb. Higher microbial richness was associated with the absence of disease progression. Natural progression of sarcoma significantly improved in ATB-R with tumor growth delay, the accumulation of CXCR3 + CD4+ T cells, and the upregulation of PD-L1 in splenic T cells after PD-L1 blockade. A. mucinophila, E. hirae and Alistipes indistinctus from responder stool samples showed restoration of anti-tumor activity of ICIs previously inhibited by antibiotics. |
| Gopalakrishnan et al., 2018 [118] | Germ-free mouse models injected with BP melanoma cells were divided into 2 parts: R group: receiving gavage from ICI responder patients NR group: receiving gavage from non-responder patients | R group showed significantly decreased tumor size, improved response to anti-PD L1 therapy, increased Faecalibacterium, and increased CD45+ and CD8+ T cells. NR group showed increased regulatory CD4+ FoxP3+ T cells and CD4+ IL-17+ T cells in the spleen, suggesting an impaired immune response. |
| Matson et al., 2018 [119] | Germ-free mice injected with B16.SIY melanoma cells were divided into 3 groups: SPF: GF mice gavaged with feces of Taconic-specific pathogen-free (SPF) mice R group: GF mice gavaged with feces of patients responding to ICI NR group: GF mice gavaged with feces of patients not responding to ICI | SPF mice did not show any change in baseline tumor growth rate. 2 out of 3 mice in the R group showed slow tumor progression compared with 1 out of 3 mice in the NR group. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, D.; Sainatham, C.; Filippov, E.; Kanagala, S.G.; Ishaq, S.M.; Jayakrishnan, T. Gut Microbiome–Colorectal Cancer Relationship. Microorganisms 2024, 12, 484. https://doi.org/10.3390/microorganisms12030484
Yadav D, Sainatham C, Filippov E, Kanagala SG, Ishaq SM, Jayakrishnan T. Gut Microbiome–Colorectal Cancer Relationship. Microorganisms. 2024; 12(3):484. https://doi.org/10.3390/microorganisms12030484
Chicago/Turabian StyleYadav, Devvrat, Chiranjeevi Sainatham, Evgenii Filippov, Sai Gautham Kanagala, Syed Murtaza Ishaq, and Thejus Jayakrishnan. 2024. "Gut Microbiome–Colorectal Cancer Relationship" Microorganisms 12, no. 3: 484. https://doi.org/10.3390/microorganisms12030484
APA StyleYadav, D., Sainatham, C., Filippov, E., Kanagala, S. G., Ishaq, S. M., & Jayakrishnan, T. (2024). Gut Microbiome–Colorectal Cancer Relationship. Microorganisms, 12(3), 484. https://doi.org/10.3390/microorganisms12030484







