Seed-Borne Bacterial Diversity of Fescue (Festuca ovina L.) and Properties Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Isolation and Identification of Seed-Borne Bacteria from Fescue Seeds
2.3. Determination of Biological Properties and Functions
2.3.1. Determination of Motility of Seed-Borne Bacteria
2.3.2. Biofilm-Forming Capacity
2.3.3. Determination of Minimum Inhibitory Concentration (MIC)
2.3.4. Measurement of Indole-3-Acetic Acid (IAA)
2.3.5. Nitrogen Fixation and Extracellular Enzyme Activity
2.4. Statistical Analysis
3. Results
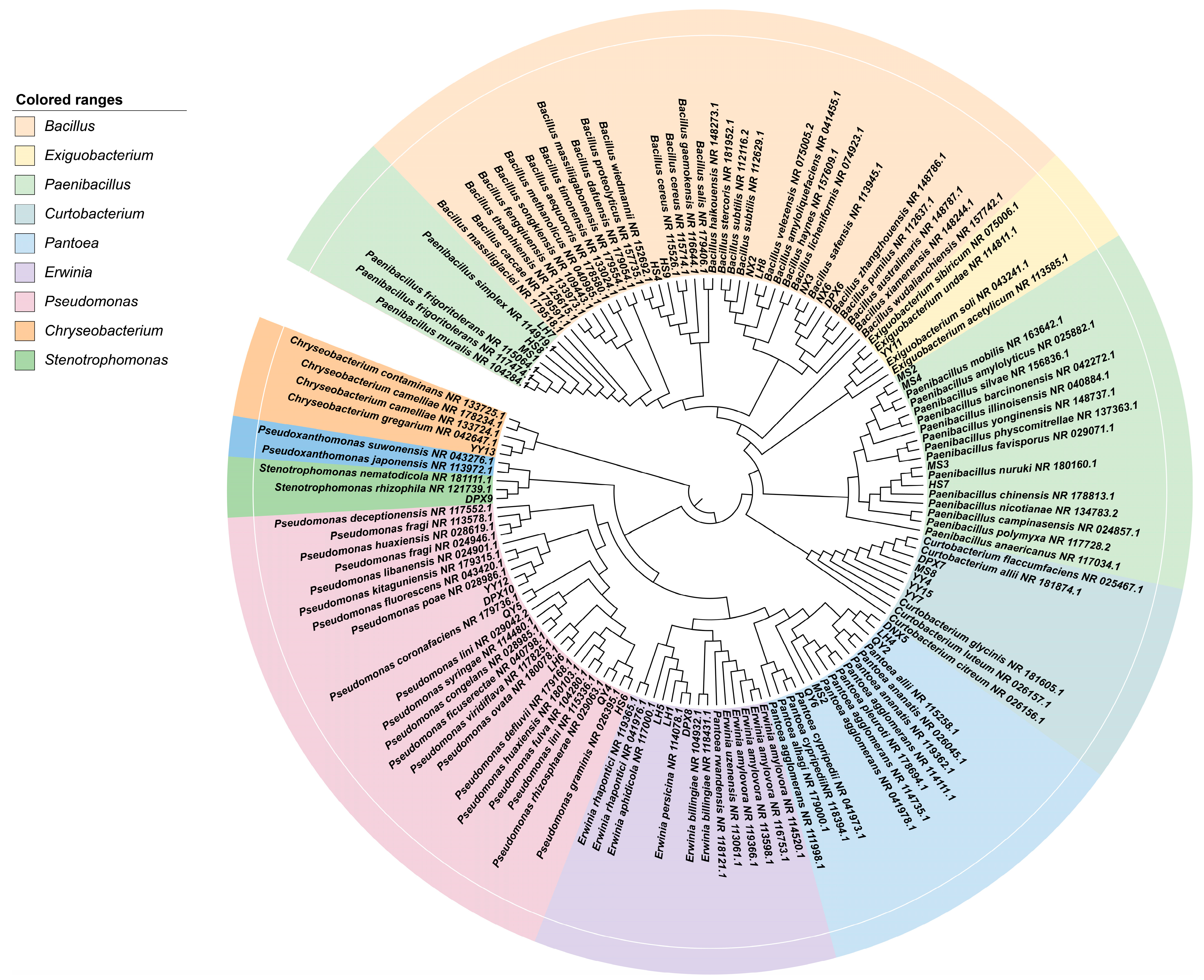
3.1. Fescue Seeds from Different Varieties as Natural Carriers of Taxonomically Diverse Culturable Seed-Borne Bacteria
3.2. Community Structure and Abundance Analysis of Seed-Borne Bacteria
3.3. IAA Production, Nitrogen Fixation Capacity, Soluble Amylase and Protease Activity
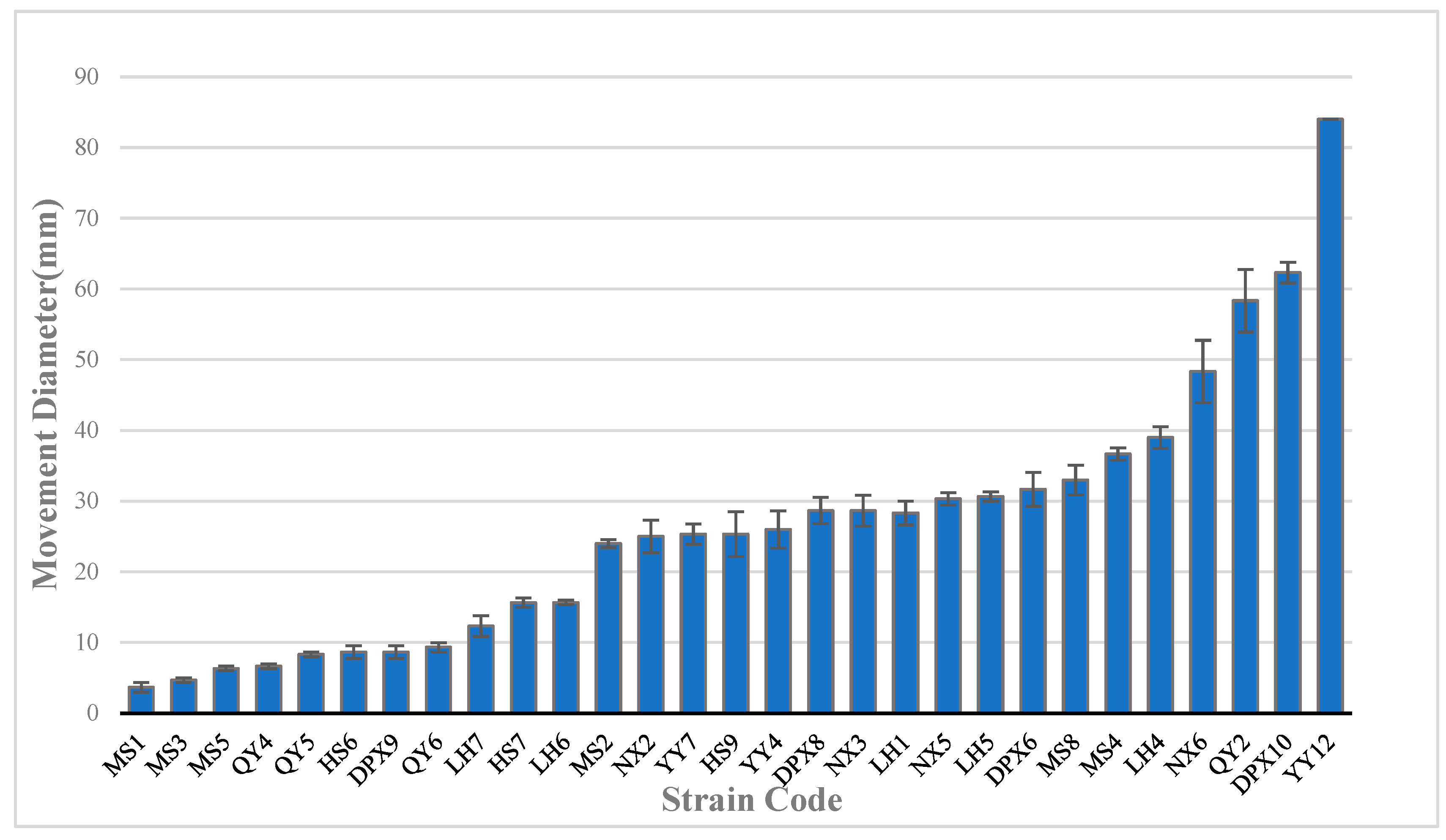
3.4. Motility of Seed-Borne Bacteria
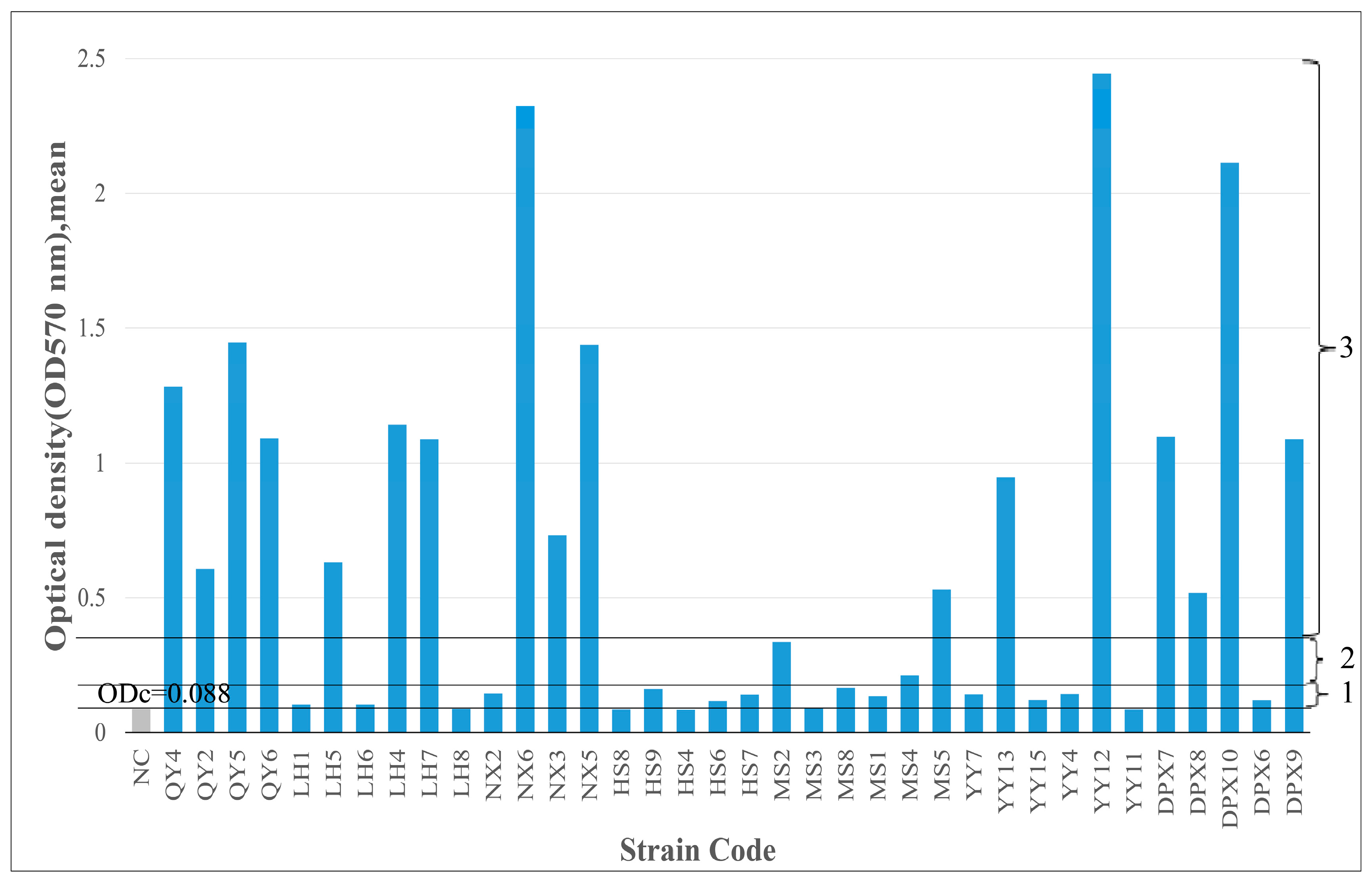
3.5. Biofilm-Forming Ability
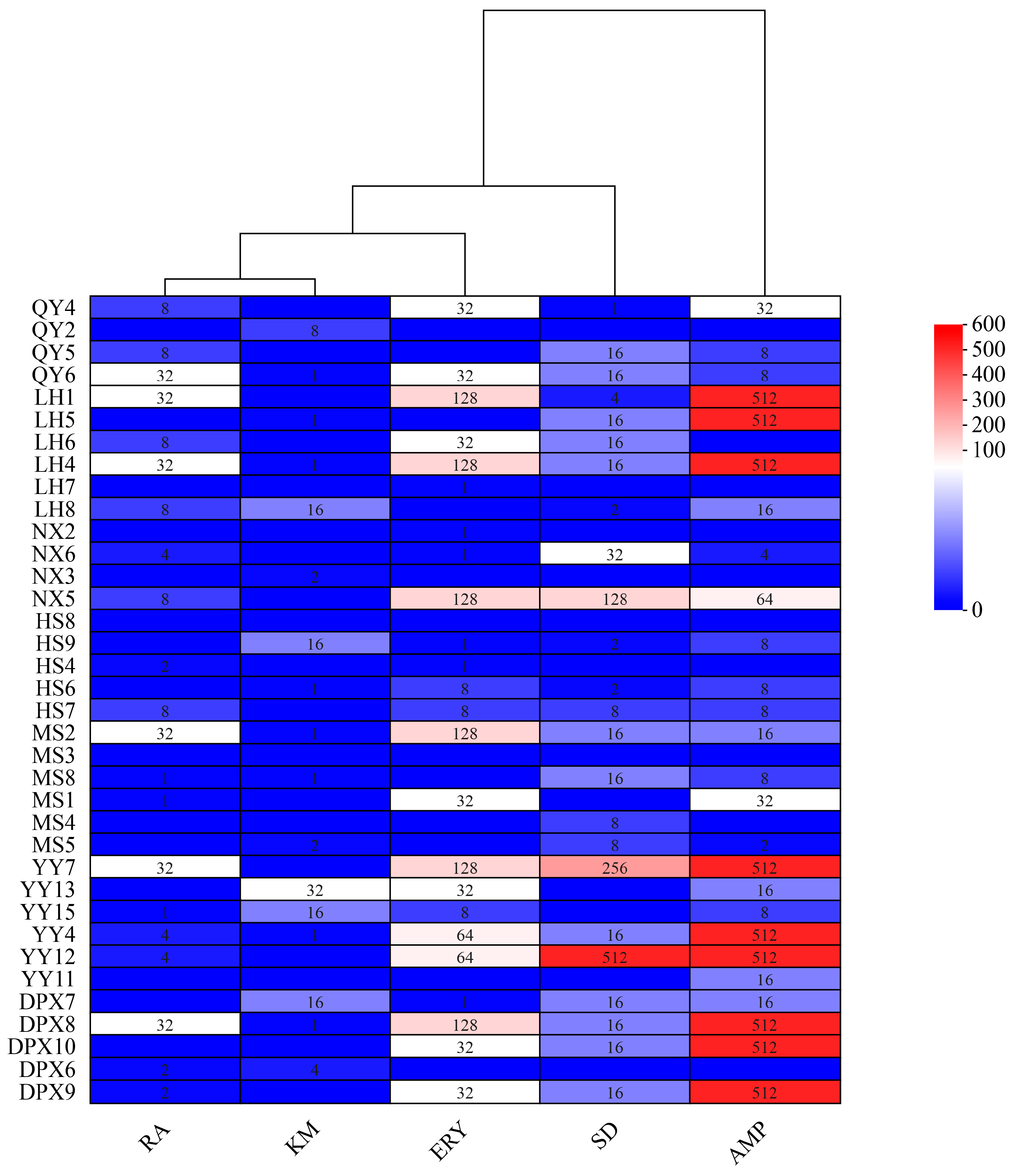
3.6. Antibiotic Resistance of Seed-Borne Bacteria
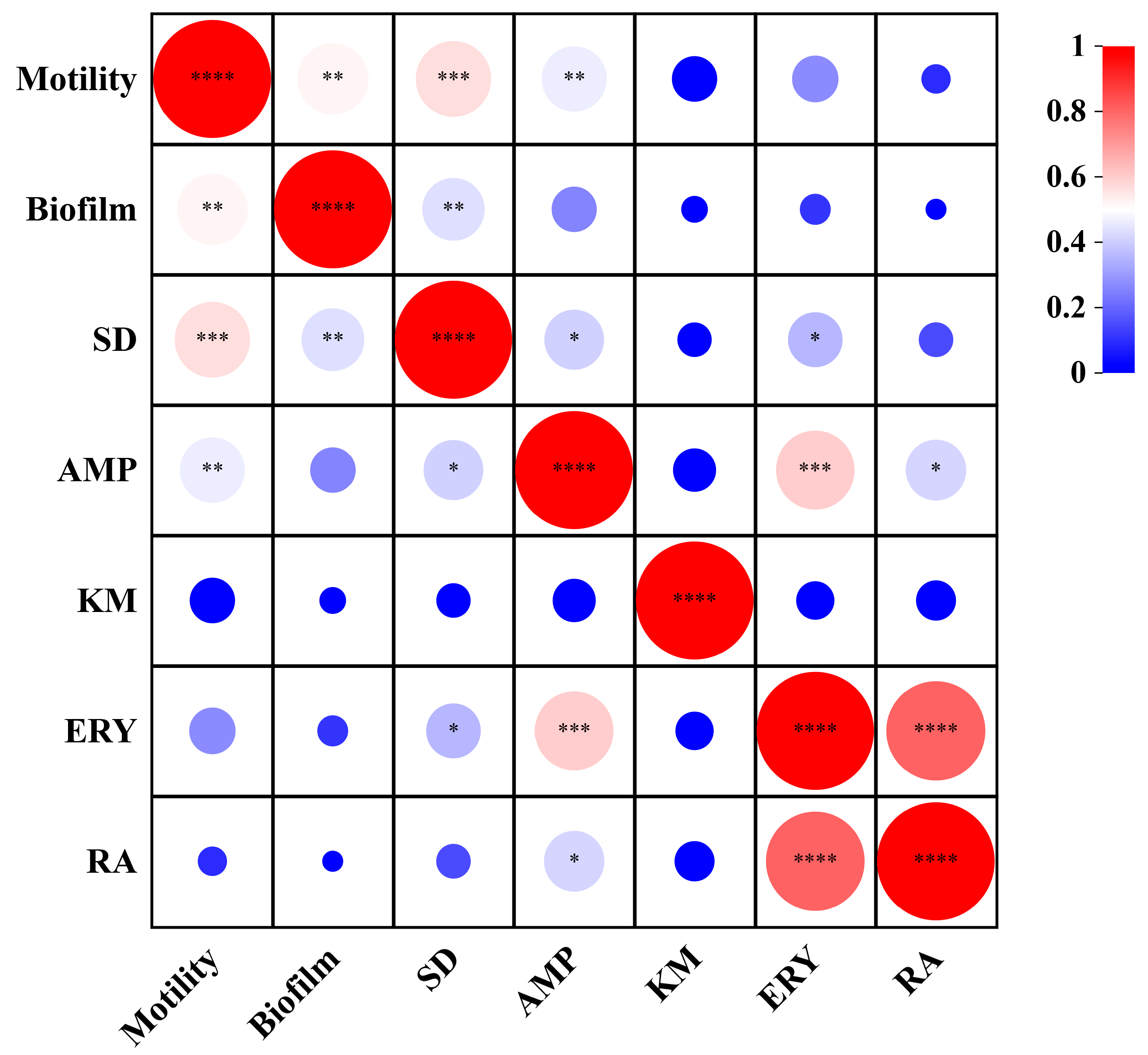
3.7. Correlation Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, M.; Tian, P.; Gao, M.; Li, M. The Promotion of Festuca Sinensis under Heavy Metal Treatment Mediated by Epichloë Endophyte. Agronomy 2021, 11, 2049. [Google Scholar] [CrossRef]
- Tian, P.; Kuang, Y.; Nan, Z.B. The characteristics of Festuca sinensis and its breeding potential. Acta Pratacult. Sin. 2015, 32, 1079–1087. [Google Scholar]
- Sheffer, K.M.; Dunn, J.H.; Minner, D.D. Summer Drought Response and Rooting Depth of Three Cool-Season Turfgrasses. HortScience 1987, 22, 296–297. [Google Scholar] [CrossRef]
- Nelson, E.B. Microbial Dynamics and Interactions in the Spermosphere. Annu. Rev. Phytopathol. 2004, 42, 271–309. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.L.; Zhang, S.; Tsolis, R.M.; Kingsley, R.A.; Garry Adams, L.; Bäumler, A.J. Animal Models of Salmonella Infections: Enteritis versus Typhoid Fever. Microbes Infect. 2001, 3, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Miliute, I.; Buzaite, O.; Baniulis, D.; Stanys, V. Bacterial Endophytes in Agricultural Crops and Their Role in Stress Tolerance: A Review. ZemdirZemdirb.-Agric. 2015, 102, 465–478. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Beauchamp, C.J. A Review of Issues Related to Measuring Colonization of Plant Roots by Bacteria. Can. J. Microbiol. 1992, 38, 1219–1232. [Google Scholar] [CrossRef]
- Cope-Selby, N.; Cookson, A.; Squance, M.; Donnison, I.; Flavell, R.; Farrar, K. Endophytic Bacteria in Miscanthus Seed: Implications for Germination, Vertical Inheritance of Endophytes, Plant Evolution and Breeding. GCB Bioenergy 2017, 9, 57–77. [Google Scholar] [CrossRef]
- Ishida, A.; Furuya, T. Diversity and Characteristics of Culturable Endophytic Bacteria from Passiflora edulis Seeds. MicrobiologyOpen 2021, 10, e1226. [Google Scholar] [CrossRef]
- Singh, R.; Pandey, K.D.; Singh, M.; Singh, S.K.; Hashem, A.; Al-Arjani, A.-B.F.; Abd_Allah, E.F.; Singh, P.K.; Kumar, A. Isolation and Characterization of Endophytes Bacterial Strains of Momordica charantia L. and Their Possible Approach in Stress Management. Microorganisms 2022, 10, 290. [Google Scholar] [CrossRef]
- Barea, J.M.; Pozo, M.J.; Azcón, R.; Azcón-Aguilar, C. Microbial Co-Operation in the Rhizosphere. J. Exp. Bot. 2005, 56, 1761–1778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Q.; Ying, G.G.; Pan, C.G.; Liu, Y.S.; Zhao, J.L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.L.; Cui, H.L.; Su, J.Q.; Penuelas, J.; Zhu, Y.G. Antibiotic Resistomes in Plant Microbiomes. Trends Plant Sci. 2019, 24, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Cotrufo, M.F. Grassland Soil Carbon Sequestration: Current Understanding, Challenges, and Solutions. Science 2022, 377, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Microbiological Effects of Sublethal Levels of Antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Tomida, J.; Kawamura, Y. Responses of Pseudomonas aeruginosa to Antimicrobials. Front. Microbiol. 2014, 4, 422. [Google Scholar] [CrossRef]
- Wu, S.; Liu, J.; Liu, C.; Yang, A.; Qiao, J. Quorum Sensing for Population-Level Control of Bacteria and Potential Therapeutic Applications. Cell. Mol. Life Sci. 2020, 77, 1319–1343. [Google Scholar] [CrossRef]
- Ciofu, O.; Moser, C.; Jensen, P.Ø.; Høiby, N. Tolerance and Resistance of Microbial Biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef]
- Majed, R.; Faille, C.; Kallassy, M.; Gohar, M. Bacillus cereus Biofilms—Same, Only Different. Front. Microbiol. 2016, 7, 1054. [Google Scholar] [CrossRef]
- Okshevsky, M.; Louw, M.G.; Lamela, E.O.; Nilsson, M.; Tolker-Nielsen, T.; Meyer, R.L. A Transposon Mutant Library of Bacillus cereus ATCC 10987 Reveals Novel Genes Required for Biofilm Formation and Implicates Motility as an Important Factor for Pellicle-biofilm Formation. MicrobiologyOpen 2018, 7, e00552. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Briandet, R.; Kovács, Á.T. Bacillus cereus Sensu Lato Biofilm Formation and Its Ecological Importance. Biofilm 2022, 4, 100070. [Google Scholar] [CrossRef] [PubMed]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef]
- Minamino, T.; Namba, K. Self-Assembly and Type III Protein Export of the Bacterial Flagellum. Microb. Physiol. 2004, 7, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Taylor, T.B.; Bates, J.; Buckling, A. Using Experimental Evolution to Explore Natural Patterns between Bacterial Motility and Resistance to Bacteriophages. ISME J. 2011, 5, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Samad, T.; Billings, N.; Birjiniuk, A.; Crouzier, T.; Doyle, P.S.; Ribbeck, K. Swimming Bacteria Promote Dispersal of Non-Motile Staphylococcal Species. ISME J. 2017, 11, 1933–1937. [Google Scholar] [CrossRef]
- Ayangbenro, A.; Babalola, O. A New Strategy for Heavy Metal Polluted Environments: A Review of Microbial Biosorbents. Int. J. Environ. Res. Public Health 2017, 14, 94. [Google Scholar] [CrossRef]
- López, J.L.; Alvarez, F.; Príncipe, A.; Salas, M.E.; Lozano, M.J.; Draghi, W.O.; Jofré, E.; Lagares, A. Isolation, Taxonomic Analysis, and Phenotypic Characterization of Bacterial Endophytes Present in Alfalfa (Medicago sativa) Seeds. J. Biotechnol. 2018, 267, 55–62. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Huang, R.; Yao, B. Culturable seed-borne bacteria of lucerne imported from Europe and North America and their pathogenicity to plants and animals. Acta Pratacult. Sin. 2023, 32, 161–172. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.; Bergen, M.; English, C.; Elmore, M.; Kharwar, R.N.; White, J.F. Bacterial Endophytes from Rice Cut Grass (Leersia oryzoides L.) Increase Growth, Promote Root Gravitropic Response, Stimulate Root Hair Formation, and Protect Rice Seedlings from Disease. Plant Soil 2018, 422, 223–238. [Google Scholar] [CrossRef]
- Qu, H.Y.; Li, X.P.; Meng, L.Y.; Liu, J.F.; Li, J.R.; Chen, J.R. Effect of Inhibitors on the Motilities of Vibrio parahaemolyticus. J. Food Sci. Biotechnol. 2014, 33, 480–485. [Google Scholar]
- Reisner, A.; Krogfelt, K.A.; Klein, B.M.; Zechner, E.L.; Molin, S. In Vitro Biofilm Formation of Commensal and Pathogenic Escherichia coli Strains: Impact of Environmental and Genetic Factors. J. Bacteriol. 2006, 188, 3572–3581. [Google Scholar] [CrossRef] [PubMed]
- Viksne, R.; Racenis, K.; Broks, R.; Balode, A.O.; Kise, L.; Kroica, J. In Vitro Assessment of Biofilm Production, Antibacterial Resistance of Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Obtained from Tonsillar Crypts of Healthy Adults. Microorganisms 2023, 11, 258. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of Minimum Inhibitory Concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The Minimum Inhibitory Concentration of Antibiotics: Methods, Interpretation, Clinical Relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.A.; Weber, R.P. Colorimetric Estimation of Indoleacetic Acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.D.; Li, Z.D.; Chen, X.R. Identification, pathogen inhibiting and nitrogen fixation of endophytic bacterium Z19 of Polygonum viviparum. Microbiol. China 2014, 41, 267–273. [Google Scholar] [CrossRef]
- Pal, G.; Kumar, K.; Verma, A.; Verma, S.K. Seed Inhabiting Bacterial Endophytes of Maize Promote Seedling Establishment and Provide Protection against Fungal Disease. Microbiol. Res. 2022, 255, 126926. [Google Scholar] [CrossRef]
- Truyens, S.; Jambon, I.; Croes, S.; Janssen, J.; Weyens, N.; Mench, M.; Carleer, R.; Cuypers, A.; Vangronsveld, J. The Effect of Long-Term Cd and Ni Exposure on Seed Endophytes of Agrostis capillaris and Their Potential Application in Phytoremediation of Metal-Contaminated Soils. Int. J. Phytoremediat. 2014, 16, 643–659. [Google Scholar] [CrossRef]
- Rosenblueth, M.; López-López, A.; Martínez, J.; Rogel, M.A.; Toledo, I.; Martínez-Romero, E. Seed bacterial endophytes: Common genera, seed-to-seed variability and their possible role in plants. Acta Hortic. 2012, 938, 39–48. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Hardoim, C.C.P.; Van Overbeek, L.S.; Van Elsas, J.D. Dynamics of Seed-Borne Rice Endophytes on Early Plant Growth Stages. PLoS ONE 2012, 7, e30438. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zuo, S.; Zou, Y.; Wang, J.; Song, W. Investigation on Diversity and Population Succession Dynamics of Endophytic Bacteria from Seeds of Maize (Zea mays L., Nongda108) at Different Growth Stages. Ann. Microbiol. 2013, 63, 71–79. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and Diversity of Seed Associated Endophytes in Zea across Boundaries of Evolution, Ethnography and Ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, Y.; Luo, Y.; Liu, J.; Tian, P.; Nan, Z.; Zhou, Q. The Microbiota Diversity of Festuca sinensis Seeds in Qinghai-Tibet Plateau and Their Relationship with Environments. Front. Microbiol. 2022, 13, 956489. [Google Scholar] [CrossRef]
- Rezki, S.; Campion, C.; Simoneau, P.; Jacques, M.-A.; Shade, A.; Barret, M. Assembly of Seed-Associated Microbial Communities within and across Successive Plant Generations. Plant Soil 2018, 422, 67–79. [Google Scholar] [CrossRef]
- Gibbons, S.M.; Caporaso, J.G.; Pirrung, M.; Field, D.; Knight, R.; Gilbert, J.A. Evidence for a Persistent Microbial Seed Bank throughout the Global Ocean. Proc. Natl. Acad. Sci. USA 2013, 110, 4651–4655. [Google Scholar] [CrossRef]
- Thomas, D.C.; Vandegrift, R.; Ludden, A.; Carroll, G.C.; Roy, B.A. Spatial Ecology of the Fungal Genus Xylaria in a Tropical Cloud Forest. Biotropica 2016, 48, 381–393. [Google Scholar] [CrossRef]
- Farrar, K.; Bryant, D.; Cope-Selby, N. Understanding and Engineering Beneficial Plant–Microbe Interactions: Plant Growth Promotion in Energy Crops. Plant Biotechnol. J. 2014, 12, 1193–1206. [Google Scholar] [CrossRef]
- Kuklinsky-Sobral, J.; Araújo, W.L.; Mendes, R.; Geraldi, I.O.; Pizzirani-Kleiner, A.A.; Azevedo, J.L. Isolation and Characterization of Soybean-associated Bacteria and Their Potential for Plant Growth Promotion. Environ. Microbiol. 2004, 6, 1244–1251. [Google Scholar] [CrossRef]
- Dias, A.C.F.; Costa, F.E.C.; Andreote, F.D.; Lacava, P.T.; Teixeira, M.A.; Assumpção, L.C.; Araújo, W.L.; Azevedo, J.L.; Melo, I.S. Isolation of Micropropagated Strawberry Endophytic Bacteria and Assessment of Their Potential for Plant Growth Promotion. World J. Microbiol. Biotechnol. 2009, 25, 189–195. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Botina, S.G.; Netrusov, A.I. Bacteria Associated with Orchid Roots and Microbial Production of Auxin. Microbiol. Res. 2007, 162, 69–76. [Google Scholar] [CrossRef]
- Raina, J.-B.; Fernandez, V.; Lambert, B.; Stocker, R.; Seymour, J.R. The Role of Microbial Motility and Chemotaxis in Symbiosis. Nat. Rev. Microbiol. 2019, 17, 284–294. [Google Scholar] [CrossRef]
- Wiles, T.J.; Schlomann, B.H.; Wall, E.S.; Betancourt, R.; Parthasarathy, R.; Guillemin, K. Swimming Motility of a Gut Bacterial Symbiont Promotes Resistance to Intestinal Expulsion and Enhances Inflammation. PLoS Biol. 2020, 18, e3000661. [Google Scholar] [CrossRef]
- Penesyan, A.; Paulsen, I.T.; Kjelleberg, S.; Gillings, M.R. Three Faces of Biofilms: A Microbial Lifestyle, a Nascent Multicellular Organism, and an Incubator for Diversity. npj Biofilms Microbiomes 2021, 7, 80. [Google Scholar] [CrossRef]
- Liu, F.; Ying, G.-G.; Tao, R.; Zhao, J.-L.; Yang, J.-F.; Zhao, L.-F. Effects of Six Selected Antibiotics on Plant Growth and Soil Microbial and Enzymatic Activities. Environ. Pollut. 2009, 157, 1636–1642. [Google Scholar] [CrossRef]
- Banin, E.; Hughes, D.; Kuipers, O.P. Editorial: Bacterial Pathogens, Antibiotics and Antibiotic Resistance. FEMS Microbiol. Rev. 2017, 41, 450–452. [Google Scholar] [CrossRef]

| Variety | Production Region | Production Year | Longitude (E) | Latitude (N) | Seed Provider |
|---|---|---|---|---|---|
| F. arundinacea ‘horizon’ | Beijing, China | 2022 | 116°23′51″ | 39°54′24″ | Beijing Clover Company Beijing, China |
| F. arundinacea ‘bharal’ | Beijing, China | 2022 | 116°23′51″ | 39°54′24″ | Beijing Clover Company Beijing, China |
| F. arundinacea ‘road-Fire’ | Beijing, China | 2022 | 116°23′51″ | 39°54′24″ | Beijing Clover Company Beijing, China |
| F. sinensis | Ningxia, China | 2020 | 106°28′ | 36°01′ | Lanzhou University Lanzhou, China |
| F. sinensis | Qinghai, China | 2020 | 103°05′ | 39°15′ | Beijing Clover Company Beijing, China |
| F. kryloviana | Qinghai, China | 2020 | 103°05′ | 39°15′ | Lanzhou University Lanzhou, China |
| F. rubra ‘dream-God’ | Beijing, China | 2022 | 116°23′51″ | 39°54′24″ | Beijing Clover Company Beijing, China |
| Isolate Code | Gram Strain Reaction | Similar Strain | Similarity (%) | Genebank Accession |
|---|---|---|---|---|
| QY4 | − | Pseudomonas sp. | 100 | OR858852 |
| QY2 | − | Pantoea sp. | 100 | OR858858 |
| QY5 | − | Pseudomonas sp. | 100 | OR858859 |
| QY6 | − | Pantoea sp. | 100 | OR858860 |
| LH1 | + | Erwinia sp. | 100 | OR858877 |
| LH5 | − | Erwinia sp. | 100 | OR858878 |
| LH4 | − | Pantoea sp. | 100 | OR858879 |
| LH6 | + | Pseudomonas sp. | 99 | OR858880 |
| LH7 | + | Paenibacillus sp. | 100 | OR858881 |
| LH8 | + | Bacillus sp. | 100 | OR858882 |
| NX2 | + | Bacillus sp. | 100 | OR858865 |
| NX6 | − | Bacillus sp. | 100 | OR858866 |
| NX3 | − | Bacillus sp. | 100 | OR858867 |
| NX5 | + | Pantoea sp. | 86 | OR858868 |
| HS8 | + | Paenibacillus sp. | 100 | OR858847 |
| HS9 | − | Bacillus sp. | 100 | OR858848 |
| HS4 | + | Bacillus sp. | 100 | OR858849 |
| HS6 | − | Pseudomonas sp. | 100 | OR858850 |
| HS7 | + | Paenibacillus sp. | 95 | OR858851 |
| MS8 | − | Curtobacterium sp. | 100 | OR858856 |
| MS2 | − | Pantoea sp. | 100 | OR858869 |
| MS3 | + | Paenibacillus sp. | 95 | OR858870 |
| MS1 | + | Paenibacillus sp. | 100 | OR858871 |
| MS4 | − | Paenibacillus sp. | 100 | OR858873 |
| MS5 | − | Paenibacillus sp. | 100 | OR858872 |
| YY7 | − | Curtobacterium sp. | 100 | OR858853 |
| YY13 | + | Chryseobacterium sp. | 100 | OR858876 |
| YY15 | + | Curtobacterium sp. | 100 | OR858854 |
| YY4 | − | Curtobacterium sp. | 100 | OR858855 |
| YY12 | + | Pseudomonas sp. | 91 | OR858875 |
| YY11 | + | Exiguobacterium sp. | 100 | OR858874 |
| DPX7 | − | Curtobacterium sp. | 100 | OR858857 |
| DPX8 | − | Erwinia sp. | 100 | OR858861 |
| DPX10 | − | Pseudomonas sp. | 100 | OR858862 |
| DPX6 | − | Bacillus sp. | 100 | OR858863 |
| DPX9 | − | Stenotrophomonas sp. | 100 | OR858864 |
| Isolates | IAA (μg/mL; M ± SE) | Nitrogen Fixation | Enzyme | |
|---|---|---|---|---|
| Amylase | Protease | |||
| QY2 | − | + | − | + |
| QY4 | 24.13 ± 0.050 | + | + | − |
| QY5 | − | + | − | − |
| QY6 | 40.10 ± 0.233 | + | + | + |
| LH1 | − | − | − | − |
| LH4 | 39.48 ± 0.337 | − | + | − |
| LH5 | 25.44 ± 0.657 | − | + | − |
| LH6 | − | − | − | − |
| LH7 | − | − | − | + |
| LH8 | − | − | − | − |
| NX2 | − | + | − | − |
| NX3 | − | + | − | − |
| NX5 | 33.49 ± 0.387 | + | + | + |
| NX6 | − | + | − | − |
| HS4 | − | − | − | − |
| HS6 | − | + | − | − |
| HS7 | − | + | − | − |
| HS8 | 39.11 ± 0.106 | + | − | + |
| HS9 | 40.88 ± 0.709 | + | + | + |
| MS1 | 21.57 ± 0.381 | + | − | − |
| MS2 | − | + | − | − |
| MS3 | − | + | − | − |
| MS4 | 19.53 ± 0.456 | + | − | − |
| MS5 | − | + | − | − |
| MS8 | − | − | − | − |
| YY4 | − | + | − | + |
| YY7 | 27.38 ± 0.403 | − | − | + |
| YY11 | 22.63 ± 0.535 | + | − | − |
| YY12 | 33.88 ± 0.334 | + | + | + |
| YY13 | 23.46 ± 0.46 | + | − | − |
| YY15 | 20.48 ± 0.403 | + | − | − |
| DPX6 | − | + | − | − |
| DPX7 | 34.51 ± 0.414 | + | + | + |
| DPX8 | − | − | − | − |
| DPX9 | − | + | − | − |
| DPX10 | − | − | − | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Xie, J.; Yang, J.; Hou, X.; He, L.; Zhang, Z. Seed-Borne Bacterial Diversity of Fescue (Festuca ovina L.) and Properties Study. Microorganisms 2024, 12, 329. https://doi.org/10.3390/microorganisms12020329
Zhu S, Xie J, Yang J, Hou X, He L, Zhang Z. Seed-Borne Bacterial Diversity of Fescue (Festuca ovina L.) and Properties Study. Microorganisms. 2024; 12(2):329. https://doi.org/10.3390/microorganisms12020329
Chicago/Turabian StyleZhu, Shaowei, Jinjing Xie, Jie Yang, Xuan Hou, Linxin He, and Zhenfen Zhang. 2024. "Seed-Borne Bacterial Diversity of Fescue (Festuca ovina L.) and Properties Study" Microorganisms 12, no. 2: 329. https://doi.org/10.3390/microorganisms12020329
APA StyleZhu, S., Xie, J., Yang, J., Hou, X., He, L., & Zhang, Z. (2024). Seed-Borne Bacterial Diversity of Fescue (Festuca ovina L.) and Properties Study. Microorganisms, 12(2), 329. https://doi.org/10.3390/microorganisms12020329






