Lactobacilli in COVID-19: A Systematic Review Based on Next-Generation Sequencing Studies
Abstract
1. Introduction
2. Materials and Methods
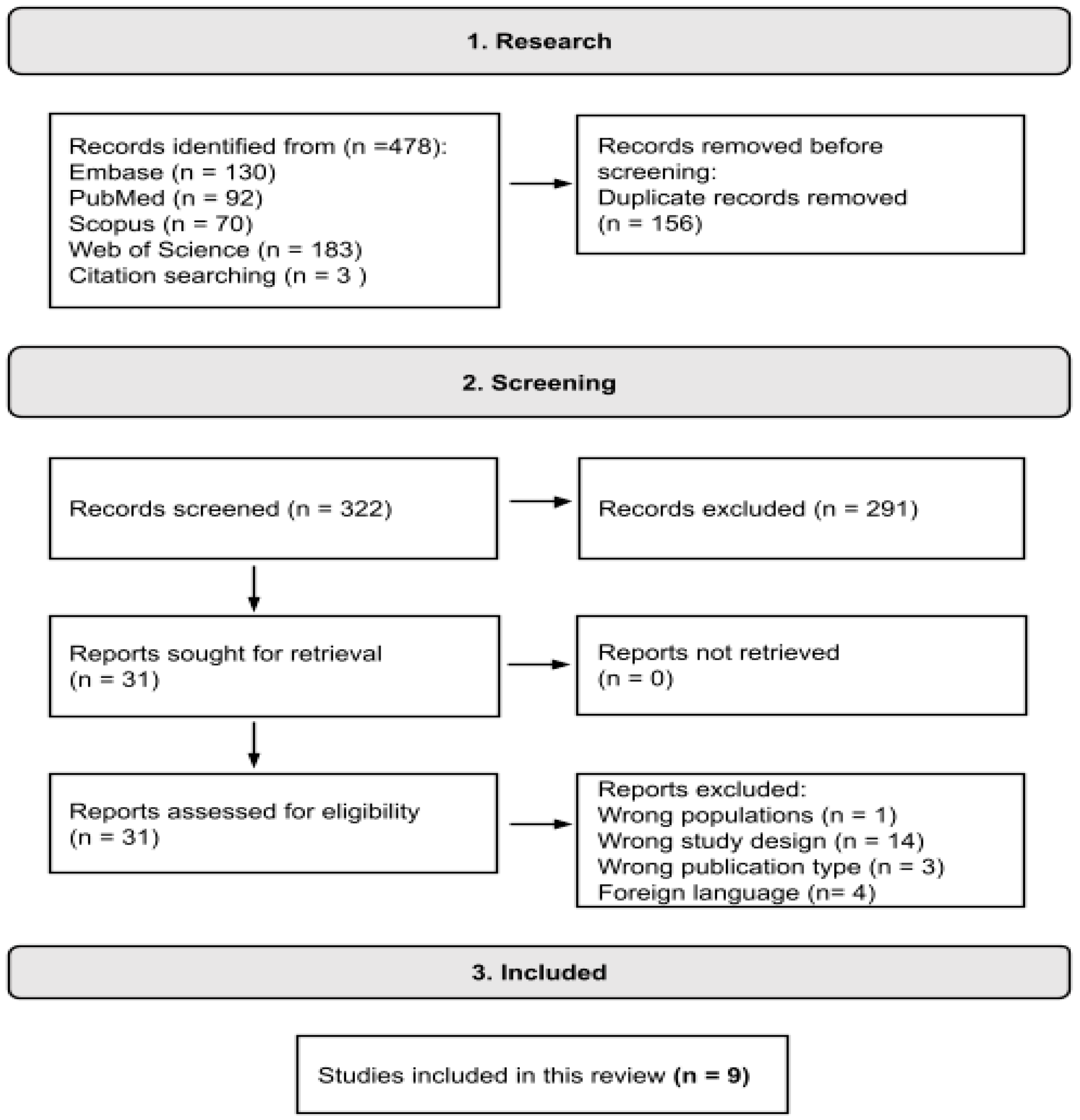
2.1. Search Strategy and Selection Criteria
2.2. Data Extraction and Analysis
3. Results
3.1. Selected Studies
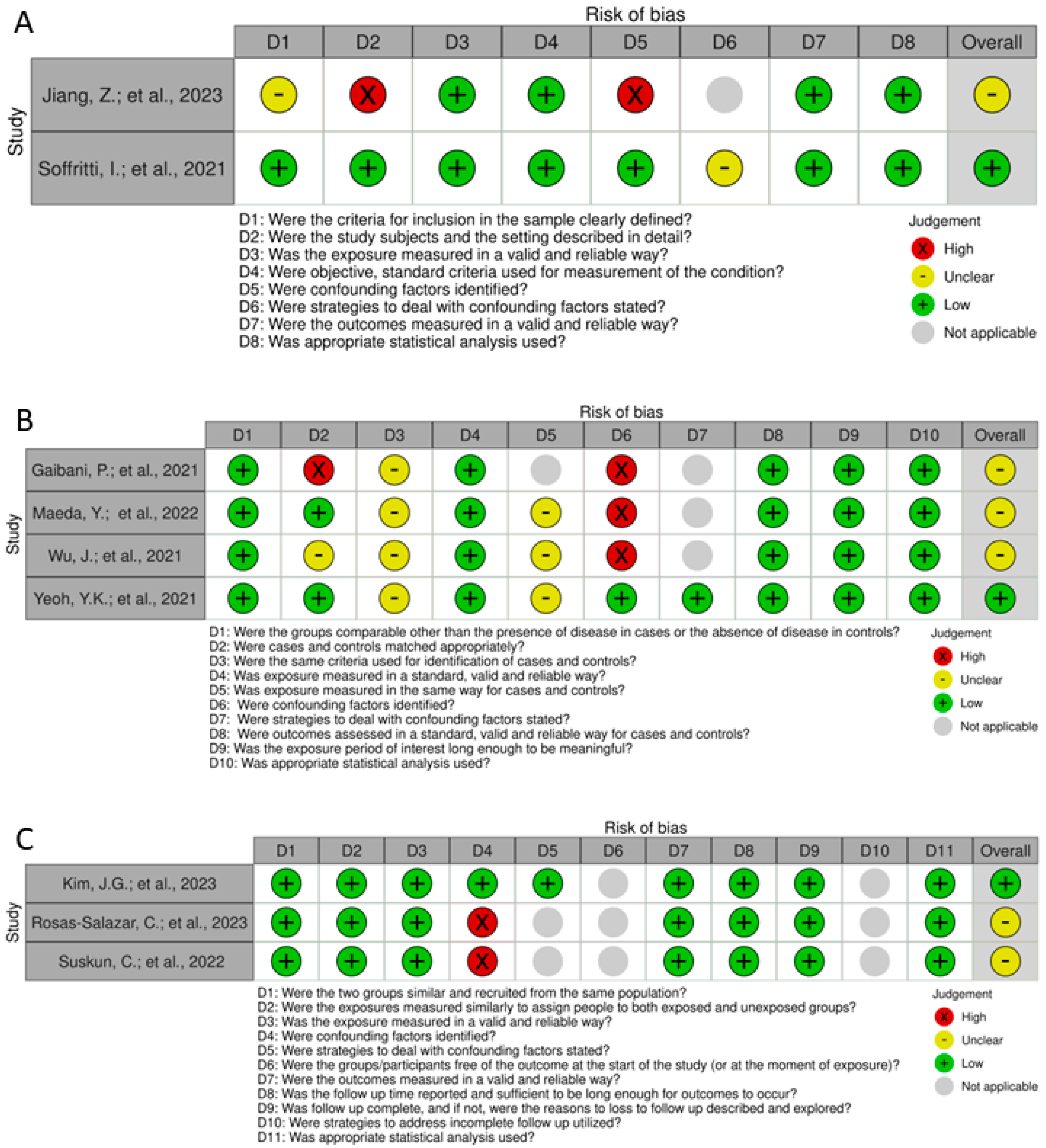
3.2. Risk of Bias and Quality Assessment
4. Discussion
4.1. The Lactobacilli
4.2. Evidence in COVID-19
4.3. Risk of Bias and Quality Assessment
4.4. Evidence in Other Viral Infections
4.5. Mechanisms of Protection of Lactobacilli in COVID-19
4.5.1. Direct Antiviral Effects
4.5.2. Immunomodulatory Effects
4.5.3. Anti-Inflammatory Effects
- L. rhamnosus and Lactobacillus helveticus downregulate Th1 pro-inflammatory responses and enhance Th2 responses during Citrobacter rodentium infection [67].
- plantarum reduces NF-κB-activating factors by diminishing NF-κB-binding activity [72].
- Lactobacillus brevis prevents IRAK1 and AKT phosphorylation [73].
4.6. Comparison between Lactobacilli and Bifidobacterium in COVID-19
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ragab, D.; Salah Eldin, H.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020, 11, 1446. [Google Scholar] [CrossRef] [PubMed]
- Kalantar-Zadeh, K.; Ward, S.A.; Kalantar-Zadeh, K.; El-Omar, E.M. Considering the Effects of Microbiome and Diet on SARS-CoV-2 Infection: Nanotechnology Roles. ACS Nano 2020, 14, 5179–5182. [Google Scholar] [CrossRef]
- Rhoades, N.S.; Pinski, A.N.; Monsibais, A.N.; Jankeel, A.; Doratt, B.M.; Cinco, I.R.; Ibraim, I.; Messaoudi, I. Acute SARS-CoV-2 infection is associated with an increased abundance of bacterial pathogens, including Pseudomonas aeruginosa in the nose. Cell Rep. 2021, 36, 109637. [Google Scholar] [CrossRef]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients with COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef] [PubMed]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.Y.; Tso, E.Y.K.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.L.; Chan, P.K.S.; et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar] [CrossRef]
- Tang, L.; Gu, S.; Gong, Y.; Li, B.; Lu, H.; Li, Q.; Zhang, R.; Gao, X.; Wu, Z.; Zhang, J.; et al. Clinical Significance of the Correlation between Changes in the Major Intestinal Bacteria Species and COVID-19 Severity. Engineering 2020, 6, 1178–1184. [Google Scholar] [CrossRef]
- Rossi, M.; Martínez-Martínez, D.; Amaretti, A.; Ulrici, A.; Raimondi, S.; Moya, A. Mining metagenomic whole genome sequences revealed subdominant but constant Lactobacillus population in the human gut microbiota. Environ. Microbiol. Rep. 2016, 8, 399–406. [Google Scholar] [CrossRef]
- Heeney, D.D.; Gareau, M.G.; Marco, M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Suarez, C.J.; Banaei, N.; Pinsky, B.A. Next-Generation Sequencing for Infectious Disease Diagnosis and Management: A Report of the Association for Molecular Pathology. J. Mol. Diagn. 2015, 17, 623–634. [Google Scholar] [CrossRef]
- Shahi, S.K.; Zarei, K.; Guseva, N.V.; Mangalam, A.K. Microbiota analysis using two-step pcr and next-generation 16s rrna gene sequencing. J. Vis. Exp. 2019, 2019, e59980. [Google Scholar] [CrossRef]
- Sanschagrin, S.; Yergeau, E. Next-generation sequencing of 16S ribosomal RNA gene amplicons. J. Vis. Exp. 2014, 90, 51709. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef]
- Purushothaman, S.; Meola, M.; Egli, A. Combination of Whole Genome Sequencing and Metagenomics for Microbiological Diagnostics. Int. J. Mol. Sci. 2022, 23, 9834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lau, R.I.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Moola, S.; Munn, Z.; Tufanaru, C.; Aromataris, E.; Sears, K.; Sfetcu, R.; Currie, M.; Lisy, K.; Qureshi, R.; Mattis, P.; et al. Chapter 7: Systematic reviews of etiology and risk. In JBI Manual for Evidence Synthesis; Aromataris, E., Munn, Z., Eds.; JBI: Adelaide, Australia, 2020. [Google Scholar] [CrossRef]
- Jiang, Z.; Yang, L.; Qian, X.; Su, K.; Huang, Y.; Qu, Y.; Zhang, Z.; Liu, W. Tongue coating microbiome composition reflects disease severity in patients with COVID-19 in Nanjing, China. J. Oral Microbiol. 2023, 15, 2236429. [Google Scholar] [CrossRef]
- Kim, J.G.; Zhang, A.; Rauseo, A.M.; Goss, C.W.; Mudd, P.A.; O’Halloran, J.A.; Wang, L. The salivary and nasopharyngeal microbiomes are associated with SARS-CoV-2 infection and disease severity. J. Med. Virol. 2023, 95, e28445. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Salazar, C.; Kimura, K.S.; Shilts, M.H.; Strickland, B.A.; Freeman, M.H.; Wessinger, B.C.; Gupta, V.; Brown, H.M.; Boone, H.H.; Rajagopala, S.V.; et al. Upper respiratory tract microbiota dynamics following COVID-19 in adults. Microb. Genom. 2023, 9, mgen000957. [Google Scholar] [CrossRef]
- Soffritti, I.; D’Accolti, M.; Fabbri, C.; Passaro, A.; Manfredini, R.; Zuliani, G.; Libanore, M.; Franchi, M.; Contini, C.; Caselli, E. Oral Microbiome Dysbiosis Is Associated with Symptoms Severity and Local Immune/Inflammatory Response in COVID-19 Patients: A Cross-Sectional Study. Front. Microbiol. 2021, 12, 687513. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; D’Amico, F.; Bartoletti, M.; Lombardo, D.; Rampelli, S.; Fornaro, G.; Coladonato, S.; Siniscalchi, A.; Re, M.C.; Viale, P.; et al. The Gut Microbiota of Critically Ill Patients with COVID-19. Front. Cell. Infect. Microbiol. 2021, 11, 670424. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Motooka, D.; Kawasaki, T.; Oki, H.; Noda, Y.; Adachi, Y.; Niitsu, T.; Okamoto, S.; Tanaka, K.; Fukushima, K.; et al. Longitudinal alterations of the gut mycobiota and microbiota on COVID-19 severity. BMC Infect. Dis. 2022, 22, 572. [Google Scholar] [CrossRef]
- Suskun, C.; Kilic, O.; Yilmaz Ciftdogan, D.; Guven, S.; Karbuz, A.; Ozkaya Parlakay, A.; Kara, Y.; Kacmaz, E.; Sahin, A.; Boga, A.; et al. Intestinal microbiota composition of children with infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and multisystem inflammatory syndrome (MIS-C). Eur. J. Pediatr. 2022, 181, 3175–3191. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, X.; Jiang, G.; Tang, H.; Ming, S.; Tang, L.; Lu, J.; Guo, C.; Shan, H.; Huang, X. Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes 2021, 7, 61. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Wang, W.; Hu, H.; Zijlstra, R.T.; Zheng, J.; Gänzle, M.G. Metagenomic reconstructions of gut microbial metabolism in weanling pigs. Microbiome 2019, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Van Der Veer, C.; Hertzberger, R.Y.; Bruisten, S.M.; Tytgat, H.L.P.; Swanenburg, J.; Angelino-bart, A.D.K.; Schuren, F.; Molenaar, D.; Reid, G.; De Vries, H.; et al. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: Implications for in vivo dominance of the vaginal microbiota. Microbiome 2019, 7, 49. [Google Scholar] [CrossRef]
- Mirzayi, C.; Renson, A.; Furlanello, C.; Sansone, S.-A.; Zohra, F.; Elsafoury, S.; Geistlinger, L.; Kasselman, L.J.; Eckenrode, K.; van de Wijgert, J.; et al. Reporting guidelines for human microbiome research: The STORMS checklist. Nat. Med. 2021, 27, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Berggren, A.; Lazou Ahrén, I.; Larsson, N.; Önning, G. Randomised, double-blind and placebo-controlled study using new probiotic lactobacilli for strengthening the body immune defence against viral infections. Eur. J. Nutr. 2011, 50, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Rizzardini, G.; Eskesen, D.; Calder, P.C.; Capetti, A.; Jespersen, L.; Clerici, M. Evaluation of the immune benefits of two probiotic strains Bifidobacterium animalis ssp. lactis, BB-12® and Lactobacillus paracasei ssp. paracasei, L. casei 431® in an influenza vaccination model: A randomised, double-blind, placebo-controlled study. Br. J. Nutr. 2012, 107, 876–884. [Google Scholar] [CrossRef]
- Cox, A.J.; Pyne, D.B.; Saunders, P.U.; Fricker, P.A. Oral administration of the probiotic Lactobacillus fermentum VRI-003 and mucosal immunity in endurance athletes. Br. J. Sports Med. 2010, 44, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Merenstein, D.; Murphy, M.; Fokar, A.; Hernandez, R.K.; Park, H.; Nsouli, H.; Sanders, M.E.; Davis, B.A.; Niborski, V.; Tondu, F.; et al. Use of a fermented dairy probiotic drink containing Lactobacillus casei (DN-114 001) to decrease the rate of illness in kids: The DRINK study A patient-oriented, double-blind, cluster-randomized, placebo-controlled, clinical trial. Eur. J. Clin. Nutr. 2010, 64, 669–677. [Google Scholar] [CrossRef]
- Makino, S.; Ikegami, S.; Kume, A.; Horiuchi, H.; Sasaki, H.; Orii, N. Reducing the risk of infection in the elderly by dietary intake of yoghurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. Br. J. Nutr. 2010, 104, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Lee, Y.T.; Le Ngo, V.; Cho, Y.H.; Ko, E.J.; Hong, S.M.; Kim, K.H.; Jang, J.H.; Oh, J.S.; Park, M.K.; et al. Heat-killed Lactobacillus casei confers broad protection against influenza A virus primary infection and develops heterosubtypic immunity against future secondary infection. Sci. Rep. 2017, 7, 17360. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Nakamura, R.; Hirose, Y.; Murosaki, S.; Yamamoto, Y.; Kase, T.; Yoshikai, Y. Oral administration of heat-killed Lactobacillus plantarum L-137 enhances protection against influenza virus infection by stimulation of type I interferon production in mice. Int. Immunopharmacol. 2009, 9, 1122–1125. [Google Scholar] [CrossRef]
- Nagai, T.; Makino, S.; Ikegami, S.; Itoh, H.; Yamada, H. Effects of oral administration of yogurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1 and its exopolysaccharides against influenza virus infection in mice. Int. Immunopharmacol. 2011, 11, 2246–2250. [Google Scholar] [CrossRef]
- Park, M.K.; NGO, V.; Kwon, Y.M.; Lee, Y.T.; Yoo, S.; Cho, Y.H.; Hong, S.M.; Hwang, H.S.; Ko, E.J.; Jung, Y.J.; et al. Lactobacillus plantarum DK119 as a Probiotic Confers Protection against Influenza Virus by Modulating Innate Immunity. PLoS ONE 2013, 8, 26–29. [Google Scholar] [CrossRef]
- Kobayashi, N.; Saito, T.; Uematsu, T.; Kishi, K.; Toba, M.; Kohda, N.; Suzuki, T. Oral administration of heat-killed Lactobacillus pentosus strain b240 augments protection against influenza virus infection in mice. Int. Immunopharmacol. 2011, 11, 199–203. [Google Scholar] [CrossRef]
- Youn, H.N.; Lee, D.H.; Lee, Y.N.; Park, J.K.; Yuk, S.S.; Yang, S.Y.; Lee, H.J.; Woo, S.H.; Kim, H.M.; Lee, J.B.; et al. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antivir. Res. 2012, 93, 138–143. [Google Scholar] [CrossRef]
- Gabryszewski, S.J.; Bachar, O.; Dyer, K.D.; Percopo, C.M.; Killoran, K.E.; Domachowske, J.B.; Rosenberg, H.F. Lactobacillus-Mediated Priming of the Respiratory Mucosa Protects against Lethal Pneumovirus Infection. J. Immunol. 2011, 186, 1151–1161. [Google Scholar] [CrossRef]
- Al Kassaa, I.; Hober, D.; Hamze, M.; Chihib, N.E.; Drider, D. Antiviral Potential of Lactic Acid Bacteria and Their Bacteriocins. Probiot. Antimicrob. Proteins 2014, 6, 177–185. [Google Scholar] [CrossRef]
- Wang, Y.; Moon, A.; Huang, J.; Sun, Y.; Qiu, H.J. Antiviral Effects and Underlying Mechanisms of Probiotics as Promising Antivirals. Front. Cell. Infect. Microbiol. 2022, 12, 928050. [Google Scholar] [CrossRef] [PubMed]
- Mattar, A.F.; Teitelbaum, D.H.; Drongowski, R.A.; Yongyi, F.; Harmon, C.M.; Coran, A.G. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr. Surg. Int. 2002, 18, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.R.; Ahrne, S.; Hyde, L.; Wei, S.; Hollingsworth, M.A. Extracellular MUC3 mucin secretion follows adherence of. Gut 2003, 52, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Cole-Jeffrey, C.T.; Liu, M.; Katovich, M.J.; Raizada, M.K.; Shenoy, V. ACE2 and Microbiota:Emerging Targets for Cardiopulmonary Disease Therapy. J. Cardiovasc. Pharmacol. 2015, 66, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Anwar, F.; Altayb, H.N.; Al-Abbasi, F.A.; Al-Malki, A.L.; Kamal, M.A.; Kumar, V. Antiviral effects of probiotic metabolites on COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 4175–4184. [Google Scholar] [CrossRef] [PubMed]
- Balmeh, N.; Mahmoudi, S.; Fard, N.A. Manipulated bio antimicrobial peptides from probiotic bacteria as proposed drugs for COVID-19 disease. Inform. Med. Unlocked 2021, 23, 100515. [Google Scholar] [CrossRef]
- Rather, I.A.; Choi, S.-B.; Kamli, M.R.; Hakeem, K.R.; Sabir, J.S.M.; Park, Y.-H.; Hor, Y.-Y. Potential adjuvant therapeutic effect of Lactobacillus plantarum probio-88 postbiotics against SARS-CoV-2. Vaccines 2021, 9, 1067. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Sato, A.; Goto, A.; Nakamura, M.; Ogawa, M.; Chiba, Y.; Hemmi, J.; Kano, H.; Takeda, K.; Okumura, K.; et al. Enhanced natural killer cell activation by exopolysaccharides derived from yogurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J. Dairy Sci. 2016, 99, 915–923. [Google Scholar] [CrossRef]
- Abreu, M.T.; Fukata, M.; Arditi, M. TLR Signaling in the Gut in Health and Disease. J. Immunol. 2005, 174, 4453–4460. [Google Scholar] [CrossRef]
- Castillo, N.A.; Perdigán, G.; De Moreno De Leblanc, A. Oral administration of a probiotic Lactobacillus modulates cytokine production and TLR expression improving the immune response against Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol. 2011, 11, 177. [Google Scholar] [CrossRef]
- Weiss, D.S.; Raupach, B.; Takeda, K.; Akira, S.; Zychlinsky, A. Toll-Like Receptors Are Temporally Involved in Host Defense. J. Immunol. 2004, 172, 4463–4469. [Google Scholar] [CrossRef] [PubMed]
- Tohno, M.; Shimosato, T.; Aso, H.; Kitazawa, H. Immunobiotic Lactobacillus strains augment NLRP3 expression in newborn and adult porcine gut-associated lymphoid tissues. Vet. Immunol. Immunopathol. 2011, 144, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.M.; Valenti, V.; Rockel, C.; Hermann, C.; Pot, B.; Boneca, I.G.; Grangette, C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut 2011, 60, 1050–1059. [Google Scholar] [CrossRef]
- Amrouche, T.; Chikindas, M.L. Probiotics for immunomodulation in prevention against respiratory viral infections with special emphasis on COVID-19. AIMS Microbiol. 2022, 8, 338–356. [Google Scholar] [CrossRef]
- Maragkoudakis, P.A.; Chingwaru, W.; Gradisnik, L.; Tsakalidou, E.; Cencic, A. Lactic acid bacteria efficiently protect human and animal intestinal epithelial and immune cells from enteric virus infection. Int. J. Food Microbiol. 2010, 141, 91–97. [Google Scholar] [CrossRef]
- Synodinou, K.D.; Nikolaki, M.D.; Triantafyllou, K.; Kasti, A.N. Immunomodulatory Effects of Probiotics on COVID-19 Infection by Targeting the Gut–Lung Axis Microbial Cross-Talk. Microorganisms 2022, 10, 1764. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M. Immunomodulatory mechanisms of lactobacilli. Microb. Cell Fact. 2011, 10, S17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, N.; Caicedo, R.; Neu, J. Alive and dead Lactobacillus rhamnosus GG decrease tumor necrosis factor-α-induced interleukin-8 production in Caco-2 cells. J. Nutr. 2005, 135, 1752–1756. [Google Scholar] [CrossRef]
- Pham, M.T.; Yang, A.J.; Kao, M.; Gankhuyag, U.; Zayabaatar, E.; Jin, S.C.; Huang, C. Gut probiotic Lactobacillus rhamnosus attenuates PDE4B-mediated interleukin-6 induced by SARS-CoV-2 membrane glycoprotein. J. Nutr. Biochem. 2021, 98, 108821. [Google Scholar] [CrossRef] [PubMed]
- Tien, M.-T.; Girardin, S.E.; Regnault, B.; Le Bourhis, L.; Dillies, M.-A.; Coppée, J.-Y.; Bourdet-Sicard, R.; Sansonetti, P.J.; Pédron, T. Anti-Inflammatory Effect of Lactobacillus casei on Shigella -Infected Human Intestinal Epithelial Cells. J. Immunol. 2006, 176, 1228–1237. [Google Scholar] [CrossRef]
- Johnson-Henry, K.C.; Nadjafi, M.; Avitzur, Y.; Mitchell, D.J.; Ngan, B.Y.; Galindo-Mata, E.; Jones, N.L.; Sherman, P.M. Amelioration of the effects of Citrobacter rodentium infection in mice by pretreatment with probiotics. J. Infect. Dis. 2005, 191, 2106–2117. [Google Scholar] [CrossRef]
- Kaci, G.; Lakhdari, O.; Doré, J.; Ehrlich, S.D.; Renault, P.; Blottière, H.M.; Delorme, C. Inhibition of the NF-κB pathway in human intestinal epithelial cells by commensal Streptococcus salivarius. Appl. Environ. Microbiol. 2011, 77, 4681–4684. [Google Scholar] [CrossRef]
- Liu, Y.; Fatheree, N.Y.; Mangalat, N.; Rhoads, J.M. Lactobacillus reuteri strains reduce incidence and severity of experimental necrotizing enterocolitis via modulation of TLR4 and NF-κB signaling in the intestine. Am. J. Physiol.-Gastrointest. Liver Physiol. 2012, 302, 608–617. [Google Scholar] [CrossRef]
- Lee, J.M.; Hwang, K.T.; Jun, W.J.; Park, C.S.; Lee, M.Y. Antiinflammatory effect of lactic acid bacteria: Inhibition of cyclooxygenase-2 by suppressing nuclear factor-κB in Raw264.7 macrophage cells. J. Microbiol. Biotechnol. 2008, 18, 1683–1688. [Google Scholar]
- Sun, K.Y.; Xu, D.H.; Xie, C.; Plummer, S.; Tang, J.; Yang, X.F.; Ji, X.H. Lactobacillus paracasei modulates LPS-induced inflammatory cytokine release by monocyte-macrophages via the up-regulation of negative regulators of NF-kappaB signaling in a TLR2-dependent manner. Cytokine 2017, 92, 1–11. [Google Scholar] [CrossRef]
- Petrof, E.O.; Claud, E.C.; Sun, J.; Abramova, T.; Guo, Y.; Waypa, T.S.; He, S.M.; Nakagawa, Y.; Chang, E.B. Bacteria-free solution derived from Lactobacillus plantarum inhibits multiple NF-kappaB pathways and inhibits proteasome function. Inflamm. Bowel Dis. 2009, 15, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.E.; Hyam, S.R.; Han, M.J.; Kim, S.Y.; Lee, B.G.; Kim, D.H. Lactobacillus brevis G-101 ameliorates colitis in mice by inhibiting NF-κB, MAPK and AKT pathways and by polarizing M1 macrophages to M2-like macrophages. J. Appl. Microbiol. 2013, 115, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Shin, H.S. Antimicrobial and immunomodulatory effects of bifidobacterium strains: A review. J. Microbiol. Biotechnol. 2021, 30, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Devika, N.T.; Raman, K. Deciphering the metabolic capabilities of Bifidobacteria using genome-scale metabolic models. Sci. Rep. 2019, 9, 18222. [Google Scholar] [CrossRef]
- Fujiwara, S.; Hashiba, H.; Hirota, T.; Forstner, J.F. Proteinaceous factor(s) in culture supernatant fluids of bifidobacteria which prevents the binding of enterotoxigenic Escherichia coli to gangliotetraosylceramide. Appl. Environ. Microbiol. 1997, 63, 506–512. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Z.Y.; Dong, K.; Guo, X.K. Adhesion and immunomodulatory effects of bifidobacterium lactis HN019 on intestinal epithelial cells INT-407. World J. Gastroenterol. 2010, 16, 2283–2290. [Google Scholar] [CrossRef]
- Lievin, V.; Peiffer, I.; Hudault, S.; Rochat, F.; Brassart, D.; Neeser, J.R.; Servin, A.L. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 2000, 47, 646–652. [Google Scholar] [CrossRef]
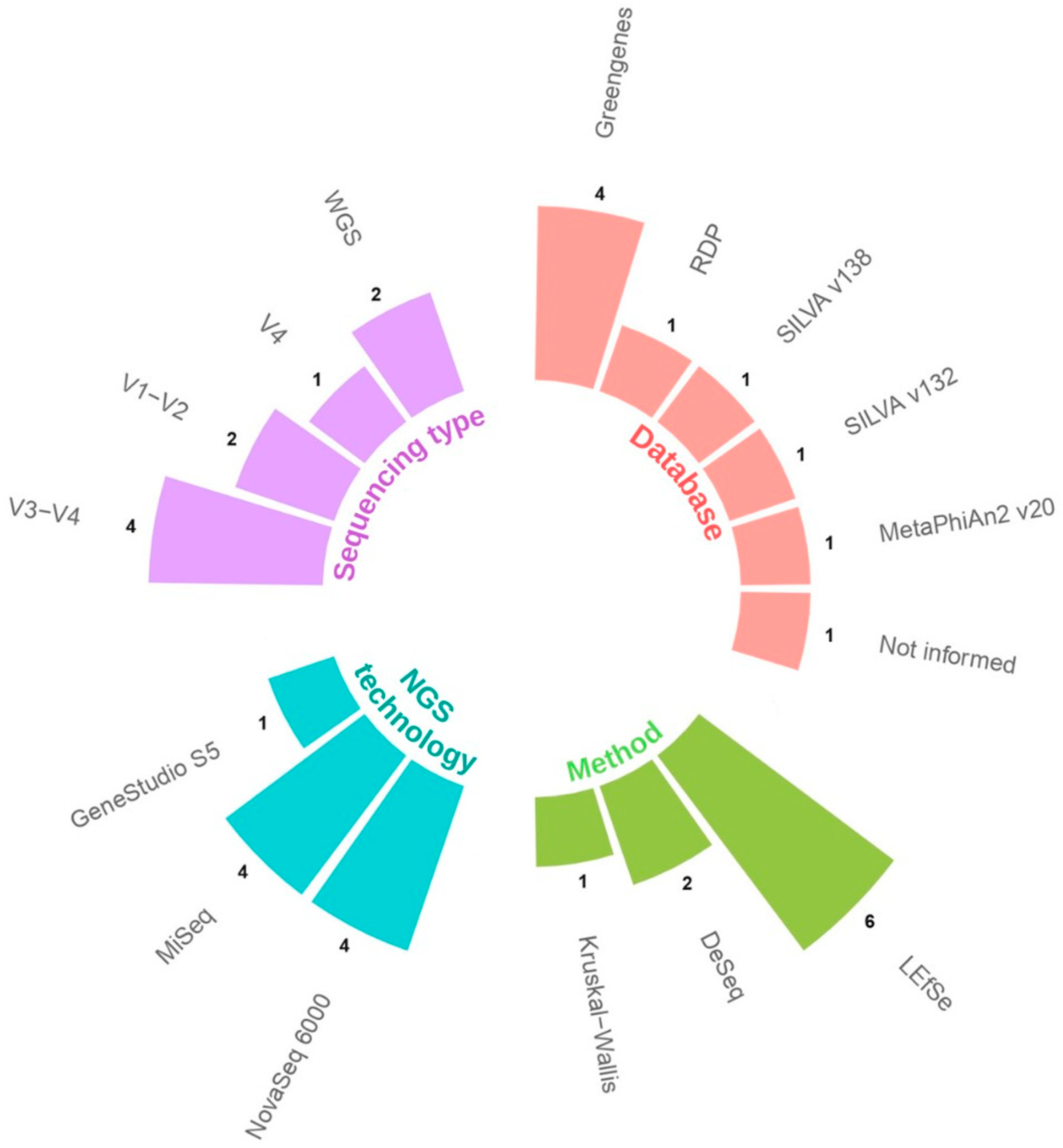
| Study | Country | Type of Study | Type of Sample | NGS Technology | Type of Sequencing | N | Groups | Abundance in COVID-19 Group | Clinical Relevance |
|---|---|---|---|---|---|---|---|---|---|
| Gaibani, P., et al., 2021 [24] | Italy | Case–control | Stool | MiSeq | V3–V4 | 138 | 69 COVID-19 patients 69 healthy controls 1 | Enriched | Associated with disease |
| Jiang, Z., et al., 2023 [20] | China | Cross-sectional | Tongue/Oral | Novaseq 6000 | V3–V4 | 130 | 49 mild to moderate COVID-19 44 severe and critical COVID-19 37 healthy control | Greater abundance in severe–critical patients. Detected as a group biomarker | Associated with severity |
| Kim, J.G., et al., 2023 [21] | USA | Cohort | Saliva and nasopharyngeal | MiSeq | V1–V2 | NI | 114 samples COVID-19 positive 30 samples COVID-19 negative | Depleted in oral samples and increased in nasopharyngeal samples from ICU patients | Associated with severity |
| Maeda, Y., et al., 2022 [25] | Japan | Case–control | Stool | MiSeq | V1–V2 | 108 | 40 severe COVID-19 38 mild COVID-19 30 healthy control | Characteristic of the severe group. Dominant relative abundance in some recovered patients. | Associated with disease severity and recovery process |
| Rosas-Salazar, C., et al., 2023 [22] | USA | Cohort | Upper Respiratory Tract | MiSeq | V4 | 48 | 24 mild-to-moderate COVID-19 patients 24 asymptomatic uninfected control | Differentially abundant. Decreased Lactobacillus jensenii in COVID-19. | Associated with disease |
| Soffritti, I., et al., 2021 [23] | Italy | Cross-sectional | Oral | Ion Gene Studio S5 | Shotgun | 75 | 39 COVID-19 patients 36 controls | Increase relative abundance. Increase Lactobacillus fermentum. | Associated with disease |
| Suskun, C., et al., 2022 [26] | Turkey | Cohort | Stool | NovaSeq 6000 | V3–V4 | 39 | 20 COVID-19 patients 19 healthy controls | Lactobacillus ruminis dominating the control group. | Not associated with disease |
| Wu, Y.J., et al., 2021 [28] | China | Case–control | Throat and stool | NovaSeq 6000 | V3–V4 | 129 | 53 COVID-19 patients 76 healthy controls | Increased in fecal samples. | Associated with disease |
| Yeoh, Y.K., et al., 2021 [27] | China | Case–control | Stool | NovaSeq 6000 | Shotgun | 178 | 100 COVID-19 patients 78 non-COVID-19 patients | Enriched Lactobacillus ruminis in recovered patients. | Associated with recovery process |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taufer, C.R.; Rampelotto, P.H. Lactobacilli in COVID-19: A Systematic Review Based on Next-Generation Sequencing Studies. Microorganisms 2024, 12, 284. https://doi.org/10.3390/microorganisms12020284
Taufer CR, Rampelotto PH. Lactobacilli in COVID-19: A Systematic Review Based on Next-Generation Sequencing Studies. Microorganisms. 2024; 12(2):284. https://doi.org/10.3390/microorganisms12020284
Chicago/Turabian StyleTaufer, Clarissa Reginato, and Pabulo Henrique Rampelotto. 2024. "Lactobacilli in COVID-19: A Systematic Review Based on Next-Generation Sequencing Studies" Microorganisms 12, no. 2: 284. https://doi.org/10.3390/microorganisms12020284
APA StyleTaufer, C. R., & Rampelotto, P. H. (2024). Lactobacilli in COVID-19: A Systematic Review Based on Next-Generation Sequencing Studies. Microorganisms, 12(2), 284. https://doi.org/10.3390/microorganisms12020284







