Integrons in the Age of Antibiotic Resistance: Evolution, Mechanisms, and Environmental Implications: A Review
Abstract
1. Introduction
2. Integrons Structure, Function and Evolution
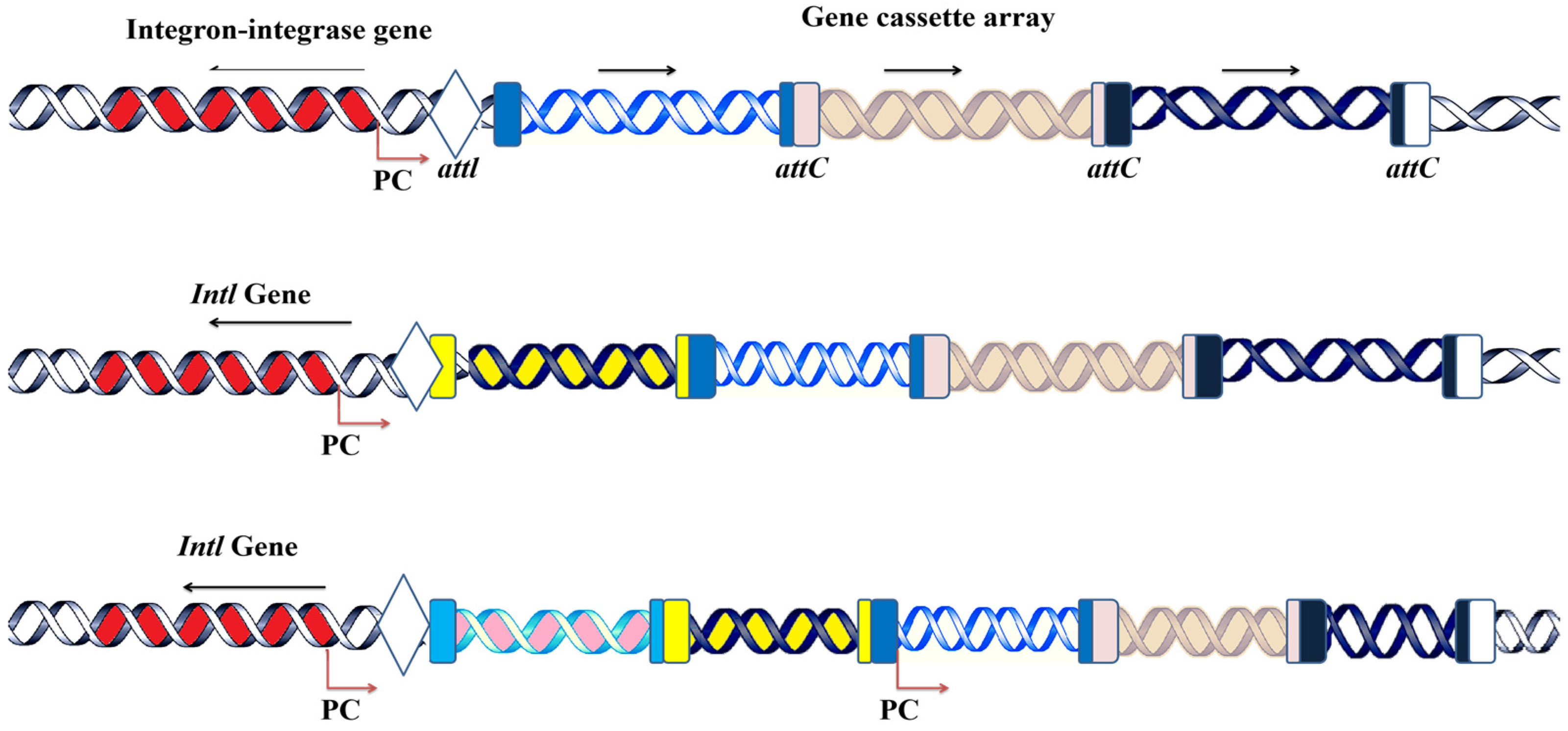
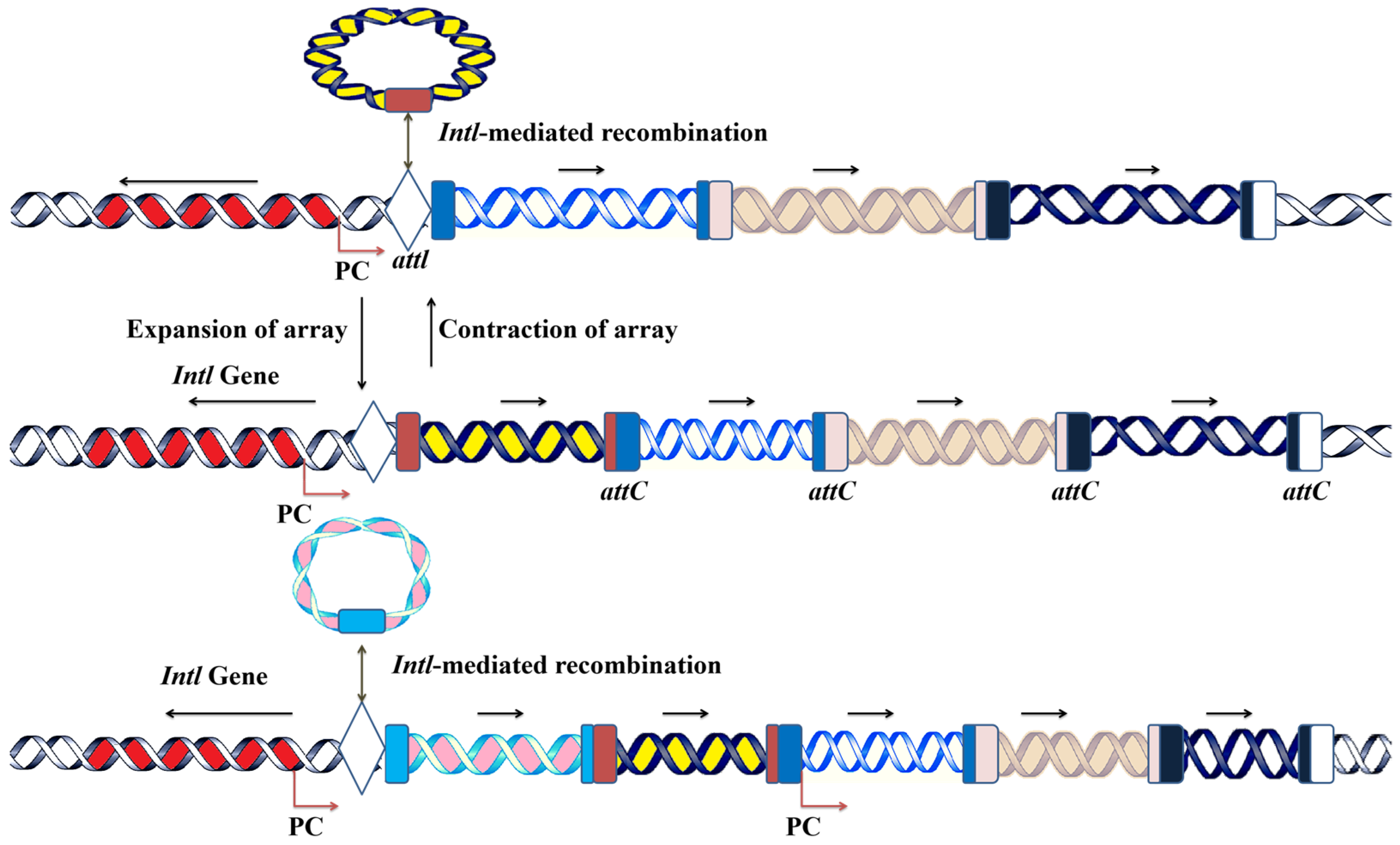
2.1. The Mechanism of Integron Functionality: Acquisition, Incorporation, and Expression of Gene Cassettes
2.2. Class 1 Integrons’ Gene Cassette Acquisition and Expression System
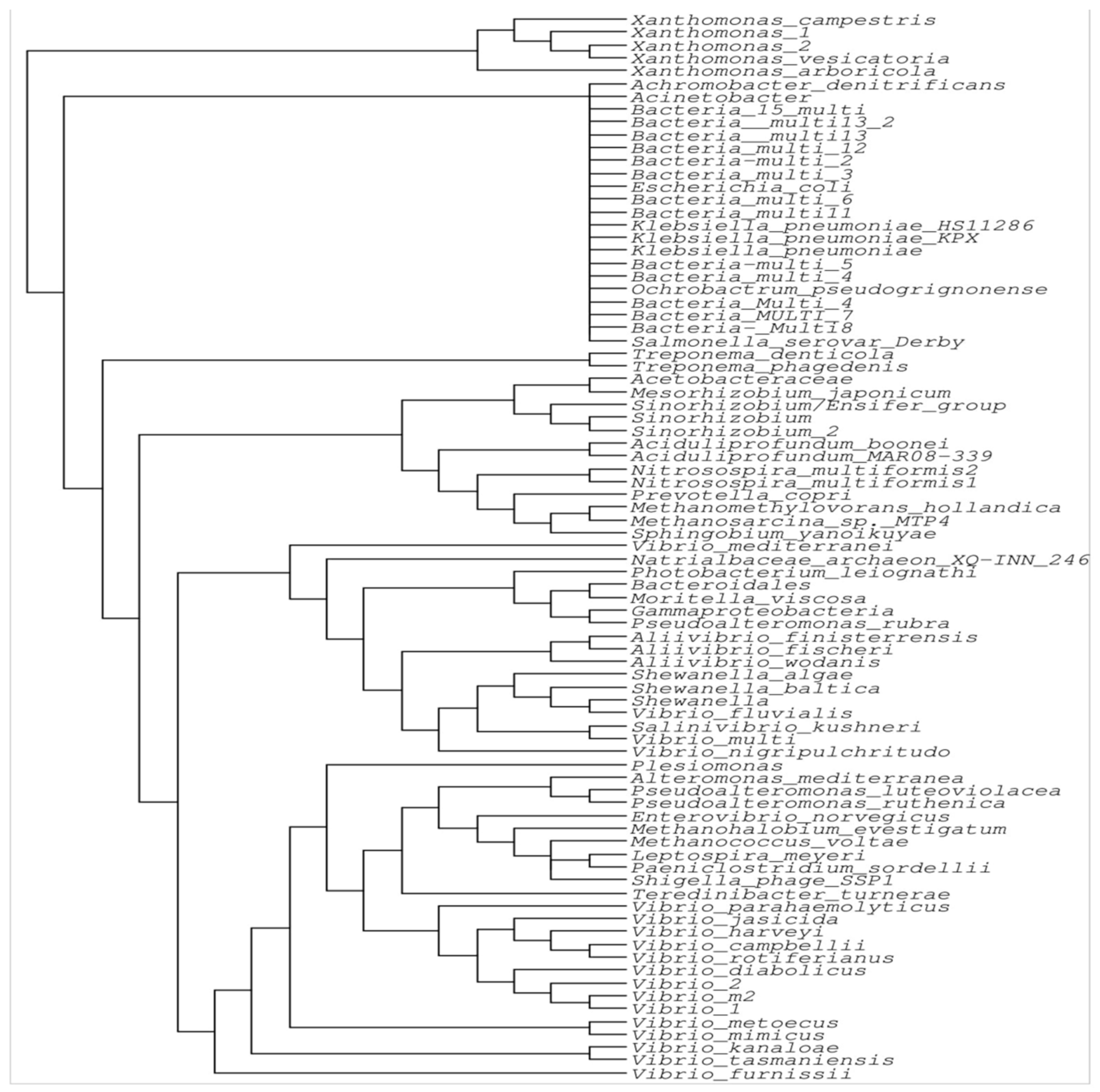
2.3. Phylogenetic Analysis of the IntI Gene and Its Evolution
3. The Role of Integrons and Gene Cassettes in Shaping Antimicrobial Resistance: Clinical Relevance and Bacterial Associations
3.1. The Role of Integrons in Shaping the Landscape of AMR
3.2. Clinically Relevant Integrons and Their Gene Cassettes
3.3. Clinically Relevant Bacteria and ARG Cassettes
| Gene Cassettes Associated with Antibiotic Resistance | Gene Cassettes | Integron Classes (CL), | Host | References |
|---|---|---|---|---|
| Erythromycin. | aadA1, aadA2, aadA5, aadB, and dfrA1 were identified, along with dfrA5, dfrA7, dfrA12, dfr14, dfrA17, dfrB2, and combinations like dfrA1-gcuC, dfrA1-aadA1, dfr17-aadA5, dfr12-gcuF-aadA2, dfrA1-sat1-aadA1, dfrA1-sat2-aadA1, estX-sat2-aadA1, and blaOXA-101-aac(6’)-Ib. | CLI, II, III | Escherichia coli | [47,48] |
| Trimethoprim, aminoglycosides, beta-lactamase, and extended spectrum. | ||||
| Beta-lactamase enzymes with extended spectrum activity, aminoglycoside antibiotics, and trimethoprim. | blaCARB-2, aadA1, aadA2, aadB, dfrA1, and dfrA7 were identified, along with combinations like dfrA1-gcuF, dfrA1-aadA1, dfr17-aadA5, dfr12-gcuF-aadA2, and sat1. | CLI, II | Acinetobacter baumannii | [19] |
| Aminoglycoside antibiotics, trimethoprim, and extended-spectrum beta-lactamases (ESBLs). | aadA, aadA1a, aadA2, aadA5, aadB, dfrA1, dfrA7, dfrA12, dfrA17, and combinations like dfrA1-gcuF, dfrA1-aadA1a, dfr17-aadA5, dfr12-gcuF-aadA2, and blaCARB-2 were identified. | CLI, II | Salmonella spp. | [49,50] |
| Extended-spectrum beta-lactamases (ESBLs), trimethoprim, and aminoglycoside antibiotics. | blaCARB-2, blaGES-1, aadA, aadA1, aadB, dfrA1, dfrA7, and gene combinations like dfrA1-gcuF, dfrA1-aadA1a, dfr17-aadA5, and dfr12-gcuF-aadA2. | CLI, II, III | Klebsiella spp. | [51,52] |
| Aminoglycosides and trimethoprim. | aadA2, aadB, and combinations like dfr17-aadA5 and dfr12-gcuF-aadA2. | CLI | Pseudomonas aeruginosa | [29,53] |
| Trimethoprim, chloramphenicol, and aminoglycosides antibiotics. | aadA1, aadA2, and combinations like dfr17-aadA5, dfr12-gcuF-aadA2, and aacA4-cmlA1 | CLI | Staphylococcus aureus | [53] |
| Trimethoprim and aminoglycosides. | aadA1a and gene combinations such as dfr12-gcuF-aadA2 and dfrA1-sat1-aadA1. | CLI | Enterococcus faecalis | [54] |
| Trimethoprim and aminoglycosides. | aadA1a, aadA2, and dfrA7, as well as gene combinations such as dfrA1-aadA1a, dfr17-aadA5, and dfr12-gcuF-aadA. | CLI | Enterobacter spp. | [55] |
4. Environmental Dissemination of Antibiotic Resistance: The Central Role of Integrons Across Ecosystems
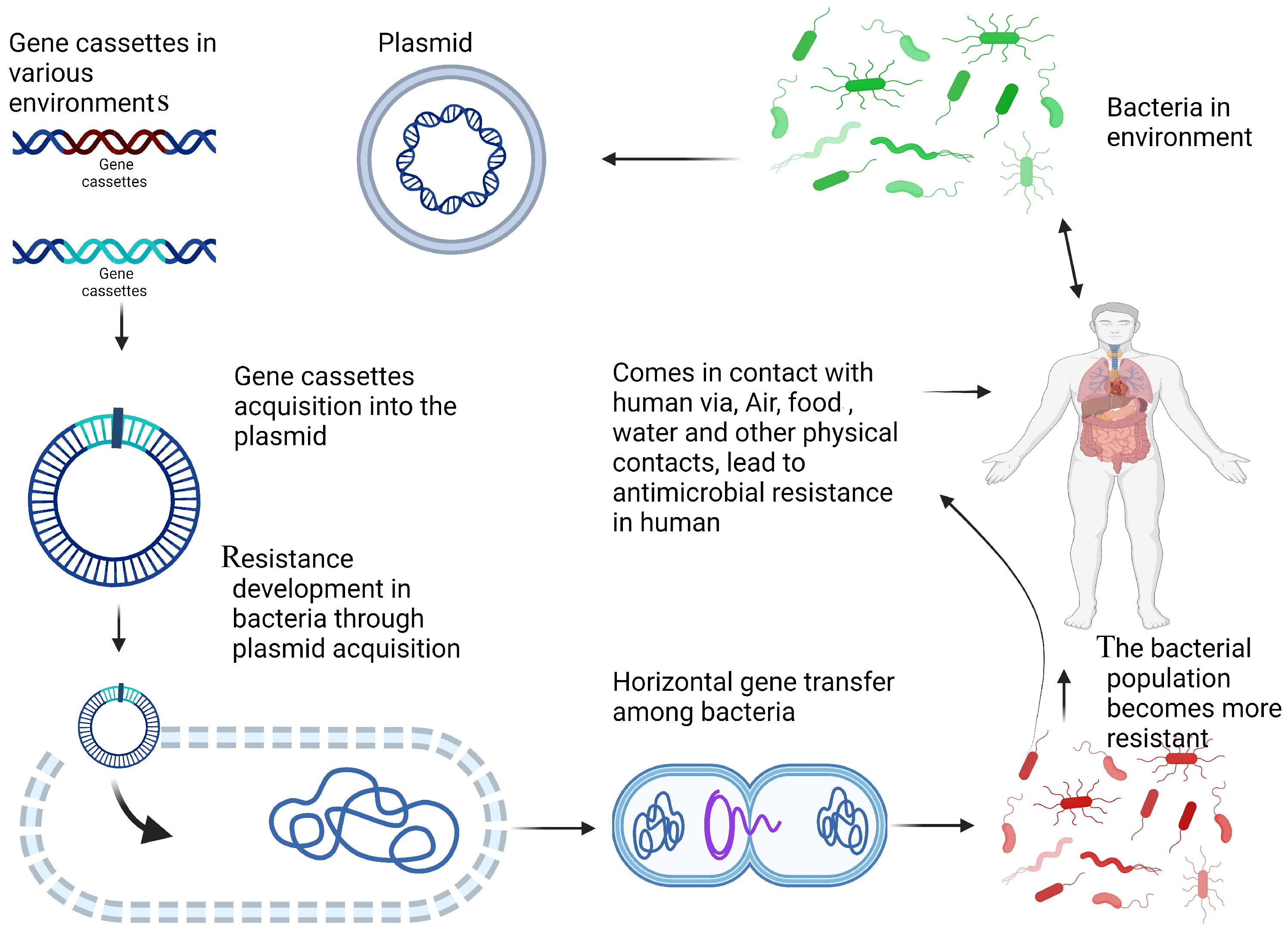
4.1. The Role of Integrons in the Dissemination of Antibiotic Resistance in the Environment
4.2. Integron Integrase Gene as an Effective Proxy for Pollution
4.3. AMR Dissemination in Wastewater
4.4. Hospital Waste Water Effluent Impact on the Dissemination of Class 1 Integrons and AMR
4.5. Fertilization and Increase in Environmental AMR and Integrons
4.6. Integrons in Marine and Freshwater Environments
5. Conclusions and Future Perspective
Future Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Böhm, M.-E.; Razavi, M.; Marathe, N.P.; Flach, C.-F.; Larsson, D.G.J. Discovery of a novel integron-borne aminoglycoside resistance gene present in clinical pathogens by screening environmental bacterial communities. Microbiome 2020, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.A.; Mir, R.A.; Qadri, H.; Dhiman, R.; Almilaibary, A.; Alkhanani, M.; Mir, M.A. Integrons in the development of antimicrobial resistance: Critical review and perspectives. Front. Microbiol. 2023, 14, 1231938. [Google Scholar] [CrossRef]
- Mitsuhashi, S.; Hashimoto, H.; Kono, M.; Morimura, M. Drug resistance of staphylococci II. Joint elimination and joint transduction of the determinants of penicillinase production and resistance to macrolide antibiotics. J. Bacteriol. 1965, 89, 988–992. [Google Scholar] [CrossRef] [PubMed]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic resistance in bacteria—A review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Helinski, D.R. A brief history of plasmids. EcoSal Plus 2022, 10, eESP00282021. [Google Scholar] [CrossRef] [PubMed]
- Tavares, R.D.S.; Fidalgo, C.; Rodrigues, E.T.; Tacão, M.; Henriques, I. Integron-associated genes are reliable indicators of antibiotic resistance in wastewater despite treatment-and seasonality-driven fluctuations. Water Res. 2024, 258, 121784. [Google Scholar] [CrossRef]
- Mustafa, S.S.; Batool, R.; Kamran, M.; Javed, H.; Jamil, N. Evaluating the Role of Wastewaters as Reservoirs of Antibiotic-Resistant ESKAPEE Bacteria Using Phenotypic and Molecular Methods. Infect. Drug Resist. 2022, 15, 5715–5728. [Google Scholar] [CrossRef]
- Alshwaikh, R.M.; Ahmed, R.Z.T. Detection of Integron Classes and Agr Group in Staphylococcus aureus Isolated from Different Clinical Samples. IBN AL-Haitham J. Pure Appl. Sci. 2024, 37, 112–128. [Google Scholar]
- Shetty, V.P.; Ahmed, R.Z.T. Integrons as the potential targets for combating multidrug resistance in Enterobacteriaceae using CRISPR-Cas9, technique. J. Appl. Microbiol. 2023, 134, lxad137. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006, 4, 608–620. [Google Scholar] [CrossRef]
- Blanco, P.; Trigo da Roza, F.; Toribio-Celestino, L.; García-Pastor, L.; Caselli, N.; Ojeda, F.; Darracq, B.; Vergara, E.; San Millan, A.; Skovgaard, O. Chromosomal Integrons are Genetically and Functionally Isolated Units of Genomes. bioRxiv 2023. [Google Scholar] [CrossRef]
- Akrami, F.; Rajabnia, M.; Pournajaf, A. Resistance integrons; A mini review. Casp. J. Intern. Med. 2019, 10, 370. [Google Scholar]
- Wang, Y.; Dagan, T. The evolution of antibiotic resistance islands occurs within the framework of plasmid lineages. Nat. Commun. 2024, 15, 4555. [Google Scholar] [CrossRef]
- Larsson, D.G.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Boucher, Y.; Labbate, M.; Koenig, J.E.; Stokes, H.W. Integrons: Mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 2007, 15, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Brtnicky, M.; Dokulilova, T.; Holatko, J.; Pecina, V.; Kintl, A.; Latal, O.; Vyhnanek, T.; Prichystalova, J.; Datta, R. Long-term effects of biochar-based organic amendments on soil microbial parameters. Agronomy 2019, 9, 747. [Google Scholar] [CrossRef]
- Ali, N.; Lin, Y.; Jiang, L.; Ali, I.; Ahmed, I.; Akhtar, K.; He, B.; Wen, R. Biochar and Manure Applications Differentially Altered the Class 1, Integrons, Antimicrobial Resistance, and Gene Cassettes Diversity in Paddy Soils. Front. Microbiol. 2022, 13, 943880. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P.R.; Stokes, H.W.; Labbate, M. Integrons: Antibiotic resistance evolution and beyond. In Bacterial Integrative Mobile Genetic Eléments; CRC Press: Boca Raton, FL, USA, 2022; pp. 53–69. [Google Scholar]
- Ploy, M.-C.; Lambert, T.; Couty, J.-P.; Denis, F. Integrons: An antibiotic resistance gene capture and expression system. Clin. Chem. Lab. Med. 2000, 38, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, A.C.W. Mobile Mercury Resistance Transposons: Surveillance and Resistance Gene Cassette Variation in Wastewater. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2022. [Google Scholar]
- Jovčić, B.; Novović, K.; Kojić, M. Molecular biology of class 1 mobile integrons. Biol. Serbica 2017, 39, 99–104. [Google Scholar]
- Hall, R.M.; Collis, C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995, 15, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Jové, T.; Da Re, S.; Denis, F.; Mazel, D.; Ploy, M.-C. Inverse correlation between promoter strength and excision activity in class 1, integrons. PLoS Genet. 2010, 6, e1000793. [Google Scholar] [CrossRef] [PubMed]
- Cambray, G.; Guerout, A.M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- Engelstädter, J.; Harms, K.; Johnsen, P.J. The evolutionary dynamics of integrons in changing environments. ISME J. 2016, 10, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, É.L.; Vicente, A.C. Integron functionality and genome innovation: An update on the subtle and smart strategy of integrase and gene cassette expression regulation. Microorganisms 2022, 10, 224. [Google Scholar] [CrossRef]
- Xu, Z.; Li, L.; Shi, L.; Shirtliff, M.E. Class 1, integron in staphylococci. Mol. Biol. Rep. 2011, 38, 5261–5279. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Geoghegan, J.L.; Tetu, S.G.; Gillings, M.R. The peril and promise of integrons: Beyond antibiotic resistance. Trends Microbiol. 2020, 28, 455–464. [Google Scholar] [CrossRef]
- Blanco, P.; Hipólito, A.; García-Pastor, L.; Trigo da Roza, F.; Toribio-Celestino, L.; Ortega, A.C.; Vergara, E.; San Millán, Á.; Escudero, J.A. Identification of promoter activity in gene-less cassettes from Vibrionaceae superintegrons. Nucleic Acids Res. 2024, 52, 2961–2976. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Shahid, M.; Singh, G.P.; Khan, H.M. Mobile genetic elements. In Beta-Lactam Resistance in Gram-Negative Bacteria: Threats and Challenges; Springer: Berlin/Heidelberg, Germany, 2022; pp. 141–152. [Google Scholar]
- De, R. Mobile genetic elements of Vibrio cholerae and the evolution of its antimicrobial resistance. Front. Trop. Dis. 2021, 2, 691604. [Google Scholar] [CrossRef]
- Makowska-Zawierucha, N.; Mokracka, J.; Małecka, M.; Balazy, P.; Chełchowski, M.; Ignatiuk, D.; Zawierucha, K. Quantification of class 1, integrons and characterization of the associated gene cassettes in the high Arctic–Interplay of humans and glaciers in shaping the aquatic resistome. Ecol. Indic. 2022, 145, 109633. [Google Scholar] [CrossRef]
- Lipszyc, A.; Szuplewska, M.; Bartosik, D. How do transposable elements activate expression of transcriptionally silent antibiotic resistance genes? Int. J. Mol. Sci. 2022, 23, 8063. [Google Scholar] [CrossRef] [PubMed]
- Gatica, J.; Tripathi, V.; Green, S.; Manaia, C.M.; Berendonk, T.; Cacace, D.; Merlin, C.; Kreuzinger, N.; Schwartz, T.; Fatta-Kassinos, D. High throughput analysis of integron gene cassettes in wastewater environments. Environ. Sci. Technol. 2016, 50, 11825–11836. [Google Scholar] [CrossRef]
- Cury, J.; Jové, T.; Touchon, M.; Néron, B.; Rocha, E.P.C. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016, 44, 4539–4550. [Google Scholar] [CrossRef]
- Fluit, A.C.; Schmitz, F.J. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 2004, 10, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Ayala Nuñez, T.; Cerbino, G.N.; Rapisardi, M.F.; Quiroga, C.; Centrón, D. Novel mobile integrons and strain-specific integrase genes within Shewanella spUnveil Multiple lateral genetic transfer events within the genus. Microorganisms 2022, 10, 1102. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A. Detection and Characterisation of Integrons, Gene Cassettes and Cassette-Located Antibiotic Resistance Genes in the Human Oral Metagenome. Ph.D. Thesis, University College London, London, UK, 2017. [Google Scholar]
- Grigaliūnas, L. Dezinfekcinių Medžiagų Poveikis Mikroorganizmams, Pasižymintiems Dauginiu Atsparumu Antibiotikams. Master’s Thesis, Lithuanian University of Health Sciences, Kaunas, Lithuania, 2020. [Google Scholar]
- Kaushik, M.; Kumar, S.; Kapoor, R.K.; Virdi, J.S.; Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 2018, 51, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Balakrishna, K.; Mukhopadhyay, C.; Kalwaje Eshwara, V. Comparison of integron mediated antimicrobial resistance in clinical isolates of Escherichia coli from urinary and bacteremic sources. BMC Microbiol. 2024, 24, 102. [Google Scholar] [CrossRef]
- Liu, C.-C.; Tang, C.Y.; Chang, K.-C.; Kuo, H.-Y.; Liou, M.-L. A comparative study of class 1, integrons in Acinetobacter baumannii. Gene 2014, 544, 75–82. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014, 5, 391. [Google Scholar] [CrossRef]
- Roy, P.K.; Ha, A.J.-W.; Mizan, M.F.R.; Hossain, M.I.; Ashrafudoulla, M.; Toushik, S.H.; Nahar, S.; Kim, Y.K.; Ha, S.-D. Effects of environmental conditions (temperature, pH, and glucose) on biofilm formation of Salmonella enterica serotype Kentucky and virulence gene expression. Poult. Sci. 2021, 100, 101209. [Google Scholar] [CrossRef] [PubMed]
- Kadlec, K.; Schwarz, S. Analysis and distribution of class 1, and class 2, integrons and associated gene cassettes among Escherichia coli isolates from swine, horses, cats and dogs collected in the BfT-GermVet monitoring study. J. Antimicrob. Chemother. 2008, 62, 469–473. [Google Scholar] [CrossRef]
- Kargar, M.; Mohammadalipour, Z.; Doosti, A.; Lorzadeh, S.; Japoni-Nejad, A. High prevalence of class 1, to 3, integrons among multidrug-resistant diarrheagenic Escherichia coli in southwest of Iran. Osong Public Health Res. Perspect. 2014, 5, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Domingues, S.; Da Silva, G.J.; Nielsen, K.M. Global dissemination patterns of common gene cassette arrays in class 1, integrons. Microbiology 2015, 161, 1313–1337. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Peixe, L. Characterization of antimicrobial resistance and class 1, and 2, integrons in Salmonella enterica isolates from different sources in Portugal. J. Antimicrob. Chemother. 2006, 58, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Karimi, E.; Ghalibafan, F.; Esfandani, A.; Arash, N.M.; Mohammadi, S.; Khaledi, A.; Akbari, H.; Khurshid, M. Antibiotic resistance pattern in Pseudomonas aeruginosa isolated from clinical samples other than burn samples in Iran. Avicenna J. Med. Biotechnol. 2021, 13, 35. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Doak, T.G.; Ye, Y. The gain and loss of chromosomal integron systems in the Treponema species. BMC Evol. Biol. 2013, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Shukla, P.; Giri, B.S.; Chowdhary, P.; Chandra, R.; Gupta, P.; Pandey, A. Prevalence and hazardous impact of pharmaceutical and personal care products and antibiotics in environment: A review on emerging contaminants. Environ. Res. 2021, 194, 110664. [Google Scholar] [CrossRef]
- Sabbagh, P.; Ebrahimzadeh-Namvar, A.; Ferdosi-Shahandashti, E.; Javanian, M.; Khafri, S.; Rajabnia, R. Molecular characterization of Staphylococcus aureus strains isolated among hospital staff nasal carriers of Babol, Iran. Casp. J. Intern. Med. 2017, 8, 311. [Google Scholar]
- Marathe, N.P.; Nagarkar, S.; Vaishampayan, A.; Rasane, M.; Samant, S.; Dohe, V.; Kagal, A.; Shouche, Y.; Deshpande, N. High prevalence of class 1, integrons in clinical isolates of methicillin-resistant Staphylococcus aureus from India. Indian J. Med. Microbiol. 2015, 33, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Michael, C.A.; Gillings, M.R.; Holmes, A.J.; Hughes, L.; Andrew, N.R.; Holley, M.P.; Stokes, H.W. Mobile gene cassettes: A fundamental resource for bacterial evolution. Am. Nat. 2004, 164, 1–12. [Google Scholar] [CrossRef]
- Brito, I.L. Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 2021, 19, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar]
- Ali, N.; Lin, Y.; Qing, Z.; Xiao, D.; Ud Din, A.; Ali, I.; Lian, T.; Chen, B.; Wen, R. The role of agriculture in the dissemination of class 1, Integrons, antimicrobial resistance, and diversity of their gene cassettes in southern China. Genes 2020, 11, 1014. [Google Scholar] [CrossRef] [PubMed]
- Goulas, A.; Belhadi, D.; Descamps, A.; Andremont, A.; Benoit, P.; Courtois, S.; Dagot, C.; Grall, N.; Makowski, D.; Nazaret, S. How effective are strategies to control the dissemination of antibiotic resistance in the environment? A systematic review. Environ. Evid. 2020, 9, 1–32. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1, integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef]
- Néron, B.; Littner, E.; Haudiquet, M.; Perrin, A.; Cury, J.; Rocha, E.P. IntegronFinder 2.0: Identification and analysis of integrons across bacteria, with a focus on antibiotic resistance in Klebsiella. Microorganisms 2022, 10, 700. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, A.-D.; Yin, X.-L.; Zhang, T. The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environ. Sci. Technol. 2017, 51, 5721–5728. [Google Scholar] [CrossRef]
- Liebert, C.A.; Hall, R.M.; Summers, A.O. Transposon Tn 21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 1999, 63, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, T.M.; Gillings, M.R.; Rajabal, V.; Paulsen, I.T.; Tetu, S.G. Horizontal gene transfer in plant microbiomes: Integrons as hotspots for cross-species gene exchange. Front. Microbiol. 2024, 15, 1338026. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Li, L.; Chen, X.; Liu, Y.-Y.; Lam, T.T.-Y.; Topp, E.; Zhang, T. Global environmental resistome: Distinction and connectivity across diverse habitats benchmarked by metagenomic analyses. Water Res. 2023, 235, 119875. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.C.A.; Ploumakis, S.; Zhang, L.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M.H. The widespread dissemination of integrons throughout bacterial communities in a riverine system. ISME J. 2018, 12, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.; Yutao, L.; Goh, S.G.; Ng, C.; Luhua, Y.; Tran, N.H.; Gin, K.Y.-H. Quaternary ammonium compounds of emerging concern: Classification, occurrence, fate, toxicity and antimicrobial resistance. J. Hazard. Mater. 2023, 445, 130393. [Google Scholar] [CrossRef]
- Reddy, S.; Kaur, K.; Barathe, P.; Shriram, V.; Govarthanan, M.; Kumar, V. Antimicrobial resistance in urban river ecosystems. Microbiol. Res. 2022, 263, 127135. [Google Scholar] [CrossRef]
- Karkman, A.; Johnson, T.A.; Lyra, C.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2016, 92, fiw014. [Google Scholar] [CrossRef]
- Zheng, W.; Huyan, J.; Tian, Z.; Zhang, Y.; Wen, X. Clinical class 1, integron-integrase gene—A promising indicator to monitor the abundance and elimination of antibiotic resistance genes in an urban wastewater treatment plant. Environ. Int. 2020, 135, 105372. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ju, F.; Beck, K.; Bürgmann, H. Differential effects of wastewater treatment plant effluents on the antibiotic resistomes of diverse river habitats. ISME J. 2023, 17, 1993–2002. [Google Scholar] [CrossRef]
- Foyle, L.; Burnett, M.; Creaser, A.; Hens, R.; Keough, J.; Madin, L.; Price, R.; Smith, H.; Stone, S.; Kinobe, R.T. Prevalence and distribution of antimicrobial resistance in effluent wastewater from animal slaughter facilities: A systematic review. Environ. Pollut. 2023, 318, 120848. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.V.; Medeiros, S.H.W.; Schneider, A.L.D.S.; dos Santos-Silva, J.C.; Fachini Souza, A.L.; Rhoden, S.A. Identification of antibiotic-resistant, gram-negative bacteria in sewage and bioaerosols from a wastewater treatment plant: A genotypic and phenotypic study. J. Water Health 2024, 22, jwh2024352. [Google Scholar] [CrossRef]
- Drane, K.; Sheehan, M.; Whelan, A.; Ariel, E.; Kinobe, R. The Role of Wastewater Treatment Plants in Dissemination of Antibiotic Resistance: Source, Measurement, Removal and Risk Assessment. Antibiotics 2024, 13, 668. [Google Scholar] [CrossRef]
- Paiva, M.C.; Avila, M.P.; Reis, M.P.; Costa, P.S.; Nardi, R.M.D.; Nascimento, A.M.A. The microbiota and abundance of the class 1, integron-integrase gene in tropical sewage treatment plant influent and activated sludge. PLoS ONE 2015, 10, e0131532. [Google Scholar] [CrossRef] [PubMed]
- Makowska, N.; Koczura, R.; Mokracka, J. Class 1, integrase, sulfonamide and tetracycline resistance genes in wastewater treatment plant and surface water. Chemosphere 2016, 144, 1665–1673. [Google Scholar] [CrossRef] [PubMed]
- Stalder, T.; Barraud, O.; Jové, T.; Casellas, M.; Gaschet, M.; Dagot, C.; Ploy, M.-C. Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. ISME J. 2014, 8, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Saima, S.; Fiaz, M.; Zafar, R.; Ahmed, I.; Arshad, M. Dissemination of antibiotic resistance in the environment. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Elsevier: Amsterdam, The Netherlands, 2020; pp. 99–116. [Google Scholar]
- Obayiuwana, A.; Ibekwe, A.M. Antibiotic resistance genes occurrence in wastewaters from selected pharmaceutical facilities in Nigeria. Water 2020, 12, 1897. [Google Scholar] [CrossRef]
- Mutuku, C.; Gazdag, Z.; Melegh, S. Occurrence of antibiotics and bacterial resistance genes in wastewater: Resistance mechanisms and antimicrobial resistance control approaches. World J. Microbiol. Biotechnol. 2022, 38, 152. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, N.; Wang, M.; Luo, M.; Peng, Y.; Li, Z.; Xu, J.; Ou, M.; Kan, B.; Li, X. The prevalence and distribution of aminoglycoside resistance genes. Biosaf. Health 2023, 5, 14–20. [Google Scholar] [CrossRef]
- Möller, V.; Östholm-Balkhed, Å.; Berild, D.; Fredriksson, M.; Gottfredsson, M.; Holmbom, M.; Järvinen, A.; Kristjansson, M.; Rydell, U.; Sönksen, U.W.; et al. Antibiotic resistance among major pathogens compared to hospital treatment guidelines and antibiotic use in Nordic hospitals 2010–2018. Infect. Dis. 2021, 53, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Kaur, K. Quaternary Ammonium Disinfectants: Current Practices and Future Perspective in Infection Control. Biomed. Pharmacol. J. 2024, 17. [Google Scholar] [CrossRef]
- Chen, S.; Fu, J.; Zhao, K.; Yang, S.; Li, C.; Penttinen, P.; Ao, X.; Liu, A.; Hu, K.; Li, J. Class 1, integron carrying qacEΔ1, gene confers resistance to disinfectant and antibiotics in Salmonella. Int. J. Food Microbiol. 2023, 404, 110319. [Google Scholar] [CrossRef] [PubMed]
- Kaviani Rad, A.; Balasundram, S.K.; Azizi, S.; Afsharyzad, Y.; Zarei, M.; Etesami, H.; Shamshiri, R.R. An overview of antibiotic resistance and abiotic stresses affecting antimicrobial resistance in agricultural soils. Int. J. Environ. Res. Public Health 2022, 19, 4666. [Google Scholar] [CrossRef]
- Cytryn, E. The soil resistome: The anthropogenic, the native, and the unknown. Soil Biol. Biochem. 2013, 63, 18–23. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Zhao, Y.; Zhu, D.; Gillings, M.; Penuelas, J.; Ok, Y.S.; Capon, A.; Banwart, S. Soil biota, antimicrobial resistance and planetary health. Environ. Int. 2019, 131, 105059. [Google Scholar] [CrossRef] [PubMed]
- Alaali, Z.; Thani, A.S.B. Patterns of antimicrobial resistance observed in the Middle East: Environmental and health care retrospectives. Sci. Total Environ. 2020, 740, 140089. [Google Scholar] [CrossRef] [PubMed]
- Thanner, S.; Drissner, D.; Walsh, F. Antimicrobial resistance in agriculture. mBio 2016, 7, 10–1128. [Google Scholar] [CrossRef]
- Wu, J.; Guo, S.; Li, K.; Li, Z.; Xu, P.; Jones, D.L.; Wang, J.; Zou, J. Effect of fertilizer type on antibiotic resistance genes by reshaping the bacterial community and soil properties. Chemosphere 2023, 336, 139272. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Y.; Li, X.; Zhang, Y.; Ye, J.; Huang, H.; Zhu, C. Temporal effects of repeated application of biogas slurry on soil antibiotic resistance genes and their potential bacterial hosts. Environ. Pollut. 2020, 258, 113652. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ling, N.; Guo, J.; Ruan, Y.; Wang, M.; Shen, Q.; Guo, S. Dynamics of the antibiotic resistome in agricultural soils amended with different sources of animal manures over three consecutive years. J. Hazard. Mater. 2021, 401, 123399. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Han, W.; Chen, S.; Dong, W.; Qiao, M.; Hu, C.; Liu, B. Fifteen-year application of manure and chemical fertilizers differently impacts soil ARGs and microbial community structure. Front. Microbiol. 2020, 11, 62. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet. Sci. 2020, 7, 110. [Google Scholar] [CrossRef]
- Tampere, M. Impact of Slurry Fertilization on Nutrient Leaching and on the Abundance of Antibiotic Resistance Genes in Agricultural Soil. Ph.D. Thesis, Eesti Maaülikool, Tartu, Estonia, 2017. [Google Scholar]
- Liu, C.; Li, G.; Qin, X.; Xu, Y.; Wang, J.; Wu, G.; Feng, H.; Ye, J.; Zhu, C.; Li, X.; et al. Profiles of antibiotic-and heavy metal-related resistance genes in animal manure revealed using a metagenomic analysis. Ecotoxicol. Environ. Saf. 2022, 239, 113655. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, G.; Tian, F.; Chen, H.; Liu, W.; Li, M.; Wang, S. Antibiotic resistance genes and bacterial community on the surfaces of five cultivars of fresh tomatoes. Ecotoxicology 2021, 30, 1550–1558. [Google Scholar] [CrossRef]
- Nõlvak, H.; Truu, M.; Kanger, K.; Tampere, M.; Espenberg, M.; Loit, E.; Raave, H.; Truu, J. Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron-integrase genes in agricultural grassland soil. Sci. Total Environ. 2016, 562, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Mi, J.; Wang, Y.; Ma, B.; Zou, Y.; Liao, X.; Liang, J.B.; Wu, Y. Occurrence and contamination profiles of antibiotic resistance genes from swine manure to receiving environments in Guangdong Province southern China. Ecotoxicol. Environ. Saf. 2019, 173, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, H.; Shi, R.; Lv, J.; Li, B.; Yang, F.; Zheng, X.; Xu, J. Antibiotic resistance genes in different animal manures and their derived organic fertilizer. Environ. Sci. Eur. 2020, 32, 1–10. [Google Scholar] [CrossRef]
- Sanz, C.; Casado, M.; Navarro-Martin, L.; Cañameras, N.; Carazo, N.; Matamoros, V.; Bayona, J.M.; Piña, B. Implications of the use of organic fertilizers for antibiotic resistance gene distribution in agricultural soils and fresh food products. A plot-scale study. Sci. Total Environ. 2022, 815, 151973. [Google Scholar] [CrossRef]
- Carvalho, P.N.; Basto, M.C.P.; Almeida, C.M.R.; Brix, H. A review of plant–pharmaceutical interactions: From uptake and effects in crop plants to phytoremediation in constructed wetlands. Environ. Sci. Pollut. Res. 2014, 21, 11729–11763. [Google Scholar] [CrossRef]
- Pan, M.; Chu, L.M. Fate of antibiotics in soil and their uptake by edible crops. Sci. Total Environ. 2017, 599, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Lenzi, A.; Marvasi, M.; Baldi, A. ; Baldi, A. Agronomic practices to limit pre-and post-harvest contamination and proliferation of human pathogenic Enterobacteriaceae in vegetable produce. Food Control 2021, 119, 107486. [Google Scholar] [CrossRef]
- Iwu, C.D.; Okoh, A.I. Preharvest transmission routes of fresh produce associated bacterial pathogens with outbreak potentials: A review. Int. J. Environ. Res. Public Health 2019, 16, 4407. [Google Scholar] [CrossRef]
- Mostafidi, M.; Sanjabi, M.R.; Shirkhan, F.; Zahedi, M.T. A review of recent trends in the development of the microbial safety of fruits and vegetables. Trends Food Sci. Technol. 2020, 103, 321–332. [Google Scholar] [CrossRef]
- Asfaw, T.; Genetu, D.; Shenkute, D.; Shenkutie, T.T.; Amare, Y.E.; Yitayew, B. Foodborne pathogens and antimicrobial resistance in Ethiopia: An urgent call for action on “one health”. Infect. Drug Resist. 2022, 15, 5265–5274. [Google Scholar] [CrossRef]
- Duffy, B.; Holliger, E.; Walsh, F. Streptomycin use in apple orchards did not increase abundance of mobile resistance genes. FEMS Microbiol. Lett. 2014, 350, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Ramírez, S.; Ghaly, T.; Gillings, M. ; Gillings, M. Non-clinical settings–the understudied facet of antimicrobial drug resistance. Environ. Microbiol. 2021, 23, 7271–7274. [Google Scholar] [CrossRef] [PubMed]
- Abella, J.; Fahy, A.; Duran, R.; Cagnon, C. Integron diversity in bacterial communities of freshwater sediments at different contamination levels. FEMS Microbiol. Ecol. 2015, 91, fiv140. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R. DNA as a pollutant: The clinical class 1, integron. Curr. Pollut. Rep. 2018, 4, 49–55. [Google Scholar] [CrossRef]
- Elsaied, H.; Stokes, H.W.; Yoshioka, H.; Mitani, Y.; Maruyama, A. Novel integrons and gene cassettes from a Cascadian submarine gas-hydrate-bearing core. FEMS Microbiol. Ecol. 2014, 87, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, T.M.; Tetu, S.G.; Penesyan, A.; Qi, Q.; Rajabal, V.; Gillings, M.R. Discovery of integrons in Archaea: Platforms for cross-domain gene transfer. Sci. Adv. 2022, 8, eabq6376. [Google Scholar] [CrossRef] [PubMed]
- Abella, J.; Bielen, A.; Huang, L.; Delmont, T.O.; Vujaklija, D.; Duran, R.; Cagnon, C. Integron diversity in marine environments. Environ. Sci. Pollut. Res. 2015, 22, 15360–15369. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Qi, Y.; Zhang, X.-X.; Ma, L. Xenogenetic evolutionary of integrons promotes the environmental pollution of antibiotic resistance genes—Challenges, progress and prospects. Water Res. 2023, 231, 119629. [Google Scholar] [CrossRef]
- Zahir, M.W.S. Epidemiology and Molecular Characterization of Emerging Antimicrobial Resistant Strains of Acinetobacter Baumannii in Saudi Arabia. Ph.D. Thesis, King Abdulaziz University, Jeddah, Saudi Arabia, 2019. [Google Scholar]
- Corno, G.; Ghaly, T.; Sabatino, R.; Eckert, E.M.; Galafassi, S.; Gillings, M.R.; Di Cesare, A. Class 1, integron and related antimicrobial resistance gene dynamics along a complex freshwater system affected by different anthropogenic pressures. Environ. Pollut. 2023, 316, 120601. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J. Continental-scale spatio-temporal distribution of antibiotic resistance genes in coastal waters along coastline of China. Chemosphere 2020, 247, 125908. [Google Scholar] [CrossRef] [PubMed]
- Di Cesare, A.; Sabatino, R.; Sbaffi, T.; Fontaneto, D.; Brambilla, D.; Beghi, A.; Pandolfi, F.; Borlandelli, C.; Fortino, D.; Biccai, G. Anthropogenic pollution drives the bacterial resistome in a complex freshwater ecosystem. Chemosphere 2023, 331, 138800. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Z.; Song, W.; Du, L.; Ye, C.; Zhao, B.; Liu, W.; Deng, D.; Pan, Y.; Lin, H. Metagenomic insights into the abundance and composition of resistance genes in aquatic environments: Influence of stratification and geography. Environ. Int. 2019, 127, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I.; Mackie, R.I. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 2007, 271, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Adenaya, A.; Berger, M.; Brinkhoff, T.; Ribas-Ribas, M.; Wurl, O. Usage of antibiotics in aquaculture and the impact on coastal waters. Mar. Pollut. Bull. 2023, 188, 114645. [Google Scholar] [CrossRef] [PubMed]
- Ullah, R.; Yasir, M.; Bibi, F.; Abujamel, T.S.; Hashem, A.M.; Sohrab, S.S.; Al-Ansari, A.; Al-Sofyani, A.A.; Al-Ghamdi, A.K.; Al-Sieni, A. Taxonomic diversity of antimicrobial-resistant bacteria and genes in the Red Sea coast. Sci. Total Environ. 2019, 677, 474–483. [Google Scholar] [CrossRef]
- Gao, Q.; Li, Y.; Qi, Z.; Yue, Y.; Min, M.; Peng, S.; Shi, Z.; Gao, Y. Diverse and abundant antibiotic resistance genes from mariculture sites of China’s coastline. Sci. Total Environ. 2018, 630, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Yin, G.; Liu, M.; Chen, C.; Jiang, Y.; Hou, L.; Zheng, Y. A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 2021, 777, 146009. [Google Scholar] [CrossRef]
- Wu, J.; Su, Y.; Deng, Y.; Guo, Z.; Cheng, C.; Ma, H.; Liu, G.; Xu, L.; Feng, J. Spatial and temporal variation of antibiotic resistance in marine fish cage-culture area of Guangdong, China. Environ. Pollut. 2019, 246, 463–471. [Google Scholar] [CrossRef] [PubMed]
- He, L.-X.; He, L.-Y.; Gao, F.-Z.; Zhang, M.; Chen, J.; Jia, W.-L.; Ye, P.; Jia, Y.-W.; Hong, B.; Liu, S.-S. Mariculture affects antibiotic resistome and microbiome in the coastal environment. J. Hazard. Mater. 2023, 452, 131208. [Google Scholar] [CrossRef]
- Partridge, S.R.; Tsafnat, G.; Coiera, E.; Iredell, J.R. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 2009, 33, 757–784. [Google Scholar] [CrossRef] [PubMed]
- Almutrafy, A. Antibiotic Resistance Genes of Class 1, Integrons in Chicken Microbiomes Modulated by Prebiotics. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2021. [Google Scholar]
- Shintani, M.; Vestergaard, G.; Milaković, M.; Kublik, S.; Smalla, K.; Schloter, M.; Udiković-Kolić, N. Integrons, transposons and IS elements promote diversification of multidrug resistance plasmids and adaptation of their hosts to antibiotic pollutants from pharmaceutical companies. Environ. Microbiol. 2023, 25, 3035–3051. [Google Scholar] [CrossRef] [PubMed]
- Subirats, J.; Sharpe, H.; Tai, V.; Fruci, M.; Topp, E. Metagenome meta-analysis reveals an increase in the abundance of some multidrug efflux pumps and mobile genetic elements in chemically polluted environments. Appl. Environ. Microbiol. 2023, 89, e01047-23. [Google Scholar] [CrossRef] [PubMed]
- Stokes, H.W.; Holmes, A.J.; Nield, B.S.; Holley, M.P.; Nevalainen, K.H.; Mabbutt, B.C.; Gillings, M.R. Gene cassette PCR: Sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 2001, 67, 5240–5246. [Google Scholar] [CrossRef] [PubMed]


| Fertilizer Types | Effects on AMR Genes | Percent Increase | Reference |
|---|---|---|---|
| Manure and bio-organic fertilizer application | Aminoglycosides, beta-lactamases, chloramphenicol, macrolide-lincosamide-streptograminB (MLSB), multidrug, sulfonamide, tetracycline, vancomycin resistance genes | 116% | [90] |
| Mineral fertilizer (NH4NO3), cattle slurry, and cattle slurry digestate amendment | Tetracycline, sulfonamides, macrolides, integrase gene copy number increased | 83%, 20%, 64%, 83%, log copies/gm soil | [96] |
| Composted manure | Aminoglycoside, bacitracin, chloramphenicol, sulfonamide, tetracycline, and multidrug resistance was present in higher abundances than the other resistance genes | 24% increase in total abundance | [97] |
| Cattle slurry digestate | TetA, blaCTX-M, blaOXA2, qnrS, intI1, and intI2 | 104–105 copies/gm soil and (1.2 × 109 copies/gm soil) | [98,99] |
| Swine manure | ARGs (ermB, qnrS, acc(6′)-Ib, tetM, tetO, and tetQ) tetQ and tetW, and ermB and ermF | 3.01 × 108 to 7.18 × 1014 copies/g | [100] |
| Manure applications | CL1, QACs, sulfonamide, tetracycline, and multidrug | 109 copies/gm and 16–48% increase | [18] |
| Organic fertilizers and livestock and poultry manure | ARGs, including sul2, TetB-01, TetG-01, and TetM-01, TetK, and ermC | 12–96% | [101] |
| Organic fertilizers | IntI1, sul1, and tetM, blaTEM, and blaOXA-48, qnrS1 | 20–100-fold increase change | [102] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, N.; Ali, I.; Din, A.U.; Akhtar, K.; He, B.; Wen, R. Integrons in the Age of Antibiotic Resistance: Evolution, Mechanisms, and Environmental Implications: A Review. Microorganisms 2024, 12, 2579. https://doi.org/10.3390/microorganisms12122579
Ali N, Ali I, Din AU, Akhtar K, He B, Wen R. Integrons in the Age of Antibiotic Resistance: Evolution, Mechanisms, and Environmental Implications: A Review. Microorganisms. 2024; 12(12):2579. https://doi.org/10.3390/microorganisms12122579
Chicago/Turabian StyleAli, Niyaz, Izhar Ali, Ahmad Ud Din, Kashif Akhtar, Bing He, and Ronghui Wen. 2024. "Integrons in the Age of Antibiotic Resistance: Evolution, Mechanisms, and Environmental Implications: A Review" Microorganisms 12, no. 12: 2579. https://doi.org/10.3390/microorganisms12122579
APA StyleAli, N., Ali, I., Din, A. U., Akhtar, K., He, B., & Wen, R. (2024). Integrons in the Age of Antibiotic Resistance: Evolution, Mechanisms, and Environmental Implications: A Review. Microorganisms, 12(12), 2579. https://doi.org/10.3390/microorganisms12122579











