Geography, Antimicrobial Resistance, and Genomics of Salmonella enterica (Serotypes Newport and Anatum) from Meat in Mexico (2021–2023)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolates
2.2. Geographic Information Systems
2.3. Antibiotic Susceptibility Testing
2.4. Extraction, Whole-Genome Sequencing (WGS), and Genome Assembly
2.5. Bioinformatics Analysis
2.6. Statistical Analysis and Data Visualization
3. Results
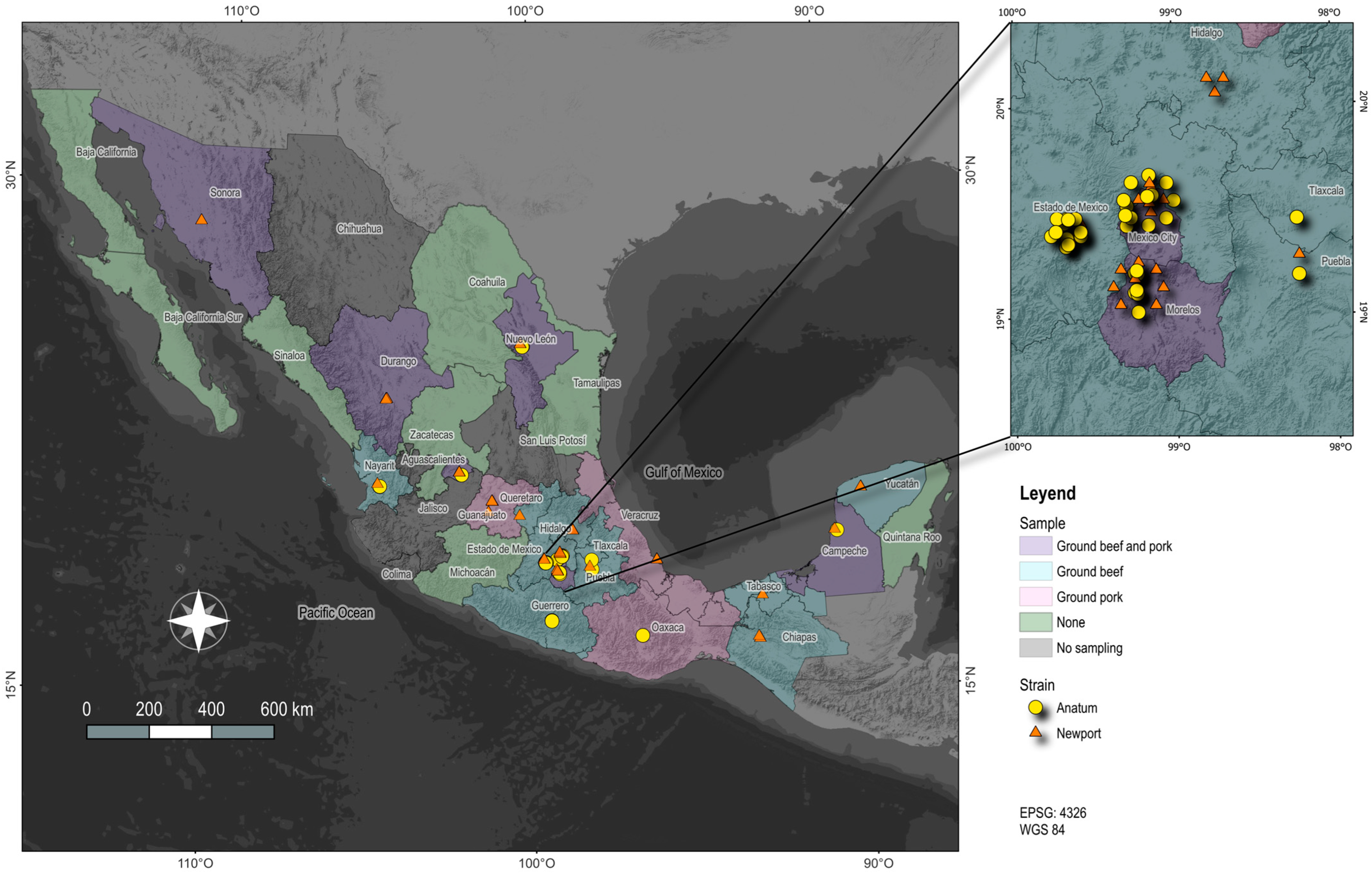
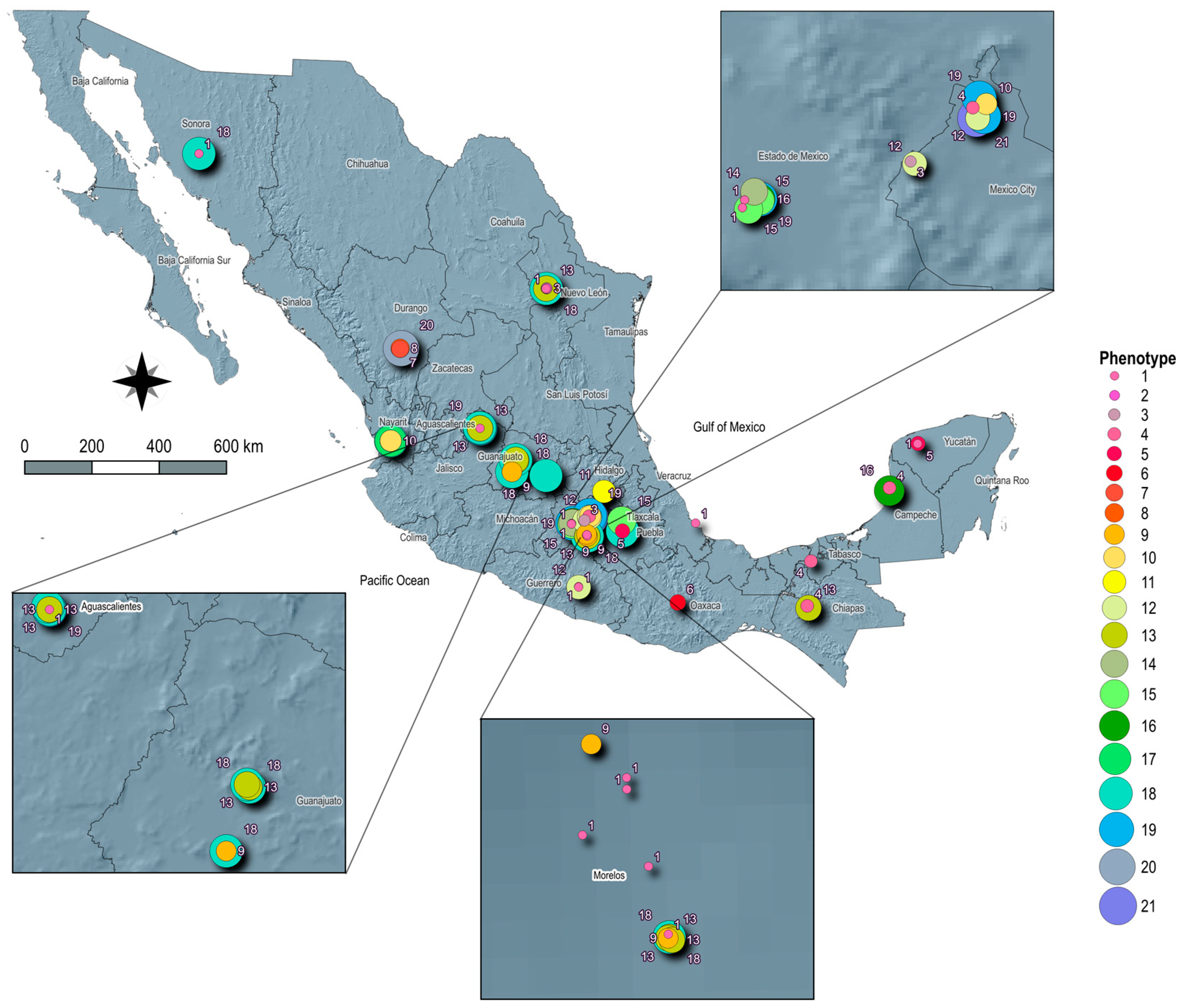
3.1. Geographic Distribution
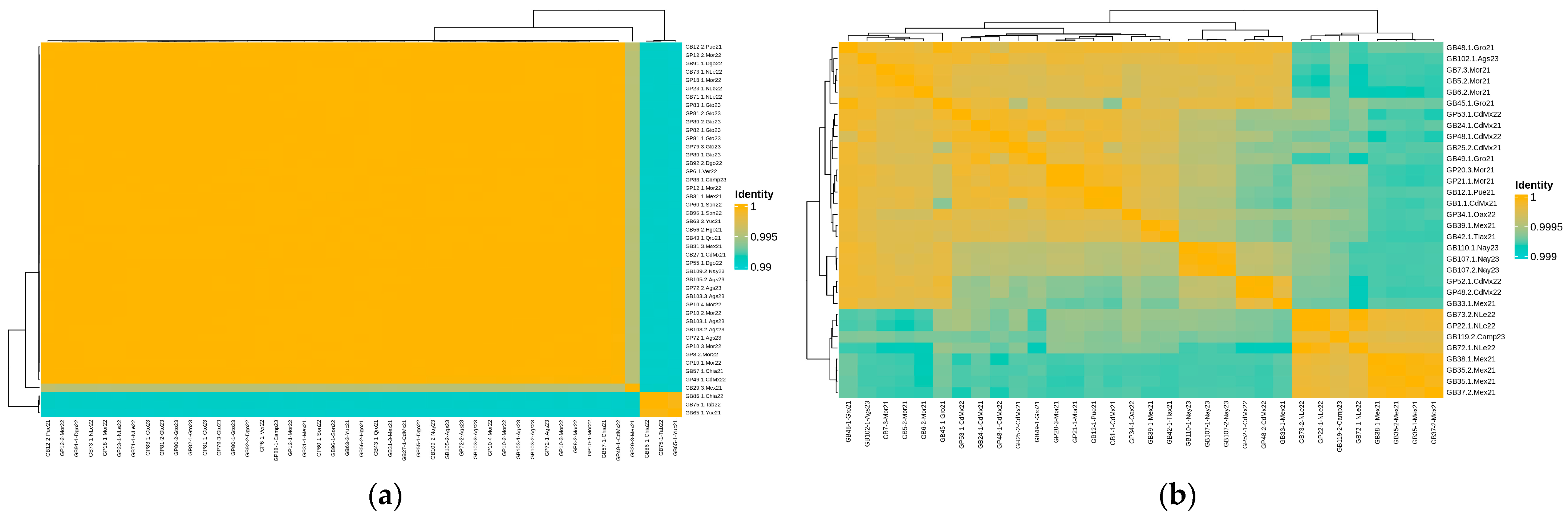
3.2. Comparative Analysis of Salmonella Newport and Anatum Genomes
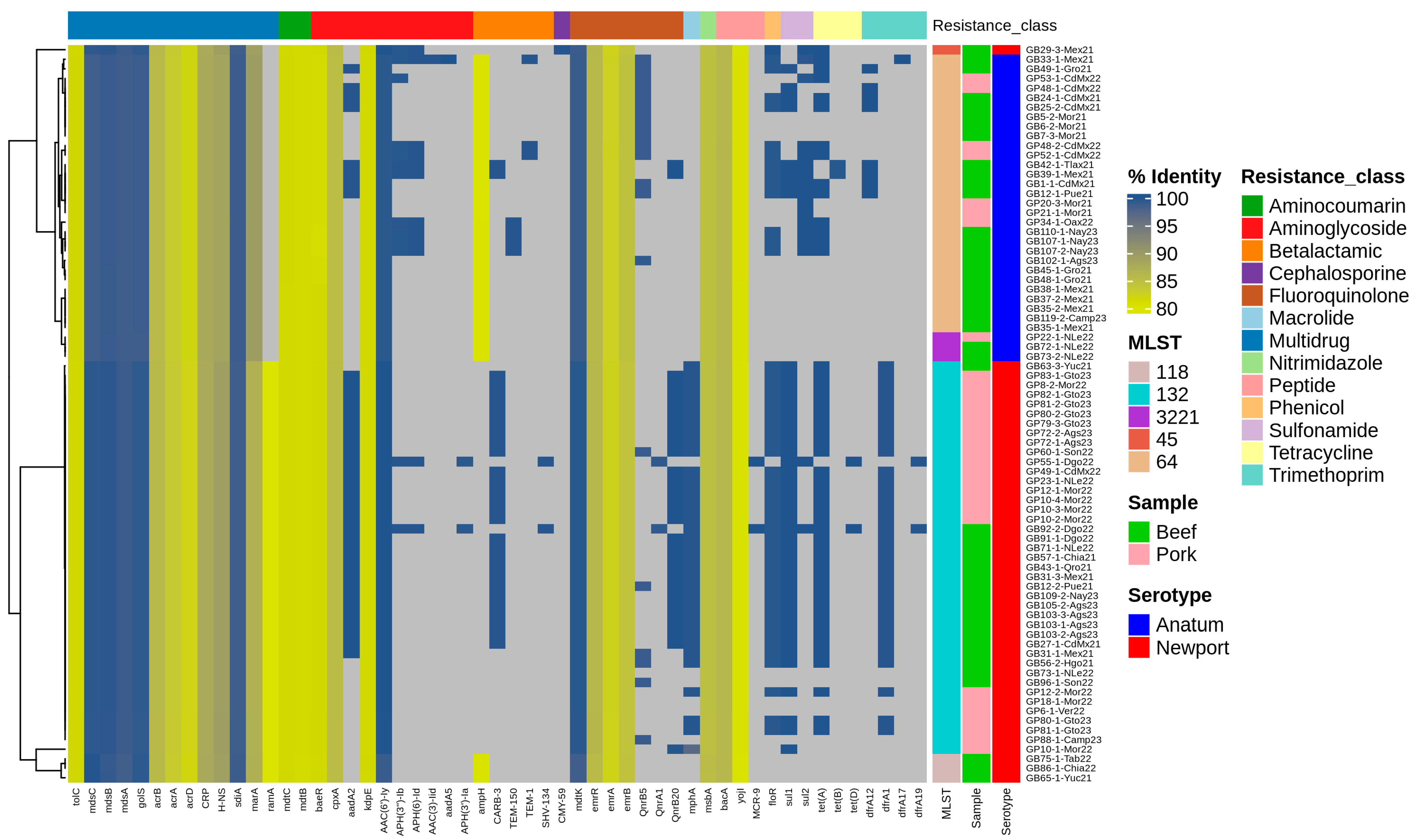
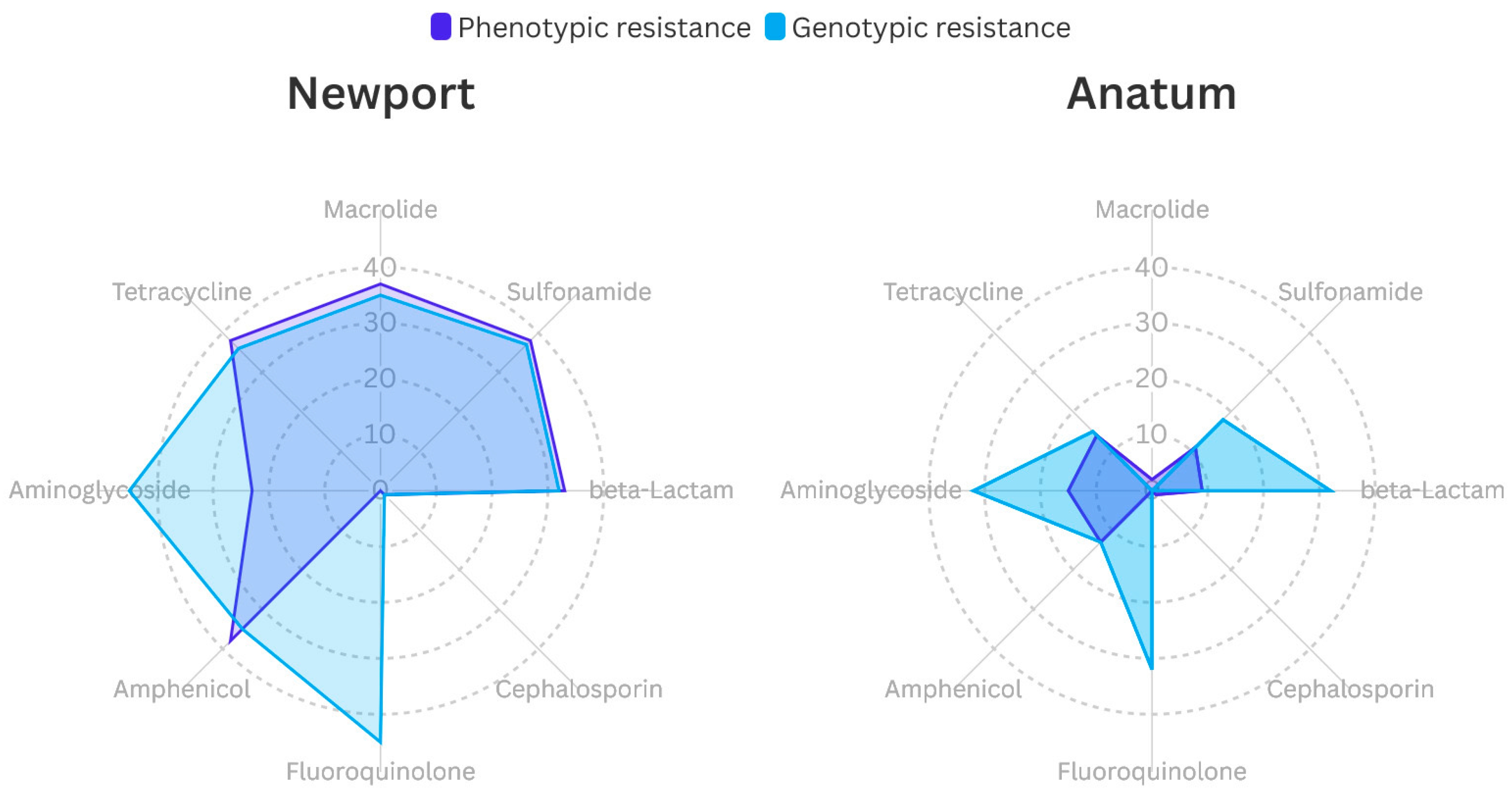
3.3. Phenotypic and Genotypic of Antimicrobial-Resistance Profiles
3.4. Pangenome Analysis of Newport and Anatum Serotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhunia, A.K. Salmonella enterica. In Foodborne Microbial Pathogens; Springer Nature: New York, NY, USA, 2018; pp. 271–287. ISBN 1572-0330. [Google Scholar]
- Lamas, A.; Miranda, J.M.; Regal, P.; Vázquez, B.; Franco, C.M.; Cepeda, A. A Comprehensive Review of Non-Enterica Subspecies of Salmonella enterica. Microbiol. Res. 2018, 206, 60–73. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Salmonella (Non-Typhoidal). Available online: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (accessed on 13 October 2024).
- He, Y.; Wang, J.; Zhang, R.; Chen, L.; Zhang, H.; Qi, X.; Chen, J. Epidemiology of Foodborne Diseases Caused by Salmonella in Zhejiang Province, China, between 2010 and 2021. Front. Public Health 2023, 11, 1127925. [Google Scholar] [CrossRef] [PubMed]
- Sima, C.M.; Buzilă, E.R.; Trofin, F.; Păduraru, D.; Luncă, C.; Duhaniuc, A.; Dorneanu, O.S.; Nastase, E.V. Emerging Strategies against Non-Typhoidal Salmonella: From Pathogenesis to Treatment. Curr. Issues Mol. Biol. 2024, 46, 7447–7472. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Dirección General de Epidemiología (DGE). Boletín Epidemiológico, Sistema Nacional de Vigilancia Epidemiológica, Sistema Único de Información [Epidemiological Bulletin, National System of Epidemiological Surveillance, Single Information System]. Available online: https://www.gob.mx/salud/acciones-y-programas/historico-boletin-epidemiologico (accessed on 7 October 2024).
- Chen, Z.; Toro, M.; Moreno-Switt, A.I.; Adell, A.D.; Delgado-Suárez, E.J.; Bonelli, R.R.; Oliveira, C.J.B.; Reyes-Jara, A.; Huang, X.; Albee, B.; et al. Unveiling the Genomic Landscape of Salmonella enterica Serotypes Typhimurium, Newport, and Infantis in Latin American Surface Waters: A Comparative Analysis. Microbiol. Spectr. 2024, 12, e0011724. [Google Scholar] [CrossRef]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-Analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same Species, Different Diseases: How and Why Typhoidal and Non-Typhoidal Salmonella Enterica Serovars Differ. Front. Microbiol. 2014, 5, 391. [Google Scholar] [CrossRef]
- Li, C.; Tyson, G.H.; Hsu, C.-H.; Harrison, L.; Strain, E.; Tran, T.-T.; Tillman, G.E.; Dessai, U.; McDermott, P.F.; Zhao, S. Long-Read Sequencing Reveals Evolution and Acquisition of Antimicrobial Resistance and Virulence Genes in Salmonella enterica. Front. Microbiol. 2021, 12, 777817. [Google Scholar] [CrossRef]
- Zhao, S.; Li, C.; Hsu, C.-H.; Tyson, G.H.; Strain, E.; Tate, H.; Tran, T.-T.; Abbott, J.; McDermott, P.F. Comparative Genomic Analysis of 450 Strains of Salmonella enterica Isolated from Diseased Animals. Genes 2020, 11, 1025. [Google Scholar] [CrossRef]
- Trees, E.; Carleton, H.A.; Folster, J.P.; Gieraltowski, L.; Hise, K.; Leeper, M.; Nguyen, T.-A.; Poates, A.; Sabol, A.; Tagg, K.A.; et al. Genetic Diversity in Salmonella enterica in Outbreaks of Foodborne and Zoonotic Origin in the USA in 2006–2017. Microorganisms 2024, 12, 1563. [Google Scholar] [CrossRef]
- Chen, Z.; Moreno-Switt, A.I.; Reyes-Jara, A.; Delgado Suarez, E.; Adell, A.D.; Oliveira, C.J.B.; Bonelli, R.R.; Huang, X.; Brown, E.; Allard, M.; et al. A Multicenter Genomic Epidemiological Investigation in Brazil, Chile, and Mexico Reveals the Diversity and Persistence of Salmonella Populations in Surface Waters. mBio 2024, 15, e00777-24. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.-S.; Lauderdale, T.-L.; Chang, J.-H.; Liang, S.-Y.; Tsao, C.-S.; Wei, H.L.; Wang, Y.-W.; Teng, R.-H.; Hong, Y.-P.; Chen, B.-H.; et al. Epidemiological Trends in Serotypes Distribution and Antimicrobial Resistance in Salmonella from Humans in Taiwan, 2004–2022. IJID Reg. 2024, 11, 100372. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Suárez, E.J.; García-Meneses, A.V.; Ponce-Hernández, E.A.; Ruíz-López, F.A.; Hernández-Pérez, C.F.; Ballesteros-Nova, N.E.; Soberanis-Ramos, O.; Rubio-Lozano, M.S. Long-Term Genomic Surveillance Reveals the Circulation of Clinically Significant Salmonella in Lymph Nodes and Beef Trimmings from Slaughter Cattle from a Mexican Feedlot. PLoS ONE 2024, 19, e0312275. [Google Scholar] [CrossRef]
- Karp, B.E.; Leeper, M.M.; Chen, J.C.; Tagg, K.A.; Francois Watkins, L.K.; Friedman, C.R. Multidrug-Resistant Salmonella Serotype Anatum in Travelers and Seafood from Asia, United States. Emerg. Infect. Dis. 2020, 26, 1030–1033. [Google Scholar] [CrossRef]
- Feng, Y.; Chang, Y.-J.; Pan, S.-C.; Su, L.-H.; Li, H.-C.; Yang, H.-P.; Yu, M.-J.; Chiu, C.-H. Characterization and Source Investigation of Multidrug-Resistant Salmonella Anatum from a Sustained Outbreak, Taiwan. Emerg. Infect. Dis. 2020, 26, 2951–2955. [Google Scholar] [CrossRef]
- Yilmaz, K.; Dorterler, M.E.; Demirbilek, M.; Levent, B. Pyelonephritis Caused by Salmonella Anatum: An Unusual Case. Urol. Int. 2018, 100, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Godínez-Oviedo, A.; Tamplin, M.L.; Bowman, J.P.; Hernández-Iturriaga, M. Salmonella enterica in Mexico 2000–2017: Epidemiology, Antimicrobial Resistance, and Prevalence in Food. Foodborne Pathog. Dis. 2020, 17, 98–118. [Google Scholar] [CrossRef]
- Gómez-Baltazar, A.; Godínez-Oviedo, A.; Vázquez-Marrufo, G.; Vázquez-Garcidueñas, M.S.; Hernández-Iturriaga, M. Genomic Analysis of the MLST Population Structure and Antimicrobial Resistance Genes Associated with Salmonella enterica in Mexico. Genome 2023, 66, 319–332. [Google Scholar] [CrossRef]
- Carroll, L.M.; Buehler, A.J.; Gaballa, A.; Siler, J.D.; Cummings, K.J.; Cheng, R.A.; Wiedmann, M. Monitoring the Microevolution of Salmonella enterica in Healthy Dairy Cattle Populations at the Individual Farm Level Using Whole-Genome Sequencing. Front. Microbiol. 2021, 12, 763669. [Google Scholar] [CrossRef]
- Ford, L.; Ellison, Z.; Schwensohn, C.; Griffin, I.; Birhane, M.G.; Cote, A.; Fortenberry, G.Z.; Tecle, S.; Higa, J.; Spencer, S.; et al. Strain of Multidrug-Resistant Salmonella Newport Remains Linked to Travel to Mexico and U.S. Beef Products—United States, 2021–2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 1225–1229. [Google Scholar] [CrossRef]
- Campos Granados, C.M.; del Sierra Gómez Pedroso, L.C.; Hernández-Pérez, C.F.; Ballesteros-Nova, N.E.; Rubio-Lozano, M.S.; Sánchez-Zamorano, L.M.; Delgado-Suárez, E.J. Fuertes Perfiles de Resistencia a Antibióticos En Salmonella spp. Aislada de Carne de Res Molida En El Centro de México. Vet. México OA 2023, 10. [Google Scholar] [CrossRef]
- Zavala-Norzagaray, A.A.; Aguirre, A.A.; Angulo-Zamudio, U.A.; Ley-Quiñonez, C.P.; Flores-Villaseñor, H.; León-Sicairos, N.; Velázquez-Román, J.; Elorriaga-Verplancken, F.R.; Zavala-Félix, K.A.; Hart, C.E.; et al. Isolation, Characterization, and Antimicrobial Susceptibility of Bacteria Isolated from Sea Lion (Zalophus californianus) Pups in Northwestern Mexico. J. Wildl. Dis. 2022, 58, 500–511. [Google Scholar] [CrossRef]
- Ballesteros-Nova, N.E.; Sánchez, S.; Steffani, J.L.; Sierra, L.C.; Chen, Z.; Ruíz-López, F.A.; Bell, R.L.; Reed, E.A.; Balkey, M.; Rubio-Lozano, M.S.; et al. Genomic Epidemiology of Salmonella enterica Circulating in Surface Waters Used in Agriculture and Aquaculture in Central Mexico. Appl. Environ. Microbiol. 2022, 88, e0214921. [Google Scholar] [CrossRef]
- Burris, K.P.; Simmons, O.D.; Webb, H.M.; Deese, L.M.; Moore, R.G.; Jaykus, L.-A.; Zheng, J.; Reed, E.; Ferreira, C.M.; Brown, E.W.; et al. Colonization and Internalization of Salmonella enterica and Its Prevalence in Cucumber Plants. Front. Microbiol. 2020, 11, 1135. [Google Scholar] [CrossRef]
- Delgado-Suárez, E.J.; Palós-Guitérrez, T.; Ruíz-López, F.A.; Hernández Pérez, C.F.; Ballesteros-Nova, N.E.; Soberanis-Ramos, O.; Méndez-Medina, R.D.; Allard, M.W.; Rubio-Lozano, M.S. Genomic Surveillance of Antimicrobial Resistance Shows Cattle and Poultry Are a Moderate Source of Multi-Drug Resistant Non-Typhoidal Salmonella in Mexico. PLoS ONE 2021, 16, e0243681. [Google Scholar] [CrossRef]
- Delgado-Suárez, E.J. Salmonella spp. Antibiotic Susceptibility Testing by the Kirby-Bauer Disk Diffusion Method. protocols.io 2021. [Google Scholar] [CrossRef]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th Revision (PDF); Antimicrobial Resistance Division, Global Coordinationand Partnership, Nutrition and Food Safety, Ed.; WHO: Geneva, Switzerland, 2019; ISBN 978-92-4-151552-8. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, 14th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2024; ISBN 978-1-68440-225-0. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 15 October 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the Bacterial Bioinformatics Database and Analysis Resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Deneke, C.; Brendebach, H.; Uelze, L.; Borowiak, M.; Malorny, B.; Tausch, S.H. Species-Specific Quality Control, Assembly and Contamination Detection in Microbial Isolate Sequences with AQUAMIS. Genes 2021, 12, 644. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Zhang, S.; den Bakker, H.C.; Li, S.; Chen, J.; Dinsmore, B.A.; Lane, C.; Lauer, A.C.; Fields, P.I.; Deng, X. SeqSero2: Rapid and Improved Salmonella Serotype Determination Using Whole-Genome Sequencing Data. Appl. Environ. Microbiol. 2019, 85, e01746-19. [Google Scholar] [CrossRef]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.J.; Nash, J.H.E.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef]
- Achtman, M.; Wain, J.; Weill, F.-X.; Nair, S.; Zhou, Z.; Sangal, V.; Krauland, M.G.; Hale, J.L.; Harbottle, H.; Uesbeck, A.; et al. Multilocus Sequence Typing as a Replacement for Serotyping in Salmonella enterica. PLoS Pathog. 2012, 8, e1002776. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2019, 48, D517–D525. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Gu, Z. Complex Heatmap Visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2. Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Houston, TX, USA, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Szklo, M.; Nieto, F.J. Epidemiology: Beyond the Basics, 4th ed.; Jones & Bartlett Publishers: Burlington, MA, USA, 2019; ISBN 9781284116595. [Google Scholar]
- Billah, M.M.; Rahman, M.S. Salmonella in the Environment: A Review on Ecology, Antimicrobial Resistance, Seafood Contaminations, and Human Health Implications. J. Hazard. Mater. Adv. 2024, 13, 100407. [Google Scholar] [CrossRef]
- Godínez-Oviedo, A.; Sampedro, F.; Bowman, J.P.; Garcés-Vega, F.J.; Hernández-Iturriaga, M. Risk Ranking of Food Categories Associated with Salmonella enterica Contamination in the Central Region of Mexico. Risk Anal. 2023, 43, 308–323. [Google Scholar] [CrossRef]
- Venkateswaran, N.; Swetschinski, L.R.; Fastl, C.; Di Bari, C.; Criscuolo, N.G.; Mulchandani, R.; Zhao, C.; Meštrović, T.; Ikuta, K.S.; Martins, S.B.; et al. Using Priorities between Human and Livestock Bacterial Antimicrobial Resistance (AMR) to Identify Data Gaps in Livestock AMR Surveillance. BMC Infect. Dis. 2024, 24, 1027. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Diaz, E.; Barbosa-Cardenas, C.M.; Perez-Montaño, J.A.; Gonzalez-Aguilar, D.; Pacheco-Gallardo, C.; Barba, J. Occurrence, Serotype Diversity, and Antimicrobial Resistance of Salmonella in Ground Beef at Retail Stores in Jalisco State, Mexico. J. Food Prot. 2013, 76, 2004–2010. [Google Scholar] [CrossRef]
- Rincón-Gamboa, S.M.; Poutou-Piñales, R.A.; Carrascal-Camacho, A.K. Antimicrobial Resistance of Non-Typhoid Salmonella in Meat and Meat Products. Foods 2021, 10, 1731. [Google Scholar] [CrossRef]
- Abu Hatab, A.; Cavinato, M.E.R.; Lindemer, A.; Lagerkvist, C.-J. Urban Sprawl, Food Security and Agricultural Systems in Developing Countries: A Systematic Review of the Literature. Cities 2019, 94, 129–142. [Google Scholar] [CrossRef]
- Villalpando-Guzmán, S.; Vázquez-Quiñones, C.R.; Natividad-Bonifacio, I.; Curiel-Quesada, E.; Quiñones-Ramírez, E.I.; Vázquez-Salinas, C. Frecuencia, Susceptibilidad Antimicrobiana y Patrón de Adherencia de Salmonella enterica Aislada de Carne de Pollo, Res y Cerdo de La Ciudad de México. Rev. Chil. Infectología 2017, 34, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.D.M.; Widmer, K.W.; Rivera, W.L. PCR-Based Detection and Serovar Identification of Salmonella in Retail Meat Collected from Wet Markets in Metro Manila, Philippines. PLoS ONE 2020, 15, e0239457. [Google Scholar] [CrossRef] [PubMed]
- Bonifait, L.; Thépault, A.; Baugé, L.; Rouxel, S.; Le Gall, F.; Chemaly, M. Occurrence of Salmonella in the Cattle Production in France. Microorganisms 2021, 9, 872. [Google Scholar] [CrossRef] [PubMed]
- da Cunha-Neto, A.; Carvalho, L.A.; Castro, V.S.; Barcelos, F.G.; Carvalho, R.C.T.; dos Rodrigues, D.P.; Conte-Junior, C.A.; de Figueiredo, E.E.S. Salmonella Anatum, S. Infantis and S. Schwarzengrund in Brazilian Cheeses: Occurrence and Antibiotic Resistance Profiles. Int. J. Dairy Technol. 2020, 73, 296–300. [Google Scholar] [CrossRef]
- Hugho, E.A.; Kumburu, H.H.; Thomas, K.; Lukambagire, A.S.; Wadugu, B.; Amani, N.; Kinabo, G.; Hald, T.; Mmbaga, B.T. High Diversity of Salmonella spp. from Children with Diarrhea, Food, and Environmental Sources in Kilimanjaro—Tanzania: One Health Approach. Front. Microbiol. 2024, 14, 1277019. [Google Scholar] [CrossRef]
- Marshall, K.E.; Cui, Z.; Gleason, B.L.; Hartley, C.; Wise, M.E.; Bruce, B.B.; Griffin, P.M. An Approach to Describe Salmonella Serotypes of Concern for Outbreaks: Using Burden and Trajectory of Outbreak-Related Illnesses Associated with Meat and Poultry. J. Food Prot. 2024, 87, 100331. [Google Scholar] [CrossRef]
- Jansson Mörk, M.; Karamehmedovic, N.; Hansen, A.; Nederby Öhd, J.; Lindblad, M.; Östlund, E.; Rehn, M.; Jernberg, C. Outbreak of Salmonella Newport Linked to Imported Frozen Cooked Crayfish in Dill Brine, Sweden, July to November 2019. Eurosurveillance 2022, 27, 2100918. [Google Scholar] [CrossRef]
- Ge, B.; Mukherjee, S.; Li, C.; Harrison, L.B.; Hsu, C.-H.; Tran, T.-T.; Whichard, J.M.; Dessai, U.; Singh, R.; Gilbert, J.M.; et al. Genomic Analysis of Azithromycin-Resistant Salmonella from Food Animals at Slaughter and Processing, and Retail Meats, 2011–2021, United States. Microbiol. Spectr. 2024, 12, e0348523. [Google Scholar] [CrossRef]
- Sangal, V.; Harbottle, H.; Mazzoni, C.J.; Helmuth, R.; Guerra, B.; Didelot, X.; Paglietti, B.; Rabsch, W.; Brisse, S.; Weill, F.-X.; et al. Evolution and Population Structure of Salmonella enterica Serovar Newport. J. Bacteriol. 2010, 192, 6465–6476. [Google Scholar] [CrossRef]
- Cao, G.; Meng, J.; Strain, E.; Stones, R.; Pettengill, J.; Zhao, S.; McDermott, P.; Brown, E.; Allard, M. Phylogenetics and Differentiation of Salmonella Newport Lineages by Whole Genome Sequencing. PLoS ONE 2013, 8, e55687. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, Y.; Qin, X.; Aspridou, Z.; Zheng, J.; Wang, X.; Li, Z.; Dong, Q. The Prevalence and Epidemiology of Salmonella in Retail Raw Poultry Meat in China: A Systematic Review and Meta-Analysis. Foods 2021, 10, 2757. [Google Scholar] [CrossRef] [PubMed]
- Ramtahal, M.A.; Somboro, A.M.; Amoako, D.G.; Abia, A.L.K.; Perrett, K.; Bester, L.A.; Essack, S.Y. Molecular Epidemiology of Salmonella enterica in Poultry in South Africa Using the Farm-to-Fork Approach. Int. J. Microbiol. 2022, 2022, 5121273. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.K.; Andershock, W.E.; Qian, X.; Gibbs, P.L.; Orejuela, K.; Garman, K.N.; Dunn, J.R.; Denes, T.G. Phylogeny and Genomic Characterization of Clinical Salmonella enterica Serovar Newport Collected in Tennessee. Microbiol. Spectr. 2023, 11, e0387622. [Google Scholar] [CrossRef]
- Zheng, J.; Luo, Y.; Reed, E.; Bell, R.; Brown, E.W.; Hoffmann, M. Whole-Genome Comparative Analysis of Salmonella enterica Serovar Newport Strains Reveals Lineage-Specific Divergence. Genome Biol. Evol. 2017, 9, 1047–1050. [Google Scholar] [CrossRef]
- Pouwels, K.B.; Muller-Pebody, B.; Smieszek, T.; Hopkins, S.; Robotham, J.V. Selection and Co-Selection of Antibiotic Resistances among Escherichia coli by Antibiotic Use in Primary Care: An Ecological Analysis. PLoS ONE 2019, 14, e0218134. [Google Scholar] [CrossRef]
- Engin, A.B.; Engin, E.D.; Engin, A. Effects of Co-Selection of Antibiotic-Resistance and Metal-Resistance Genes on Antibiotic-Resistance Potency of Environmental Bacteria and Related Ecological Risk Factors. Environ. Toxicol. Pharmacol. 2023, 98, 104081. [Google Scholar] [CrossRef]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial Resistance of Salmonella Isolated from Food Animals: A Review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- Murray, L.M.; Hayes, A.; Snape, J.; Kasprzyk-Hordern, B.; Gaze, W.H.; Murray, A.K. Co-Selection for Antibiotic Resistance by Environmental Contaminants. Npj Antimicrob. Resist. 2024, 2, 9. [Google Scholar] [CrossRef]
- Tack, B.; Phoba, M.-F.; Thong, P.; Lompo, P.; Hupko, C.; Desmet, S.; Martiny, D.; Mattheus, W.; Pardos de la Gandara, M.; Mbuyi-Kalonji, L.; et al. Epidemiological Cut-off Value and Antibiotic Susceptibility Test Methods for Azithromycin in a Collection of Multi-Country Invasive Non-Typhoidal Salmonella. Clin. Microbiol. Infect. 2022, 28, 1615–1623. [Google Scholar] [CrossRef]
- Konyali, D.; Guzel, M.; Soyer, Y. Genomic Characterization of Salmonella enterica Resistant to Cephalosporin, Quinolones, and Macrolides. Curr. Microbiol. 2023, 80, 344. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Suárez, E.J.; Selem-Mojica, N.; Ortiz-López, R.; Gebreyes, W.A.; Allard, M.W.; Barona-Gómez, F.; Rubio-Lozano, M.S. Whole Genome Sequencing Reveals Widespread Distribution of Typhoidal Toxin Genes and VirB/D4 Plasmids in Bovine-Associated Nontyphoidal Salmonella. Sci. Rep. 2018, 8, 9864. [Google Scholar] [CrossRef]
- Palós Gutiérrez, T.; Rubio Lozano, M.S.; Delgado Suárez, E.J.; Rosi Guzmán, N.; Soberanis Ramos, O.; Hernández Pérez, C.F.; Méndez Medina, R.D. Linfonodos y Carne Molida de Res Como Reservorios de Salmonella spp. de Importancia En Salud Pública. Rev. Mex. Cienc. Pecu. 2020, 11, 795–810. [Google Scholar] [CrossRef]
- Barrera, S.; Vázquez-Flores, S.; Needle, D.; Rodríguez-Medina, N.; Iglesias, D.; Sevigny, J.L.; Gordon, L.M.; Simpson, S.; Thomas, W.K.; Rodulfo, H.; et al. Serovars, Virulence and Antimicrobial Resistance Genes of Non-Typhoidal Salmonella Strains from Dairy Systems in Mexico. Antibiotics 2023, 12, 1662. [Google Scholar] [CrossRef] [PubMed]
- Casaux, M.L.; D’Alessandro, B.; Vignoli, R.; Fraga, M. Phenotypic and Genotypic Survey of Antibiotic Resistance in Salmonella enterica Isolates from Dairy Farms in Uruguay. Front. Vet. Sci. 2023, 10, 1055432. [Google Scholar] [CrossRef] [PubMed]
- Ben Hassena, A.; Haendiges, J.; Zormati, S.; Guermazi, S.; Gdoura, R.; Gonzalez-Escalona, N.; Siala, M. Virulence and Resistance Genes Profiles and Clonal Relationships of Non-Typhoidal Food-Borne Salmonella Strains Isolated in Tunisia by Whole Genome Sequencing. Int. J. Food Microbiol. 2021, 337, 108941. [Google Scholar] [CrossRef]
- Seribelli, A.A.; da Silva, P.; da Cruz, M.F.; de Almeida, F.; Frazão, M.R.; Medeiros, M.I.C.; dos Rodrigues, D.P.; Kich, J.D.; de Jesus Benevides, L.; de Soares, S.C.; et al. Insights about the Epidemiology of Salmonella Typhimurium Isolates from Different Sources in Brazil Using Comparative Genomics. Gut Pathog. 2021, 13, 27. [Google Scholar] [CrossRef]
- Bellil, Z.; Meyer, S.; Tilloy, V.; Mairi, A.; De Champs, C.; Barraud, O.; Touati, A. Prevalence and Genomic Investigation of Salmonella Isolates Associated with Watermelons and Their Environmental Reservoirs in Bejaia, Algeria. Foodborne Pathog. Dis. 2024. [Google Scholar] [CrossRef]
- Fontana, H.Y.Y. One Health Genomic Surveillance of Antibiotic-Resistant Nontyphoidal Salmonella enterica Serovars in Brazil. Ph.D Thesis, Universidade de São Paulo, São Paulo, Brazil, 2024. [Google Scholar]
- Robinson, E.; Travanut, M.; Fabre, L.; Larréché, S.; Ramelli, L.; Pascal, L.; Guinard, A.; Vincent, N.; Calba, C.; Meurice, L.; et al. Outbreak of Salmonella Newport Associated with Internationally Distributed Raw Goats’ Milk Cheese, France, 2018. Epidemiol. Infect. 2020, 148, e180. [Google Scholar] [CrossRef]
- Kim, H.; Kim, M.; Kim, S.; Lee, Y.M.; Shin, S.C. Characterization of Antimicrobial Resistance Genes and Virulence Factor Genes in an Arctic Permafrost Region Revealed by Metagenomics. Environ. Pollut. 2022, 294, 118634. [Google Scholar] [CrossRef]
- Kim, H.-S.; Nagore, D.; Nikaido, H. Multidrug Efflux Pump MdtBC of Escherichia coli Is Active Only as a B 2 C Heterotrimer. J. Bacteriol. 2010, 192, 1377–1386. [Google Scholar] [CrossRef] [PubMed]
- Afridi, O.K.; Ali, J.; Chang, J.H. Resistome and Microbial Profiling of Pediatric Patient’s Gut Infected with Multidrug-Resistant Diarrhoeagenic Enterobacteriaceae Using next-Generation Sequencing; the First Study from Pakistan. Libyan J. Med. 2021, 16, 1915615. [Google Scholar] [CrossRef] [PubMed]
- Górecki, K.; McEvoy, M.M. Phylogenetic Analysis Reveals an Ancient Gene Duplication as the Origin of the MdtABC Efflux Pump. PLoS ONE 2020, 15, e0228877. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, R.; Barh, D.; Weimer, B.C.; Viana, M.V.C.; Profeta, R.; Sousa, T.J.; Aburjaile, F.F.; Quino, W.; Souza, R.P.; Mestanza, O.; et al. WGS-Based Lineage and Antimicrobial Resistance Pattern of Salmonella Typhimurium Isolated during 2000–2017 in Peru. Antibiotics 2022, 11, 1170. [Google Scholar] [CrossRef]
- Gu, Y.; Kuang, X.; Sajid, A.; Wang, Y.; Zhang, Z.; Xu, Z.; Cheng, G.; Shabbir, A.B.; Yuan, Z.; Hao, H. Prevalence and Mechanism of Antimicrobial Resistance and Pathogenicity of Salmonella Isolated from Foodborne Animal in China. LWT 2023, 184, 114906. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, N.; Wang, M.; Luo, M.; Peng, Y.; Li, Z.; Xu, J.; Ou, M.; Kan, B.; Li, X.; et al. The Prevalence and Distribution of Aminoglycoside Resistance Genes. Biosaf. Health 2023, 5, 14–20. [Google Scholar] [CrossRef]
- Liu, X.; Chang, Y.; Xu, Q.; Zhang, W.; Huang, Z.; Zhang, L.; Weng, S.; Leptihn, S.; Jiang, Y.; Yu, Y.; et al. Mutation in the Two-Component Regulator BaeSR Mediates Cefiderocol Resistance and Enhances Virulence in Acinetobacter baumannii. mSystems 2023, 8, e0129122. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Vaidyanathan, V.; Mondal, A.; Rajamohan, G. Role of the Two Component Signal Transduction System CpxAR in Conferring Cefepime and Chloramphenicol Resistance in Klebsiella Pneumoniae NTUH-K2044. PLoS ONE 2012, 7, e33777. [Google Scholar] [CrossRef]
- Freeman, Z.N.; Dorus, S.; Waterfield, N.R. The KdpD/KdpE Two-Component System: Integrating K+ Homeostasis and Virulence. PLoS Pathog. 2013, 9, e1003201. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Monte, D.F.; Lincopan, N.; Fedorka-Cray, P.J.; Landgraf, M. Current Insights on High Priority Antibiotic-Resistant Salmonella enterica in Food and Foodstuffs: A Review. Curr. Opin. Food Sci. 2019, 26, 35–46. [Google Scholar] [CrossRef]
- Sjölund-Karlsson, M.; Howie, R.L.; Blickenstaff, K.; Boerlin, P.; Ball, T.; Chalmers, G.; Duval, B.; Haro, J.; Rickert, R.; Zhao, S.; et al. Occurrence of β-Lactamase Genes Among Non-Typhi Salmonella enterica Isolated from Humans, Food Animals, and Retail Meats in the United States and Canada. Microbial. Drug Resist. 2013, 19, 191–197. [Google Scholar] [CrossRef]
- Sepp, E.; Andreson, R.; Balode, A.; Bilozor, A.; Brauer, A.; Egorova, S.; Huik, K.; Ivanova, M.; Kaftyreva, L.; Kõljalg, S.; et al. Phenotypic and Molecular Epidemiology of ESBL-, AmpC-, and Carbapenemase-Producing Escherichia coli in Northern and Eastern Europe. Front. Microbiol. 2019, 10, 2465. [Google Scholar] [CrossRef] [PubMed]
- San, N.; Aung, M.S.; Urushibara, N.; San, T.; Maw, W.W.; Lwin, M.M.; Mar, T.T.; Myint, Y.Y.; Thu, P.P.; Hlaing, M.S.; et al. Genetic Diversity of CMY Beta-Lactamase Genes in Clinical Isolates of Escherichia coli in Myanmar: Identification of Three Novel Types and Updated Phylogenetic Classification of BlaCMY. Microbial. Drug Resist. 2020, 26, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Hull, D.M.; Harrell, E.; Harden, L.; Thakur, S. Multidrug Resistance and Virulence Genes Carried by Mobile Genomic Elements in Salmonella enterica Isolated from Live Food Animals, Processed, and Retail Meat in North Carolina, 2018–2019. Int. J. Food Microbiol. 2022, 378, 109821. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, W.L.; Jacobs, J.; Wong, V.; Klemm, E.J.; Deborggraeve, S.; Van Puyvelde, S. Fluoroquinolone Resistance in Salmonella: Insights by Whole-Genome Sequencing. Microb. Genom. 2018, 4, e000195. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, H.; Huang, B.; Hu, X.; Chen, Y.; Zheng, L.; Yang, L.; Deng, J.; Wang, Q. Characterization of Resistance Genes and Plasmids from Sick Children Caused by Salmonella enterica Resistance to Azithromycin in Shenzhen, China. Front. Cell. Infect. Microbiol. 2023, 13, 1116172. [Google Scholar] [CrossRef]
- Nishino, K.; Latifi, T.; Groisman, E.A. Virulence and Drug Resistance Roles of Multidrug Efflux Systems of Salmonella enterica Serovar Typhimurium. Mol. Microbiol. 2006, 59, 126–141. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (WOAH). WOAH List of Antimicrobial Agents of Veterinary Importance; World Organisation for Animal Health (WOAH): Paris, France, 2024. [Google Scholar]
- The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2019–2020. EFSA J. 2022, 20, e07209. [CrossRef]
- Schmerold, I.; van Geijlswijk, I.; Gehring, R. European Regulations on the Use of Antibiotics in Veterinary Medicine. Eur. J. Pharm. Sci. 2023, 189, 106473. [Google Scholar] [CrossRef]
- European Medicines Agency (EMA). Advice on the Designation of Antimicrobials or Groups of Antimicrobials Reserved for Treatment of Certain Infections in Humans—In Relation to Implementing Measures Under Article 37(5) of Regulation (EU) 2019/6 on Veterinary Medicinal Products; European Medicines Agency (EMA): Amsterdam, The Netherlands, 2022. [Google Scholar]
- Secretaría de Agricultura y Desarrollo Rural (SADER). Acuerdo Por El Que Se Establece La Clasificación y Prescripción de Los Productos Farmacéuticos Veterinarios Por El Nivel de Riesgo de Sus Ingredientes Activos; Diario Oficial de la Nación: Mexico City, Mexico, 2017.
- Chaudhari, R.; Singh, K.; Kodgire, P. Biochemical and Molecular Mechanisms of Antibiotic Resistance in Salmonella spp. Res. Microbiol. 2023, 174, 103985. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, A.C.; Stogios, P.J.; Koteva, K.; Skarina, T.; Evdokimova, E.; Savchenko, A.; Wright, G.D. The Evolution of Substrate Discrimination in Macrolide Antibiotic Resistance Enzymes. Nat. Commun. 2018, 9, 112. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Martínez-Puchol, S.; Palma, N.; Horna, G.; Ruiz-Roldán, L.; Pons, M.J.; Ruiz, J. Macrolide Resistance Mechanisms in Enterobacteriaceae: Focus on Azithromycin. Crit. Rev. Microbiol. 2017, 43, 1–30. [Google Scholar] [CrossRef]
- Nuanmuang, N.; Leekitcharoenphon, P.; Njage, P.M.K.; Gmeiner, A.; Aarestrup, F.M. An Overview of Antimicrobial Resistance Profiles of Publicly Available Salmonella Genomes with Sufficient Quality and Metadata. Foodborne Pathog. Dis. 2023, 20, 405–413. [Google Scholar] [CrossRef]
- Xie, M.; Chen, K.; Chan, E.W.; Chen, S. Identification and Genetic Characterization of Two Conjugative Plasmids That Confer Azithromycin Resistance in Salmonella. Emerg. Microbes Infect. 2022, 11, 1049–1057. [Google Scholar] [CrossRef]
- Ivanova, M.; Ovsepian, A.; Leekitcharoenphon, P.; Seyfarth, A.M.; Mordhorst, H.; Otani, S.; Koeberl-Jelovcan, S.; Milanov, M.; Kompes, G.; Liapi, M.; et al. Azithromycin Resistance in Escherichia coli and Salmonella from Food-Producing Animals and Meat in Europe. J. Antimicrob. Chemother. 2024, 79, 1657–1667. [Google Scholar] [CrossRef]
- Delgado-Suárez, E.J.; Ortíz-López, R.; Gebreyes, W.A.; Allard, M.W.; Barona-Gómez, F.; Rubio-Lozano, M.S. Genomic Surveillance Links Livestock Production with the Emergence and Spread of Multi-Drug Resistant Non-Typhoidal Salmonella in Mexico. J. Microbiol. 2019, 57, 271–280. [Google Scholar] [CrossRef]
- Kadlec, K.; Kehrenberg, C.; Schwarz, S. Efflux-Mediated Resistance to Florfenicol and/or Chloramphenicol in Bordetella Bronchiseptica: Identification of a Novel Chloramphenicol Exporter. J. Antimicrob. Chemother. 2006, 59, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular Basis of Bacterial Resistance to Chloramphenicol and Florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef]
- Sánchez-Osuna, M.; Cortés, P.; Barbé, J.; Erill, I. Origin of the Mobile Di-Hydro-Pteroate Synthase Gene Determining Sulfonamide Resistance in Clinical Isolates. Front. Microbiol. 2019, 9, 3332. [Google Scholar] [CrossRef]
- Ambrose, S.J.; Hall, R.M. DfrA Trimethoprim Resistance Genes Found in Gram-Negative Bacteria: Compilation and Unambiguous Numbering. J. Antimicrob. Chemother. 2021, 76, 2748–2756. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Orsi, R.H.; Carroll, L.M.; Kovac, J.; Ou, H.; Zhang, H.; Wiedmann, M. Serotype-Specific Evolutionary Patterns of Antimicrobial-Resistant Salmonella enterica. BMC Evol. Biol. 2019, 19, 132. [Google Scholar] [CrossRef] [PubMed]
- Pavelquesi, S.L.S.; de Oliveira Ferreira, A.C.A.; Rodrigues, A.R.M.; de Souza Silva, C.M.; Orsi, D.C.; da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, W.; Zhang, S.; Hu, H.; Yuan, Y.; Dong, J.; Chen, L.; Ma, Y.; Yang, T.; Zhou, L.; et al. A Pangenome Analysis Pipeline Provides Insights into Functional Gene Identification in Rice. Genome Biol. 2023, 24, 19. [Google Scholar] [CrossRef] [PubMed]
- Rouli, L.; Merhej, V.; Fournier, P.-E.; Raoult, D. The Bacterial Pangenome as a New Tool for Analysing Pathogenic Bacteria. New Microbes New Infect. 2015, 7, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Gu, C.; Kim, H.U.; Lee, S.Y. Current Status of Pan-Genome Analysis for Pathogenic Bacteria. Curr. Opin. Biotechnol. 2020, 63, 54–62. [Google Scholar] [CrossRef]
- Bawn, M.; Alikhan, N.-F.; Thilliez, G.; Kirkwood, M.; Wheeler, N.E.; Petrovska, L.; Dallman, T.J.; Adriaenssens, E.M.; Hall, N.; Kingsley, R.A. Evolution of Salmonella enterica Serotype Typhimurium Driven by Anthropogenic Selection and Niche Adaptation. PLoS Genet. 2020, 16, e1008850. [Google Scholar] [CrossRef]
- Zhou, Z.; Lundstrøm, I.; Tran-Dien, A.; Duchêne, S.; Alikhan, N.-F.; Sergeant, M.J.; Langridge, G.; Fotakis, A.K.; Nair, S.; Stenøien, H.K.; et al. Pan-Genome Analysis of Ancient and Modern Salmonella enterica Demonstrates Genomic Stability of the Invasive Para C Lineage for Millennia. Curr. Biol. 2018, 28, 2420–2428.e10. [Google Scholar] [CrossRef]
- Seif, Y.; Kavvas, E.; Lachance, J.-C.; Yurkovich, J.T.; Nuccio, S.-P.; Fang, X.; Catoiu, E.; Raffatellu, M.; Palsson, B.O.; Monk, J.M. Genome-Scale Metabolic Reconstructions of Multiple Salmonella Strains Reveal Serovar-Specific Metabolic Traits. Nat. Commun. 2018, 9, 3771. [Google Scholar] [CrossRef]
- Park, C.J.; Andam, C.P. Distinct but Intertwined Evolutionary Histories of Multiple Salmonella enterica Subspecies. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Price, M.N.; Wetmore, K.M.; Waters, R.J.; Callaghan, M.; Ray, J.; Liu, H.; Kuehl, J.V.; Melnyk, R.A.; Lamson, J.S.; Suh, Y.; et al. Mutant Phenotypes for Thousands of Bacterial Genes of Unknown Function. Nature 2018, 557, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, N.F.; Gerloff, D.L.; Uetz, P. Protein Domains of Unknown Function Are Essential in Bacteria. mBio 2014, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, V.W.; Mallampalli, V.K.P.S.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. Impact of Membrane Phospholipid Alterations in Escherichia coli on Cellular Function and Bacterial Stress Adaptation. J. Bacteriol. 2017, 199, e00849-16. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, G.L.; Hensel, M. Manipulating Cellular Transport and Immune Responses: Dynamic Interactions between Intracellular Salmonella enterica and Its Host Cells. Cell. Microbiol. 2006, 8, 728–737. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynoso, E.C.; Delgado-Suárez, E.J.; Hernández-Pérez, C.F.; Chavarin-Pineda, Y.; Godoy-Lozano, E.E.; Fierros-Zárate, G.; Aguilar-Vera, O.A.; Castillo-Ramírez, S.; Gómez-Pedroso, L.d.C.S.; Sánchez-Zamorano, L.M. Geography, Antimicrobial Resistance, and Genomics of Salmonella enterica (Serotypes Newport and Anatum) from Meat in Mexico (2021–2023). Microorganisms 2024, 12, 2485. https://doi.org/10.3390/microorganisms12122485
Reynoso EC, Delgado-Suárez EJ, Hernández-Pérez CF, Chavarin-Pineda Y, Godoy-Lozano EE, Fierros-Zárate G, Aguilar-Vera OA, Castillo-Ramírez S, Gómez-Pedroso LdCS, Sánchez-Zamorano LM. Geography, Antimicrobial Resistance, and Genomics of Salmonella enterica (Serotypes Newport and Anatum) from Meat in Mexico (2021–2023). Microorganisms. 2024; 12(12):2485. https://doi.org/10.3390/microorganisms12122485
Chicago/Turabian StyleReynoso, Eduardo Canek, Enrique Jesús Delgado-Suárez, Cindy Fabiola Hernández-Pérez, Yaselda Chavarin-Pineda, Elizabeth Ernestina Godoy-Lozano, Geny Fierros-Zárate, Omar Alejandro Aguilar-Vera, Santiago Castillo-Ramírez, Luz del Carmen Sierra Gómez-Pedroso, and Luisa María Sánchez-Zamorano. 2024. "Geography, Antimicrobial Resistance, and Genomics of Salmonella enterica (Serotypes Newport and Anatum) from Meat in Mexico (2021–2023)" Microorganisms 12, no. 12: 2485. https://doi.org/10.3390/microorganisms12122485
APA StyleReynoso, E. C., Delgado-Suárez, E. J., Hernández-Pérez, C. F., Chavarin-Pineda, Y., Godoy-Lozano, E. E., Fierros-Zárate, G., Aguilar-Vera, O. A., Castillo-Ramírez, S., Gómez-Pedroso, L. d. C. S., & Sánchez-Zamorano, L. M. (2024). Geography, Antimicrobial Resistance, and Genomics of Salmonella enterica (Serotypes Newport and Anatum) from Meat in Mexico (2021–2023). Microorganisms, 12(12), 2485. https://doi.org/10.3390/microorganisms12122485






