The Sweet Cherry Tree Genotype Restricts the Aggressiveness of the Wood Decay Fungi Cytospora sorbicola and Calosphaeria pulchella
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sweet Cherry Orchards Survey and Sampling
2.2. Molecular Identification of Cyt. sorbicola
2.3. Growth and Cultivation Conditions of Cyt. sorbicola and Cal. pulchella
2.4. In Planta Infection Assays
2.5. Design and Validation of the Cyt. sorbicola and Cal. pulchella Identification System by PCR-HRM
2.6. Recovery of Cyt. sorbicola and Cal. pulchella from Wood Pieces
2.7. Statistical Analysis
3. Results
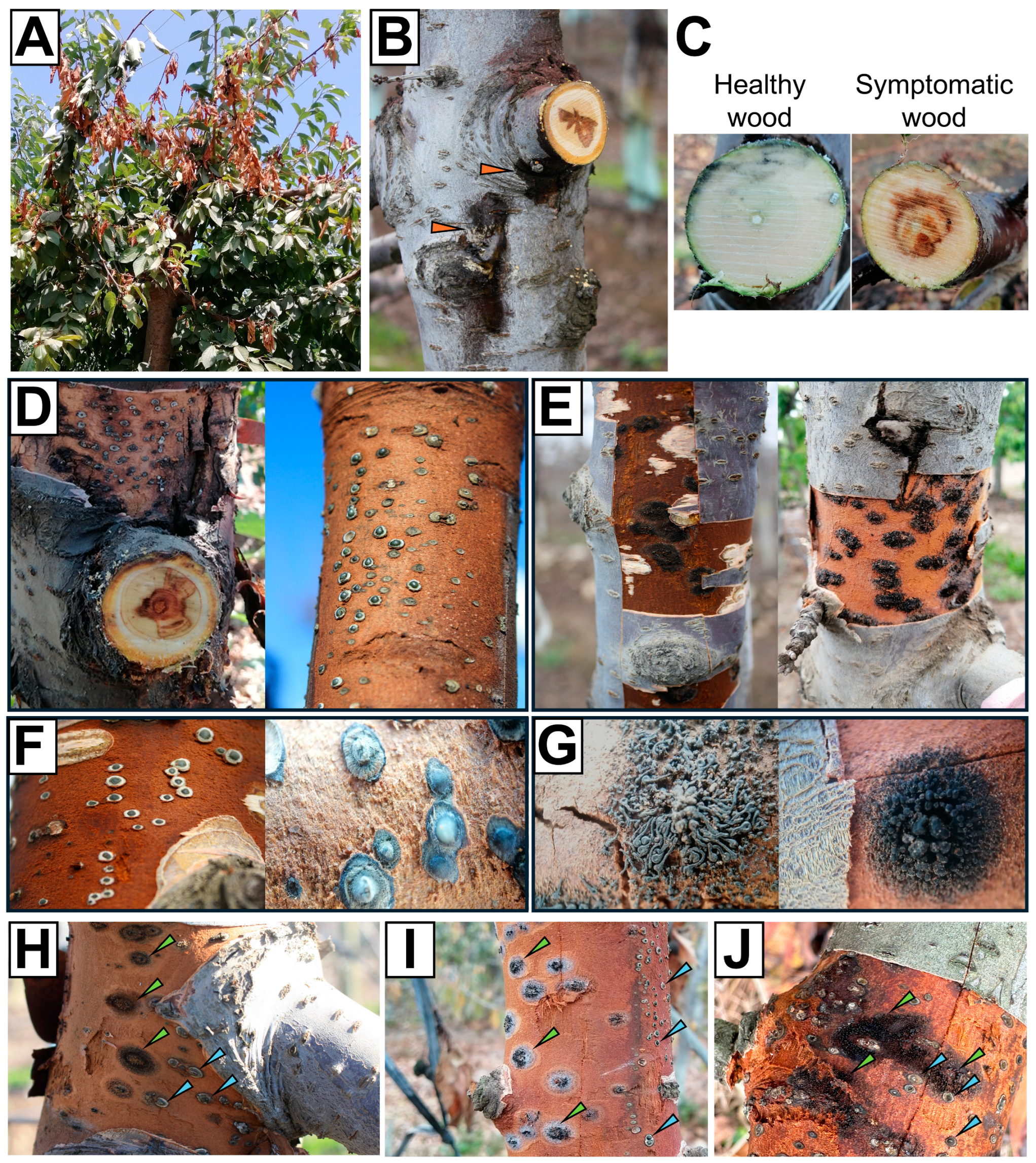
3.1. Cyt. sorbicola and Cal. pulchella Are Widespread in Sweet Cherry Orchards from the Chilean Central Valley
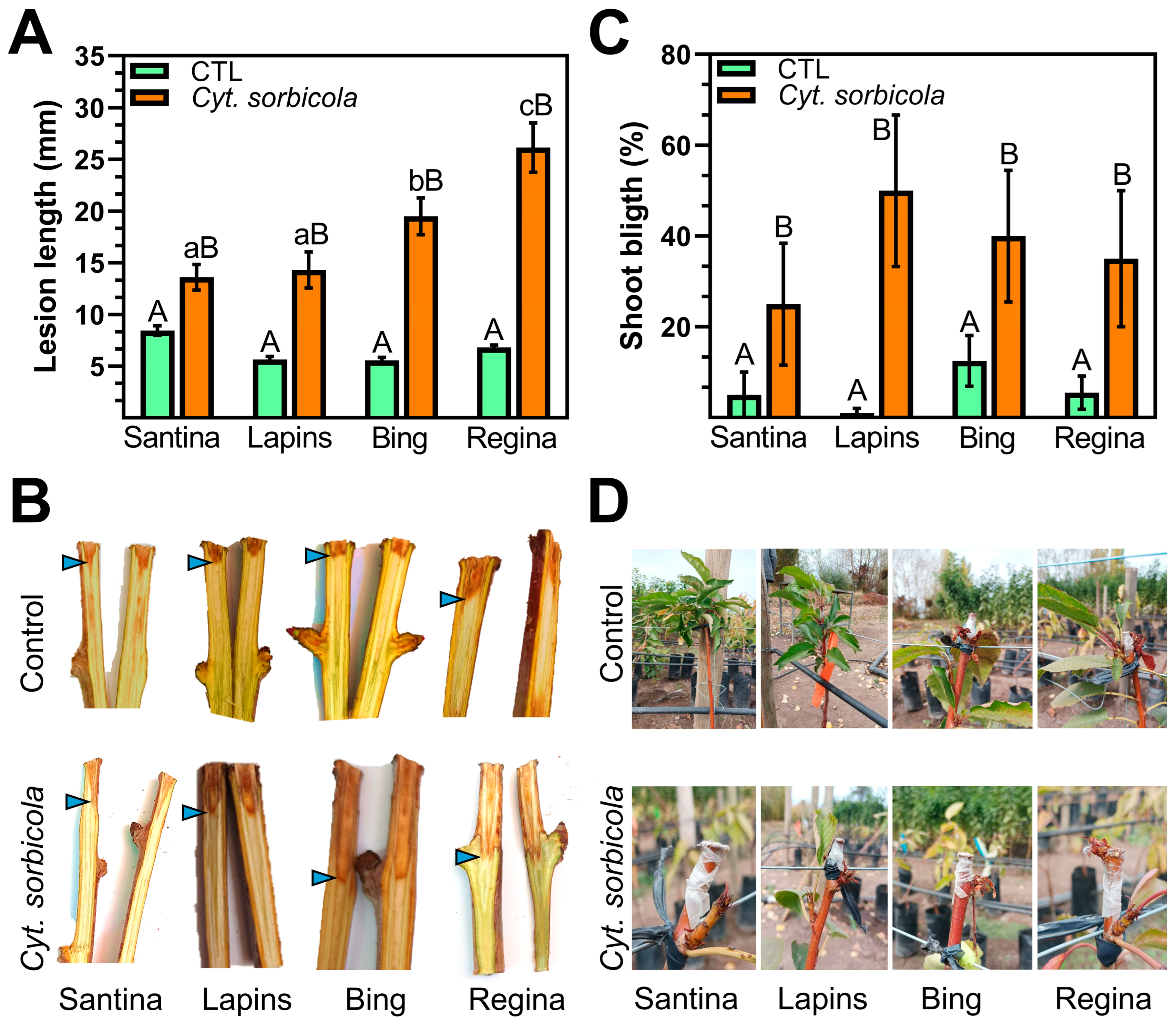
3.2. Regina Is the Sweet Cherry Variety Most Susceptible to Cyt. sorbicola and Cal. pulchella
3.3. Implementation of a System for Cyt. sorbicola and Cal. pulchella Identification by PCR-HRM
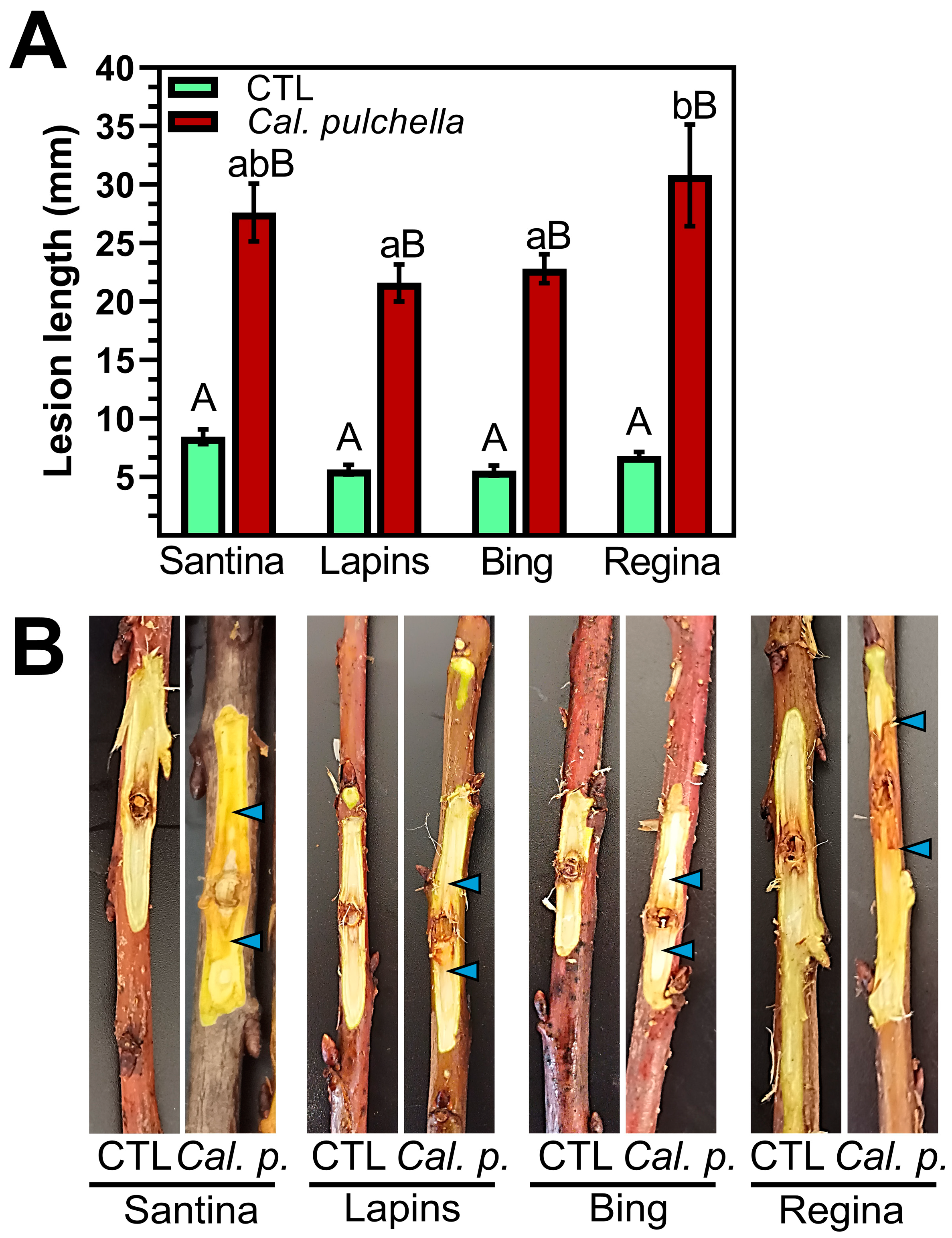
3.4. Cal. pulchella Colonizes Sweet Cherry Wood More Effectively than Cyt. sorbicola
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Auger, J.; Pozo, L.; Rubilar, M.; Briceño, N.; Osorio-Navarro, C.; Esterio, M. First Report of canker and branch dieback of sweet cherry trees caused by Calosphaeria pulchella in Chile. Plant Dis. 2020, 105, 217. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Holland, L.A.; Nouri, M.T.; Travadon, R.; Abramians, A.; Michailides, T.J.; Trouillas, F.P. Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with the descriptions of ten new species and one new combination. IMA Fungus 2018, 9, 333–370. [Google Scholar] [CrossRef]
- Li, S.; Travadon, R.; Trouillas, F.P. Seasonal Susceptibility of Sweet Cherry Pruning Wounds to Calosphaeria pulchella, Cytospora sorbicola, and Eutypa lata. Plant Dis. 2023, 107, 3517–3522. [Google Scholar] [CrossRef]
- Muñoz-Reyes, V.; Castro, J.F.; Cisterna-Oyarce, V.; Santelices, C.; Carrasco, J.; Guerra, M.; France, R.A.; Claret, M.; Millas, P. First report of canker caused by Cytospora sorbicola on Sweet Cherry (Prunus avium) in Chile. Plant Dis. 2023, 108, 524. [Google Scholar] [CrossRef]
- Li, S.; Travadon, R.; Nouri, M.T.; Trouillas, F.P. Determining the Main Infection Courts in Sweet Cherry Trees of the Canker Pathogens Calosphaeria pulchella, Cytospora sorbicola and Eutypa lata. Plant Dis. 2024, 108, 1695–1702. [Google Scholar] [CrossRef]
- Pan, M.; Zhu, H.; Bonthond, G.; Tian, C.; Fan, X. High Diversity of Cytospora Associated With Canker and Dieback of Rosaceae in China, With 10 New Species Described. Front. Plant Sci. 2020, 11, 690. [Google Scholar] [CrossRef]
- Trouillas, F.P.; Peduto, F.; Lorber, J.D.; Sosnowski, M.R.; Grant, J.; Coates, W.W.; Anderson, K.K.; Caprile, J.; Gubler, W.D. Calosphaeria Canker of Sweet Cherry Caused by Calosphaeria pulchella in California and South Australia. Plant Dis. 2012, 96, 648–658. [Google Scholar] [CrossRef]
- iQonsulting. Sweet Cherry Book 2024; iQonsulting: Santiago, Chile, 2024; Available online: http://www.iqonsulting.com/yb/ (accessed on 6 August 2024).
- Oficina de Estudios y Políticas Agrarias (ODEPA). Informe Superficie Frutícola Regional 2024. Available online: https://www.odepa.gob.cl/estadisticas-del-sector/catastros-fruticolas (accessed on 6 August 2024).
- De Salvador, F.R.; Pititto, A.; Giorgioni, M.; Folini, L.; Bassi, G.; Longo, L. Performance of ‘Lapins’ sweet cherry on several rootstocks in Italy. Acta Hortic. 2008, 795, 311–316. [Google Scholar] [CrossRef]
- Csihon, Á.; Bicskei, K.; Dremák, P.; Gonda, I. Evaluation of the growing and fruit bearing characteristics of the ‘Lapins’ sweet cherry cultivar grafted on rootstocks with different vigor. Int. J. Hortic. Sci. 2017, 23, 15–18. [Google Scholar] [CrossRef]
- Marroni, M.V.; Casonato, S.; Pitman, A.R.; Visnovsky, S.B.; Beresford, R.M.; Jones, E.E. Review of Pseudomonas species causing bacterial canker of Prunus species with emphasis on sweet cherry (Prunus avium) in New Zealand. Eur. J. Plant Pathol. 2024, 168, 297–314. [Google Scholar] [CrossRef]
- Travadon, R.; Lawrence, D.P.; Li, S.; Trouillas, F.P. Evaluation of Biological Control Agents for the Protection of Almond Pruning Wounds Against Infection by Fungal Canker Pathogens. Phytopathology 2023, 113, 1417–1427. [Google Scholar] [CrossRef]
- Holland, L.A.; Travadon, R.; Lawrence, D.P.; Nouri, M.T.; Trouillas, F. Evaluation of Pruning Wound Protection Products for the Management of Almond Canker Diseases in California. Plant Dis. 2021, 105, 3368–3375. [Google Scholar] [CrossRef]
- Travadon, R.; Lawrence, D.P.; Li, S.; Trouillas, F.P. Field evaluation of biological wound treatments for the management of almond, cherry, and grapevine fungal canker diseases. Biol. Control. 2023, 185, 105292. [Google Scholar] [CrossRef]
- Li, S.; Travadon, R.; Trouillas, F.P. Effects of Temperature on Spore Germination and Mycelial Growth of Calosphaeria pulchella, Cytospora sorbicola, and Eutypa lata Isolates Associated with Sweet Cherry Canker Diseases. Plant Dis. 2023, 107, 3448–3456. [Google Scholar] [CrossRef]
- Avenot, H.F.; Jaime-Frias, R.; Travadon, R.; Holland, L.A.; Lawrence, D.P.; Trouillas, F.P. Development of PCR-Based Assays for Rapid and Reliable Detection and Identification of Canker-Causing Pathogens from Symptomatic Almond Trees. Phytopathology 2022, 112, 1710–1722. [Google Scholar] [CrossRef]
- Stewart, J.E.; Miller, S.T.; Zink, F.A.; Caballero, J.I.; Tembrock, L.R. Genetic and Phenotypic Characterization of the Fungal Pathogen Cytospora plurivora from Western Colorado Peach Orchards and the Development of a ddPCR Assay for Detection and Quantification. Phytopathology 2022, 112, 917–928. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.; Zhu, S.; Li, J.; Chang, Y.; Huang, L. Development and Application of a LAMP Assay for the Detection of the Latent Apple Tree Pathogen Valsa mali. Plant Dis. 2021, 105, 1065–1071. [Google Scholar] [CrossRef]
- Słomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. High Resolution Melting (HRM) for High-Throughput Genotyping—Limitations and Caveats in Practical Case Studies. Int. J. Mol. Sci. 2017, 18, 2316. [Google Scholar] [CrossRef]
- Tong, S.Y.; Giffard, P.M. Microbiological applications of high-resolution melting analysis. J. Clin. Microbiol. 2012, 50, 3418–3421. [Google Scholar] [CrossRef]
- Chatzidimopoulos, M.; Ganopoulos, I.; Moraitou-Daponta, E.; Lioliopoulou, F.; Ntantali, O.; Panagiotaki, P.; Vellios, E.K. High-Resolution Melting (HRM) Analysis Reveals Genotypic Differentiation of Venturia inaequalis Populations in Greece. Front. Ecol. Evol. 2019, 7, 489. [Google Scholar] [CrossRef]
- Ratti, M.F.; Farrer, R.A.; Cano, L.M.; Faedda, R.; Goss, E.M. Evaluation of High-Resolution Melting for Rapid Differentiation of Phytophthora Hybrids and Their Parental Species. Plant Dis. 2019, 103, 2295–2304. [Google Scholar] [CrossRef]
- Wang, N.Y.; Gama, A.B.; Marin, M.V.; Peres, N.A. Development of a Multiplex High-Throughput Diagnostic Assay for the Detection of Strawberry Crown Rot Diseases Using High-Resolution Melting Analysis. Phytopathology 2021, 111, 1470–1483. [Google Scholar] [CrossRef]
- Druml, B.; Cichna-Markl, M. High resolution melting (HRM) analysis of DNA—its role and potential in food analysis. Food Chem. 2014, 158, 245–254. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Series. 1999, 41, 95–98. [Google Scholar]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hanifeh, S.; Zafari, D.; Soleimani, M.J.; Arzanlou, M. Multigene phylogeny, morphology, and pathogenicity trials reveal novel Cytospora species involved in perennial canker disease of apple trees in Iran. Fungal Biol. 2022, 126, 707–726. [Google Scholar] [CrossRef]
- Grinbergs, D.; Chilian, J.; Isla, M.; Otarola, J. First report of Calosphaeria pulchella causing cankers in peach (Prunus persica) in Chile. Plant Dis. 2023, 107, 3314. [Google Scholar] [CrossRef]
- Untergasser, A.; Ruijter, J.M.; Benes, V.; van den Hoff, M.J.B. Web-based LinRegPCR: Application for the visualization and analysis of (RT)-qPCR amplification and melting data. BMC Bioinform. 2021, 22, 398. [Google Scholar] [CrossRef]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; González, L.; Tablada, M.; Robledo, C.W. InfoStat; InfoStat Group, FCA: London, UK; Universidad Nacional de Córdoba: Córdoba, Argentina, 2015. [Google Scholar]
- Takácsné Hájos, M.; Nyéki, J.; Veres, E.; Fieszl, C.; Szabó, Z. Organoleptic evaluation of sweet cherry varieties. Int. J. Hortic. Sci. 2011, 17, 7–13. [Google Scholar] [CrossRef]
- Vavoura, M.V.; Badeka, A.V.; Kontakos, S.; Kontominas, M.G. Characterization of four popular sweet cherry cultivars grown in Greece by volatile compound and physicochemical data analysis and sensory evaluation. Molecules 2015, 20, 1922–1940. [Google Scholar] [CrossRef]
- Measham, P.F.; Quentin, A.G.; MacNair, N. Climate, winter chill, and decision-making in sweet cherry production. HortScience 2014, 49, 254–259. [Google Scholar] [CrossRef]
- Mgbechi-Ezeri, J.; Porter, L.; Johnson, K.B.; Oraguzie, N. Assessment of sweet cherry (Prunus avium L.) genotypes for response to bacterial canker disease. Euphytica 2017, 213, 145. [Google Scholar] [CrossRef]
- Garrett, C.M.E. Screening Prunus rootstocks for resistance to bacterial canker, caused by Pseudomonas morsprunorum. J. Hortic. Sci. Biotechnol. 1979, 54, 189–193. [Google Scholar] [CrossRef]
- Beluzán, F.; Miarnau, X.; Torguet, L.; Zazurca, L.; Abad-Campos, P.; Luque, J.; Armengol, J. Susceptibility of Almond (Prunus dulcis) Cultivars to Twig Canker and Shoot Blight Caused by Diaporthe amygdali. Plant Dis. 2022, 106, 1890–1897. [Google Scholar] [CrossRef]
- Mohan, S.K.; Bijman, V.P. Susceptibility of Prunus Species to Erwinia amylovora. Acta Hortic. 1999, 489, 145–148. [Google Scholar] [CrossRef]
- Dodds, P.N.; Chen, J.; Outram, M.A. Pathogen perception and signaling in plant immunity. Plant cell 2024, 36, 1465–1481. [Google Scholar] [CrossRef]
- Trdá, L.; Boutrot, F.; Claverie, J.; Brulé, D.; Dorey, S.; Poinssot, B. Perception of pathogenic or beneficial bacteria and their evasion of host immunity: Pattern recognition receptors in the frontline. Front. Plant Sci. 2015, 6, 219. [Google Scholar] [CrossRef]
- Saur, I.M.L.; Hückelhoven, R. Recognition and defence of plant-infecting fungal pathogens. J. Plant Physiol. 2021, 256, 153324. [Google Scholar] [CrossRef]
- Correa, F.; Beltrán, M.F.; Millas, P.; Moreno, Z.; Hinrichsen, P.; Meza, P.; Sagredo, B. Genome Sequence Resources of Pseudomonas syringae Strains Isolated from Sweet Cherry Orchards in Chile. Mol. Plant Microbe Interact. 2022, 35, 933–937. [Google Scholar] [CrossRef]
- Beltrán, M.F.; Osorio, V.; Lemus, G.; Millas, P.; France, A.; Correa, F.; Sagredo, B. Bacterial community associated with canker disease from sweet cherry orchards of central valley of Chile presents high resistance to copper. Chil. J. Agric. Res. 2021, 81, 378–389. [Google Scholar] [CrossRef]
- Villalobos-González, L.; Carreras, C.; Beltrán, M.F.; Figueroa, F.; Rubilar-Hernández, C.; Opazo, I.; Toro, G.; Salvatierra, A.; Sagredo, B.; Pizarro, L.; et al. Sweet Cherry Plants Prioritize Their Response to Cope with Summer Drought, Overshadowing the Defense Response to Pseudomonas syringae pv. syringae. Plants 2024, 13, 1737. [Google Scholar] [CrossRef]
- Ririe, K.M.; Rasmussen, R.P.; Wittwer, C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997, 245, 154–160. [Google Scholar] [CrossRef]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef]
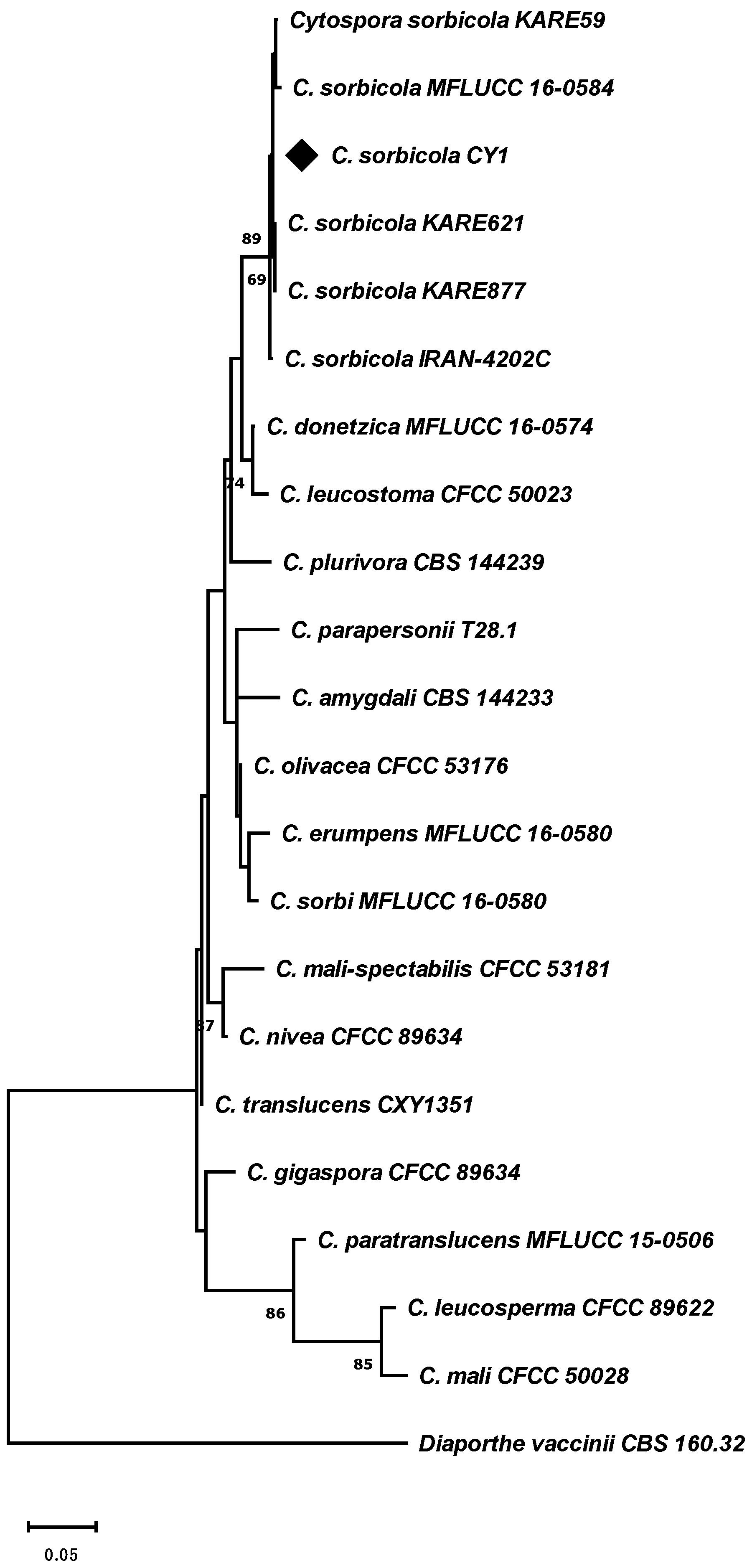


| Species | Strain | Host | Origen | GenBank Accession Number | ||
|---|---|---|---|---|---|---|
| ITS | ACT | TEF-1α | ||||
| Cytospora sorbicola | KARE59 | Prunus dulcis | California, USA | MG971861 | MG972010 | MG971572 |
| Cyt. sorbi | MFLUCC 16-0631T | Sorbus aucuparia | Rusia | KY417752 | KY417718 | NA |
| Cyt. sorbicola | KARE621 | Prunus avium | California, USA | MG971811 | MG971961 | MG971526 |
| Cyt. sorbicola | KARE877 | Prunus avium | California, USA | MG971825 | MG971975 | MG971540 |
| Cyt. sorbicola | IRAN 4202C | Malus domestica | Iran | MW295654 | MZ014514 | MW394148 |
| Cyt. donetzica | MFLUCC 16-0574T | Rosa sp. | Rusia | KY417731 | KY417696 | NA |
| Cyt. leucostoma | CFCC 50023 | Cornus alba | China | KR045635 | KU711003 | KU710926 |
| Cyt. plurivora | CBS 144239T | Olea europaea | California, USA | MG971861 | MG972010 | NA MG971572 |
| Cyt. parapersoonii | T28.1T | Prunus persica | USA | AF191181 | NA | NA |
| Cyt. amygdali | CBS 144233T | Prunus dulcis | California, USA | MG971853 | MG972002 | MG971659 |
| Cyt. olivacea | CFCC 53176T | Sorbus tianschanica | China | MK673068 | MK673038 | MK672955 |
| Cyt. erumpens | MFLUCC 16-0580T | Salix _ fragilis | Rusia | KY417733 | KY417699 | NA |
| Cyt. mali-spectabilis | CFCC 53181T | Malus spectabilis | China | MK673066 | MK673036 | MK672953 |
| Cyt. nivea | CFCC 89642T | Salix psammophila | China | KF765684 | KF765732 | NA |
| Cyt. translucens | CXY 1351 | Populus davidiana | China | KM034874 | NA | NA |
| Cyt. gigaspora | CFCC 89634T | Salix psammophila | China | KF765671 | KU711000 | KU710923 |
| Cyt. paratranslucens | MFLUCC 15-0506T | Populus alba var. bolleana | Rusia | KY417741 | KY417707 | NA |
| Cyt. leucosperma | CFCC 89622 | Pyrus bretschneideri | China | KR045616 | KU710988 | KU710911 |
| Cyt. mali | CFCC 50028 | Malus pumila | China | MH933641 | MH933548 | MH933513 |
| Diaporthe vaccinii | CBS 160.32 | Vaccinium macrocarpon | USA | KC343228 | JQ807297 | KC343954 |
| Locus | Primer Name | Primer Sequence (5′→3′) | Reference |
|---|---|---|---|
| Internal Transcribed Spacer | ITS1F | TCCGTAGGTGAACCTGCGG | [24] |
| ITS4 | TCCTCCGCTTATTGATATGC | [24] | |
| Actin | ACT512F | ATGTGCAAGGCCGGTTTCGC | [25] |
| ACT-783R | TACGAGTCCTTCTGGCCCAT | [25] | |
| Translation elongation factor 1-alpha | EF1-728 | CATCGAGAAGTTCGAGAAGG | [25] |
| EF1-986 | TACTTGAAGGAACCCTTACC | [25] | |
| Internal Transcribed Spacer | Calo-Cytos_For(ITS) | GGGTTGAAATGACGCTCG | This study |
| Calos-Cytos_Rev(ITS) | CTGGCATCGATGAAGAACG | This study | |
| Calo-Cytos_For-univ1(ITS) | CATTTCGCTRCGTTCTTCATC | This study | |
| Cyto_Rev-esp(ITS) | GGCCCACTAAACTCTTGTATT | This study | |
| Calos _Rev-esp(ITS) | GTCCTCTGAGCCAACTGAAA | This study |
| Cyt. sorbicola | Cal. pulchella | |||||
|---|---|---|---|---|---|---|
| 4 Days (%) | 7 Days (%) | Δ 1 (%) | 4 Days (%) | 7 Days (%) | Δ 1 (%) | |
| Santina | 14.8 | 18.5 | 20.0 | 50.9 | 63.2 | 19.5 |
| Lapins | 33.3 | 33.3 | 0.0 | 87.9 | 89.7 | 2.0 |
| Bing | 35.0 | 36.7 | 4.6 | 79.1 | 79.1 | 0.0 |
| Regina | 18.3 | 18.3 | 0.0 | 70.0 | 72.5 | 3.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osorio-Navarro, C.; Saez, C.; Durán, F.; Rubilar, M.; Reyes-Bravo, P.; Azócar, M.; Estrada, V.; Esterio, M.; Auger, J. The Sweet Cherry Tree Genotype Restricts the Aggressiveness of the Wood Decay Fungi Cytospora sorbicola and Calosphaeria pulchella. Microorganisms 2024, 12, 2456. https://doi.org/10.3390/microorganisms12122456
Osorio-Navarro C, Saez C, Durán F, Rubilar M, Reyes-Bravo P, Azócar M, Estrada V, Esterio M, Auger J. The Sweet Cherry Tree Genotype Restricts the Aggressiveness of the Wood Decay Fungi Cytospora sorbicola and Calosphaeria pulchella. Microorganisms. 2024; 12(12):2456. https://doi.org/10.3390/microorganisms12122456
Chicago/Turabian StyleOsorio-Navarro, Claudio, Constanza Saez, Felipe Durán, Mauricio Rubilar, Paula Reyes-Bravo, Madelaine Azócar, Verónica Estrada, Marcela Esterio, and Jaime Auger. 2024. "The Sweet Cherry Tree Genotype Restricts the Aggressiveness of the Wood Decay Fungi Cytospora sorbicola and Calosphaeria pulchella" Microorganisms 12, no. 12: 2456. https://doi.org/10.3390/microorganisms12122456
APA StyleOsorio-Navarro, C., Saez, C., Durán, F., Rubilar, M., Reyes-Bravo, P., Azócar, M., Estrada, V., Esterio, M., & Auger, J. (2024). The Sweet Cherry Tree Genotype Restricts the Aggressiveness of the Wood Decay Fungi Cytospora sorbicola and Calosphaeria pulchella. Microorganisms, 12(12), 2456. https://doi.org/10.3390/microorganisms12122456






