Assessing Normandy Soil Microbial Diversity for Antibacterial Activities Using Traditional Culture and iChip Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sampling
2.2. Cultivation Methods
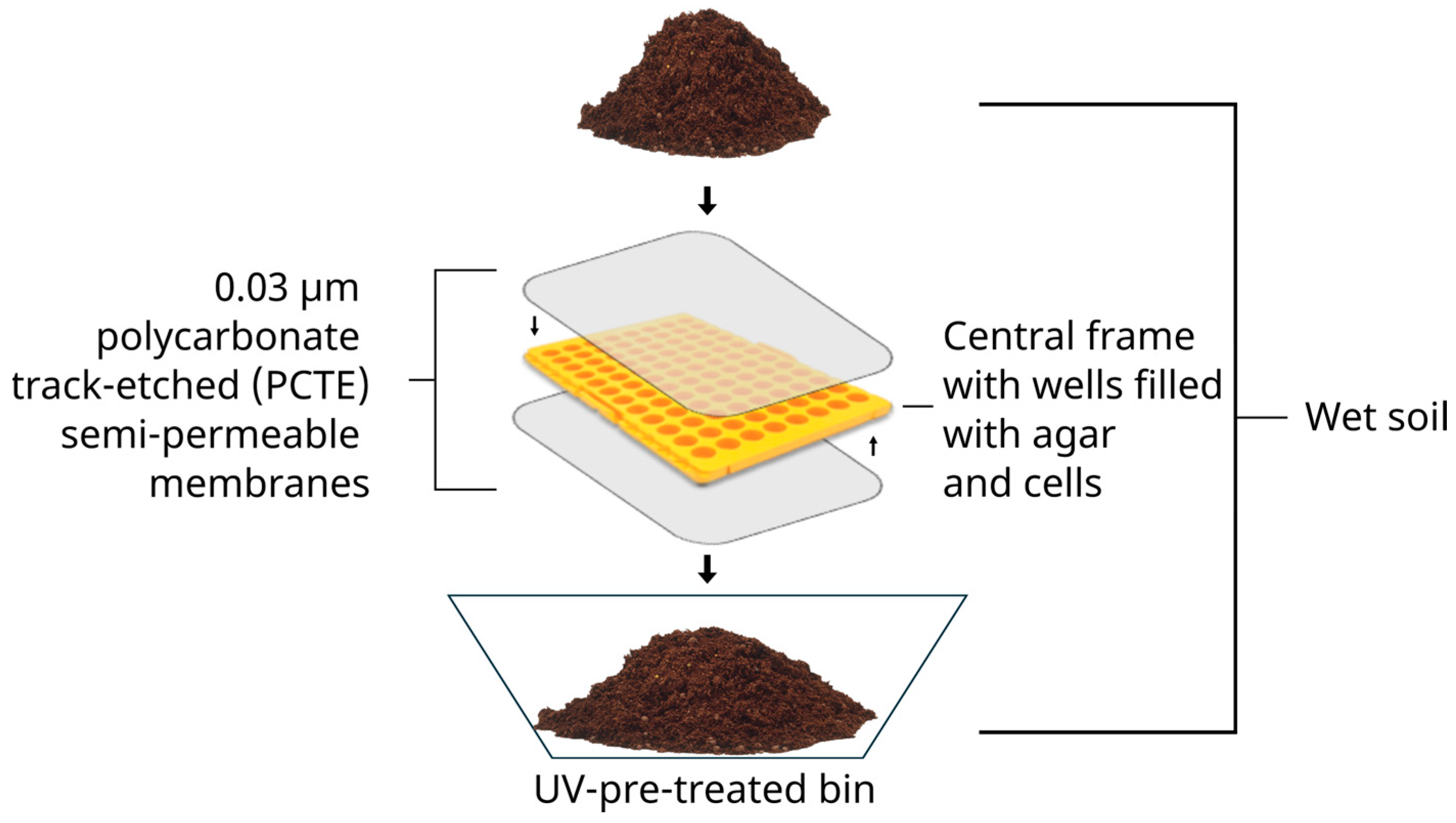
2.2.1. Isolation Chips
2.2.2. Dilution and Direct Plating (DDP)
2.3. Microbial Identification and Phylogenetic Analysis
2.3.1. MALDI-TOF Mass Spectrometry Identification
2.3.2. Molecular Identification
2.4. Indicator Strains and Culture Conditions
2.5. Antibacterial Screening
2.5.1. Deferred Antagonism Assays
2.5.2. Liquid Fermentation and Agar-Well Diffusion Assays
3. Results
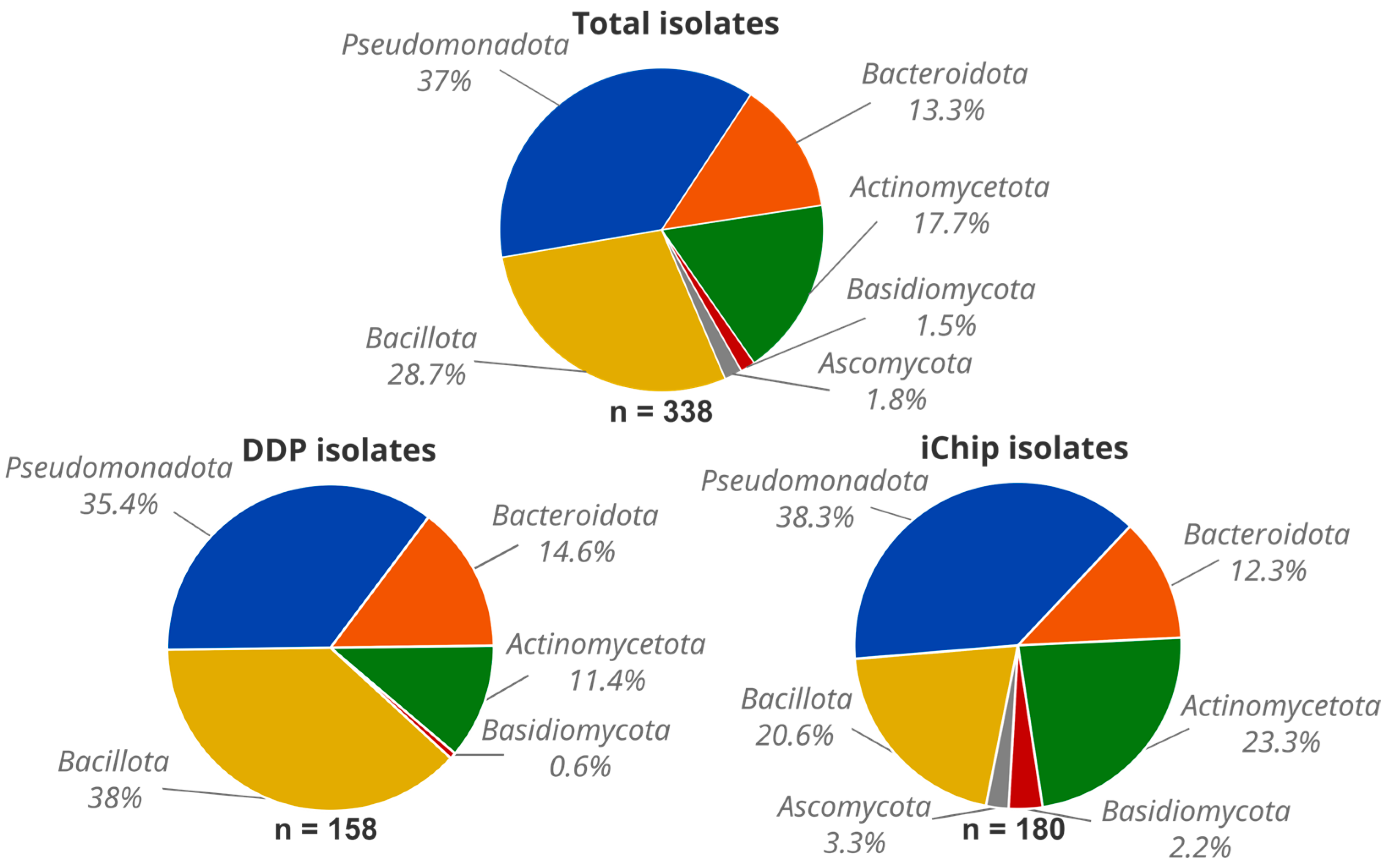
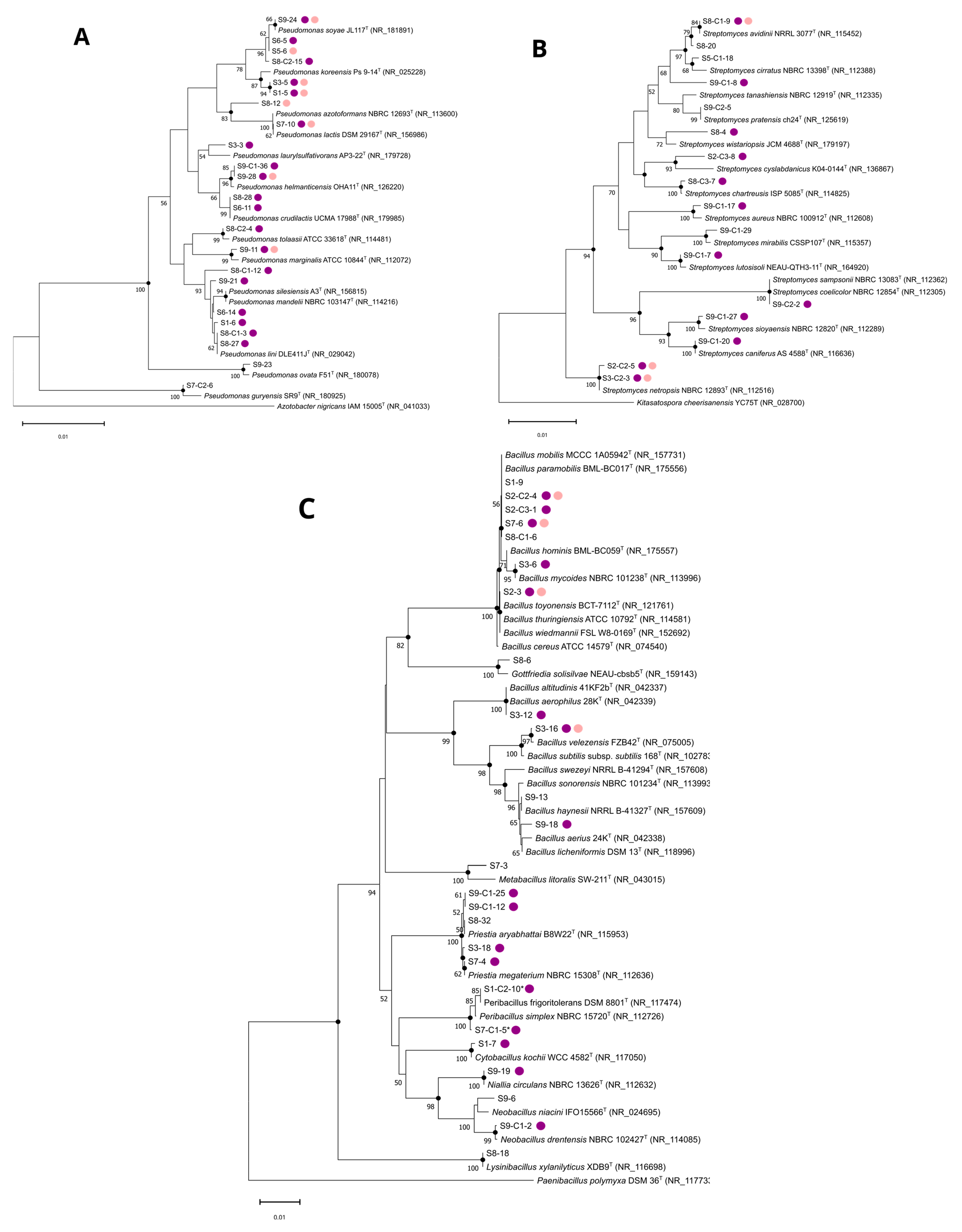
3.1. Microbial Diversity from Normandy Soils
3.2. Antibacterial Activity Screening
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- GBD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Report Government of The United Kingdom; Wellcome Trust: London, UK, 2016. [Google Scholar]
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- World Health Organization. 2021 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- World Health Organization. 2023 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Demain, A.L. Importance of Microbial Natural Products and the Need to Revitalize Their Discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef]
- Stennett, H.L.; Back, C.R.; Race, P.R. Derivation of a Precise and Consistent Timeline for Antibiotic Development. Antibiotics 2022, 11, 1237. [Google Scholar] [CrossRef]
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef]
- Ehlert, F.; Neu, H.C. In Vitro Activity of LY146032 (Daptomycin), a New Peptolide. Eur. J. Clin. Microbiol. 1987, 6, 84–90. [Google Scholar] [CrossRef]
- Lewis, K. Recover the lost art of drug discovery. Nature 2012, 485, 439–440. [Google Scholar] [CrossRef]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome Mining of Biosynthetic and Chemotherapeutic Gene Clusters in Streptomyces Bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into Secondary Metabolism from a Global Analysis of Prokaryotic Biosynthetic Gene Clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef]
- Staley, J.T.; Konopka, A. Measurement of In Situ Activities of Nonphotosynthetic Microorganisms in Aquatic and Terrestrial Habitats. Annu. Rev. Microbiol. 1985, 39, 321–346. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.-H. Phylogenetic Identification and In Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems 2018, 3, e00055-18. [Google Scholar] [CrossRef] [PubMed]
- Lewis, W.H.; Tahon, G.; Geesink, P.; Sousa, D.Z.; Ettema, T.J.G. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 2021, 19, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef]
- Schultz, J.; Modolon, F.; Rosado, A.S.; Voolstra, C.R.; Sweet, M.; Peixoto, R.S. Methods and Strategies to Uncover Coral-Associated Microbial Dark Matter. mSystems 2022, 7, e00367-22. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of Ichip for High-Throughput In Situ Cultivation of “Uncultivable” Microbial Species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A New Antibiotic Kills Pathogens without Detectable Resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Shukla, R.; Peoples, A.J.; Ludwig, K.C.; Maity, S.; Derks, M.G.N.; de Benedetti, S.; Krueger, A.M.; Vermeulen, B.J.A.; Harbig, T.; Lavore, F.; et al. A New Antibiotic from an Uncultured Bacterium Binds to an Immutable Target. Cell 2023, 186, 4059–4073.e27. [Google Scholar] [CrossRef]
- Berdy, B.; Spoering, A.L.; Ling, L.L.; Epstein, S.S. In situ cultivation of previously uncultivable microorganisms using the ichip. Nat. Protoc. 2017, 12, 2232–2242. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Fitch, W.M. Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Connon, S.A.; Giovannoni, S.J. High-Throughput Methods for Culturing Microorganisms in Very-Low-Nutrient Media Yield Diverse New Marine Isolates. Appl. Environ. Microbiol. 2002, 68, 3878–3885. [Google Scholar] [CrossRef]
- Rappé, M.S.; Connon, S.A.; Vergin, K.L.; Giovannoni, S.J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 2002, 418, 630–633. [Google Scholar] [CrossRef]
- Zengler, K.; Toledo, G.; Rappé, M.; Elkins, J.; Mathur, E.J.; Short, J.M.; Keller, M. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 2002, 99, 15681–15686. [Google Scholar] [CrossRef]
- Choi, E.J.; Nam, S.-J.; Paul, L.; Beatty, D.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Previously Uncultured Marine Bacteria Linked to Novel Alkaloid Production. Chem. Biol. 2015, 22, 1270–1279. [Google Scholar] [CrossRef]
- Janssen, P.H.; Yates, P.S.; Grinton, B.E.; Taylor, P.M.; Sait, M. Improved Culturability of Soil Bacteria and Isolation in Pure Culture of Novel Members of the Divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 2002, 68, 2391–2396. [Google Scholar] [CrossRef]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “Uncultivable” Microorganisms in Pure Culture in a Simulated Natural Environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Quigley, J.; Peoples, A.; Sarybaeva, A.; Hughes, D.; Ghiglieri, M.; Achorn, C.; Desrosiers, A.; Felix, C.; Liang, L.; Malveira, S.; et al. Novel Antimicrobials from Uncultured Bacteria Acting against Mycobacterium tuberculosis. mBio 2020, 11, e01516-20. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, D.A.; Ludwig, K.C.; Arts, M.; Marx, C.E.; Krannich, S.; Barac, P.; Kehraus, S.; Josten, M.; Henrichfreise, B.; Müller, A.; et al. Biosynthesis and Mechanism of Action of the Cell Wall Targeting Antibiotic Hypeptin. Angew. Chem. Int. Ed. 2021, 60, 13579–13586. [Google Scholar] [CrossRef] [PubMed]
- Polrot, A.; Kirby, J.R.; Olorunniji, F.J.; Birkett, J.W.; Sharples, G.P. iChip increases the success of cultivation of TBT-resistant and TBT-degrading bacteria from estuarine sediment. World J. Microbiol. Biotechnol. 2022, 38, 180. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.J.; Hugenholtz, P.; Sangwan, P.; Osborne, C.A.; Janssen, P.H. Laboratory Cultivation of Widespread and Previously Uncultured Soil Bacteria. Appl. Environ. Microbiol. 2003, 69, 7210–7215. [Google Scholar] [CrossRef] [PubMed]
- Furlong, M.A.; Singleton, D.R.; Coleman, D.C.; Whitman, W.B. Molecular and Culture-Based Analyses of Prokaryotic Communities from an Agricultural Soil and the Burrows and Casts of the Earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 2002, 68, 1265–1279. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Khulan, A.; Kim, J. Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 2019, 9, 6666. [Google Scholar] [CrossRef]
- Bahram, M.; Netherway, T.; Frioux, C.; Ferretti, P.; Coelho, L.P.; Geisen, S.; Bork, P.; Hildebrand, F. Metagenomic assessment of the global diversity and distribution of bacteria and fungi. Environ. Microbiol. 2021, 23, 316–326. [Google Scholar] [CrossRef]
- Wei, H.; Peng, C.; Yang, B.; Song, H.; Li, Q.; Jiang, L.; Wei, G.; Wang, K.; Wang, H.; Liu, S.; et al. Contrasting Soil Bacterial Community, Diversity, and Function in Two Forests in China. Front. Microbiol. 2018, 9, 1693. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- dos Santos, J.D.N.; João, S.A.; Martín, J.; Vicente, F.; Reyes, F.; Lage, O.M. iChip-Inspired Isolation, Bioactivities and Dereplication of Actinomycetota from Portuguese Beach Sediments. Microorganisms 2022, 10, 1471. [Google Scholar] [CrossRef]
- Bernardet, J.-F.; Bowman, J.P. The Genus Flavobacterium. In The Prokaryotes: Volume 7: Proteobacteria: Delta, Epsilon Subclass; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 481–531. ISBN 978-0-387-30747-3. [Google Scholar]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, M.; Tazato, N.; Handa, Y.; Umekawa, N.; Kigawa, R.; Sano, C.; Sugiyama, J. Krasilnikoviella muralis gen. nov., sp. nov., a Member of the Family Promicromonosporaceae, Isolated from the Takamatsuzuka Tumulus Stone Chamber Interior and Reclassification of Promicromonospora flava as Krasilnikoviella flava comb. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Shakir, Y.; Deng, Y.; Zhang, Y. Use of modified ichip for the cultivation of thermo-tolerant microorganisms from the hot spring. BMC Microbiol. 2023, 23, 56. [Google Scholar] [CrossRef]
- Bollmann, A.; Lewis, K.; Epstein, S.S. Incubation of Environmental Samples in a Diffusion Chamber Increases the Diversity of Recovered Isolates. Appl. Environ. Microbiol. 2007, 73, 6386–6390. [Google Scholar] [CrossRef]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An Updated Compendium of Streptomycetes Natural Products. Nucleic Acids Res. 2020, 49, D600–D604. [Google Scholar] [CrossRef] [PubMed]
- Culp, E.J.; Waglechner, N.; Wang, W.; Fiebig-Comyn, A.A.; Hsu, Y.-P.; Koteva, K.; Sychantha, D.; Coombes, B.K.; Van Nieuwenhze, M.S.; Brun, Y.V.; et al. Evolution-Guided Discovery of Antibiotics That Inhibit Peptidoglycan Remodelling. Nature 2020, 578, 582–587. [Google Scholar] [CrossRef]
- Lacey, H.J.; Rutledge, P.J. Recently Discovered Secondary Metabolites from Streptomyces Species. Molecules 2022, 27, 887. [Google Scholar] [CrossRef]
- Lafuente, I.; Sevillano, E.; Peña, N.; Cuartero, A.; Hernández, P.E.; Cintas, L.M.; Muñoz-Atienza, E.; Borrero, J. Production of Pumilarin and a Novel Circular Bacteriocin, Altitudin A, by Bacillus altitudinis ECC22, a Soil-Derived Bacteriocin Producer. Int. J. Mol. Sci. 2024, 25, 2020. [Google Scholar] [CrossRef]
- Shen, Y.; Sun, T.; Jiang, S.; Mu, S.; Li, D.; Guo, X.; Zhang, J.; Zhao, J.; Xiang, W. Streptomyces lutosisoli sp. nov., a Novel Actinomycete Isolated from Muddy Soil. Antonie Van Leeuwenhoek 2018, 111, 2403–2412. [Google Scholar] [CrossRef]
- Ramlawi, S.; Abusharkh, S.; Carroll, A.; McMullin, D.R.; Avis, T.J. Biological and Chemical Characterization of Antimicrobial Activity in Arthrobacter spp. Isolated from Disease-Suppressive Compost. J. Basic Microbiol. 2021, 61, 745–756. [Google Scholar] [CrossRef]
- Cao, Y.; Shen, Z.; Zhang, N.; Deng, X.; Thomashow, L.S.; Lidbury, I.; Liu, H.; Li, R.; Shen, Q.; Kowalchuk, G.A. Phosphorus availability influences disease-suppressive soil microbiome through plant-microbe interactions. Microbiome 2024, 12, 185. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, J.-Y.; Guo, Y.-B.; Wang, J.-H.; Wang, H.-M. Biological Control of Grapevine Crown Gall: Purification and Partial Characterisation of an Antibacterial Substance Produced by Rahnella aquatilis Strain HX2. Eur. J. Plant Pathol. 2009, 124, 427–437. [Google Scholar] [CrossRef]
- Tejman-Yarden, N.; Robinson, A.; Davidov, Y.; Shulman, A.; Varvak, A.; Reyes, F.; Rahav, G.; Nissan, I. Delftibactin-A, a Non-ribosomal Peptide with Broad Antimicrobial Activity. Front. Microbiol. 2019, 10, 2377. [Google Scholar] [CrossRef]
- Hayward, A.C.; Fegan, N.; Fegan, M.; Stirling, G.R. Stenotrophomonas and Lysobacter: Ubiquitous Plant-associated Gamma-proteobacteria of Developing Significance in Applied Microbiology. J. Appl. Microbiol. 2010, 108, 756–770. [Google Scholar] [CrossRef]
- Dahal, R.H.; Chaudhary, D.K.; Kim, D.-U.; Pandey, R.P.; Kim, J. Chryseobacterium antibioticum sp. nov. with Antimicrobial Activity against Gram-Negative Bacteria, Isolated from Arctic Soil. J. Antibiot. 2021, 74, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Tavarideh, F.; Pourahmad, F.; Nemati, M. Diversity and Antibacterial Activity of Endophytic Bacteria Associated with Medicinal Plant, Scrophularia striata. Vet. Res. Forum 2022, 13, 409–415. [Google Scholar] [CrossRef]
- Chhetri, G.; Kim, I.; Kim, J.; So, Y.; Seo, T. Chryseobacterium tagetis sp. nov., a Plant Growth Promoting Bacterium with an Antimicrobial Activity Isolated from the Roots of Medicinal Plant (Tagetes patula). J. Antibiot. 2022, 75, 312–320. [Google Scholar] [CrossRef]
- Gavriilidou, A.; Gutleben, J.; Versluis, D.; Forgiarini, F.; van Passel, M.W.J.; Ingham, C.J.; Smidt, H.; Sipkema, D. Comparative Genomic Analysis of Flavobacteriaceae: Insights into Carbohydrate Metabolism, Gliding Motility and Secondary Metabolite Biosynthesis. BMC Genom. 2020, 21, 569. [Google Scholar] [CrossRef]
- Silva, S.G.; Homsi, M.N.; Keller-Costa, T.; Rocha, U.; Costa, R. Natural product biosynthetic potential reflects macroevolutionary diversification within a widely distributed bacterial taxon. mSystems 2023, 8, e00643-23. [Google Scholar] [CrossRef]
- Liu, S.-W.; Zhai, X.-X.; Liu, D.; Liu, Y.-Y.; Sui, L.-Y.; Luo, K.-K.; Yang, Q.; Li, F.-N.; Nikandrova, A.A.; Imamutdinova, A.N.; et al. Bioprospecting of Actinobacterial Diversity and Antibacterial Secondary Metabolites from the Sediments of Four Saline Lakes on the Northern Tibetan Plateau. Microorganisms 2023, 11, 2475. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. USA 2014, 111, 7266–7271. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, Y.; Zhang, C.; Davis, K.M.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol. 2019, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Seyedsayamdost, M.R. Discovery of a Cryptic Depsipeptide from Streptomyces ghanaensis via MALDI-MS-Guided High-Throughput Elicitor Screening. Angew. Chem. Int. Ed. 2020, 59, 23005–23009. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Seyedsayamdost, M.R. Induction of Diverse Cryptic Fungal Metabolites by Steroids and Channel Blockers. Angew. Chem. Int. Ed. 2022, 61, e202204519. [Google Scholar] [CrossRef]
- Moon, K.; Xu, F.; Zhang, C.; Seyedsayamdost, M.R. Bioactivity-HiTES Unveils Cryptic Antibiotics Encoded in Actinomycete Bacteria. ACS Chem. Biol. 2019, 14, 767–774. [Google Scholar] [CrossRef]
- Moon, K.; Xu, F.; Seyedsayamdost, M.R. Cebulantin, a Cryptic Lanthipeptide Antibiotic Uncovered Using Bioactivity-Coupled HiTES. Angew. Chem. Int. Ed. 2019, 58, 5973–5977. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perrier, F.; Morice, J.; Gueulle, S.; Géry, A.; Riboulet-Bisson, E.; Garon, D.; Muller, C.; Desriac, F. Assessing Normandy Soil Microbial Diversity for Antibacterial Activities Using Traditional Culture and iChip Methods. Microorganisms 2024, 12, 2422. https://doi.org/10.3390/microorganisms12122422
Perrier F, Morice J, Gueulle S, Géry A, Riboulet-Bisson E, Garon D, Muller C, Desriac F. Assessing Normandy Soil Microbial Diversity for Antibacterial Activities Using Traditional Culture and iChip Methods. Microorganisms. 2024; 12(12):2422. https://doi.org/10.3390/microorganisms12122422
Chicago/Turabian StylePerrier, Fabien, Juliette Morice, Sabrina Gueulle, Antoine Géry, Eliette Riboulet-Bisson, David Garon, Cécile Muller, and Florie Desriac. 2024. "Assessing Normandy Soil Microbial Diversity for Antibacterial Activities Using Traditional Culture and iChip Methods" Microorganisms 12, no. 12: 2422. https://doi.org/10.3390/microorganisms12122422
APA StylePerrier, F., Morice, J., Gueulle, S., Géry, A., Riboulet-Bisson, E., Garon, D., Muller, C., & Desriac, F. (2024). Assessing Normandy Soil Microbial Diversity for Antibacterial Activities Using Traditional Culture and iChip Methods. Microorganisms, 12(12), 2422. https://doi.org/10.3390/microorganisms12122422





