Genomic and Transcriptomic Analyses Identify Two Key Glycosyltransferase Genes alhH and alhK of Exopolysaccharide Biosynthesis in Pantoea alhagi NX-11
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids and Media
2.2. Whole-Genome Sequencing and Gene Function Annotation
2.3. Transcriptome Sequencing
2.4. Quantitative Real-Time PCR Validation
2.5. Construction of Overexpression Strains
2.6. Construction of Knockout Strains
2.7. Construction of alhH and alhK Gene Complement Strain
2.8. Analysis of Yield, Molecular Weight and Monosaccharide Components of EPS
2.9. Data Analysis
3. Results and Discussion
3.1. Genome Characteristics and Functional Annotation of P. alhagi NX-11
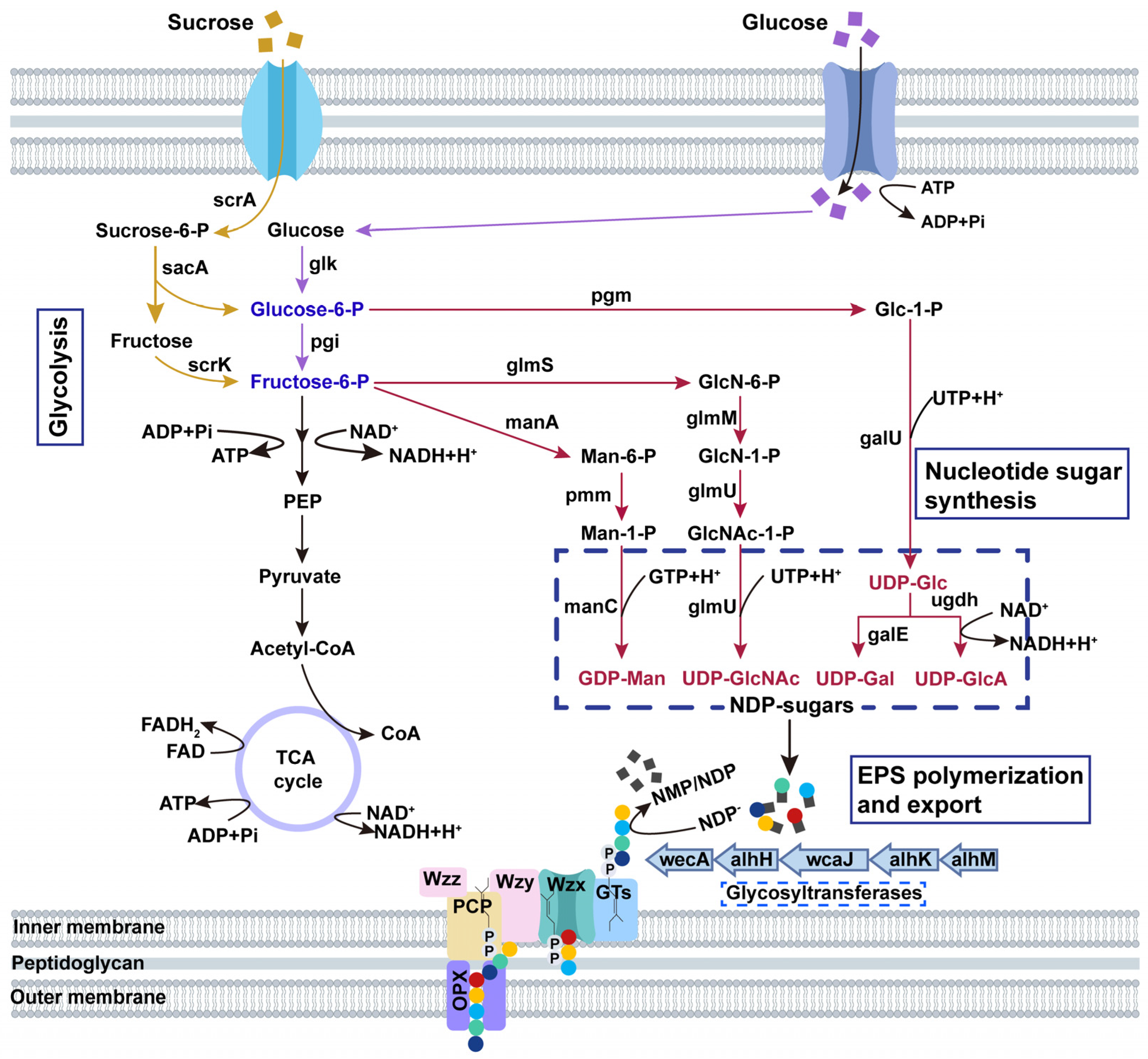
3.2. Alhagan Synthesis Gene Cluster of P. alhagi NX-11
3.3. Alhagan Biosynthesis Pathway of P. alhagi NX-11
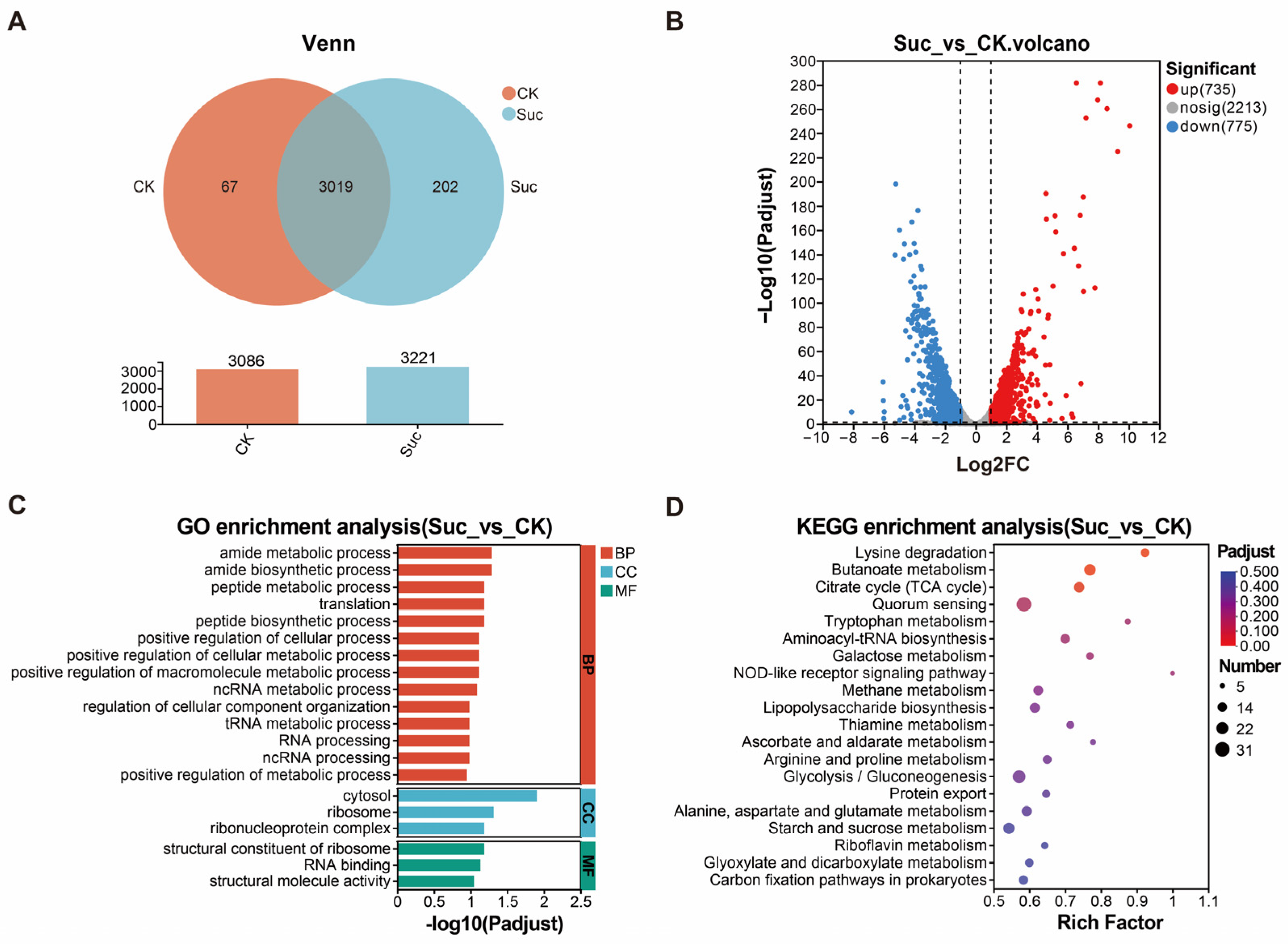
3.4. Transcriptome Sequencing and Differential Gene Enrichment Analysis
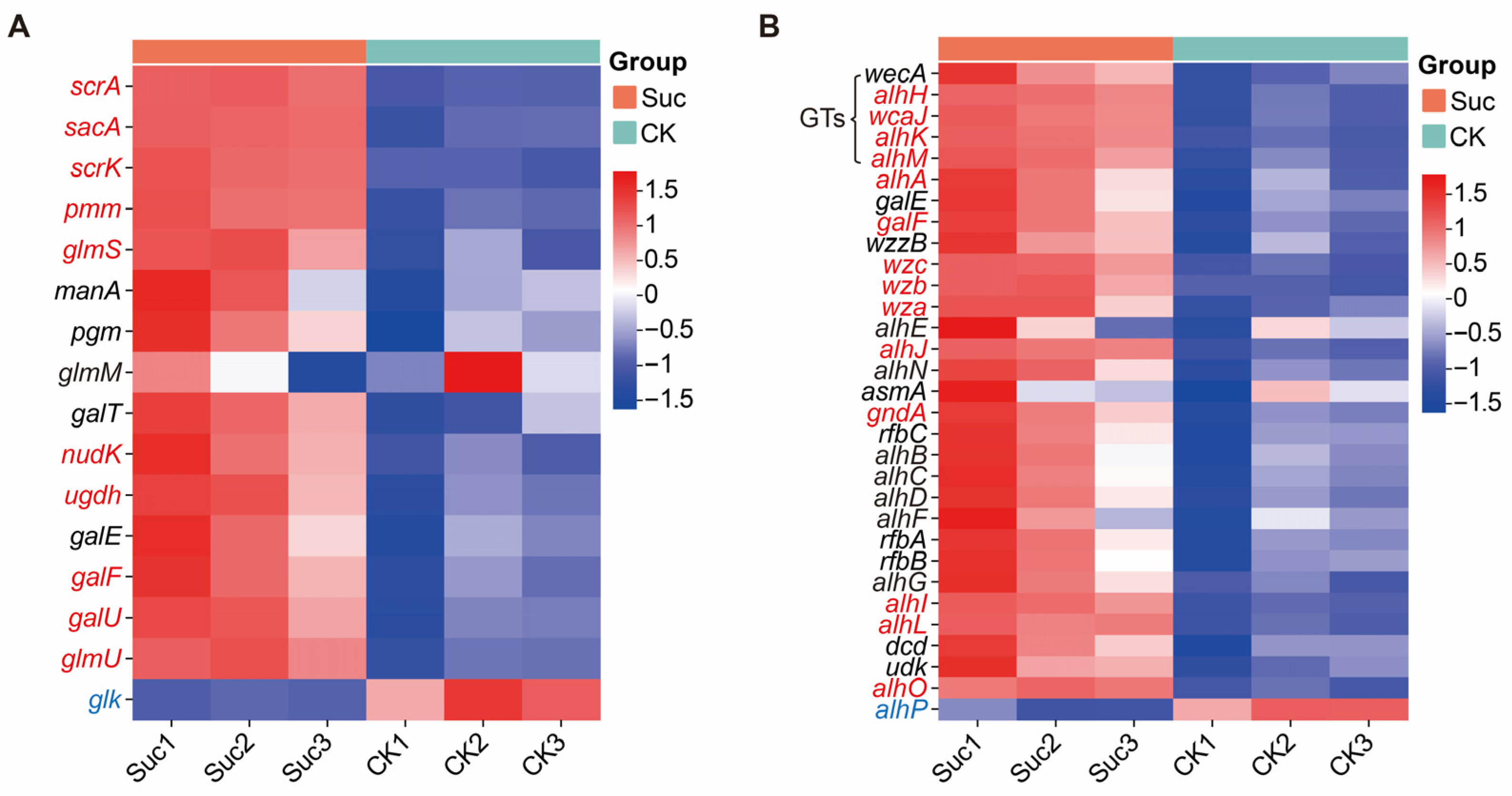
3.5. Analysis of Alhagan Biosynthesis Genes and qPCR Verification
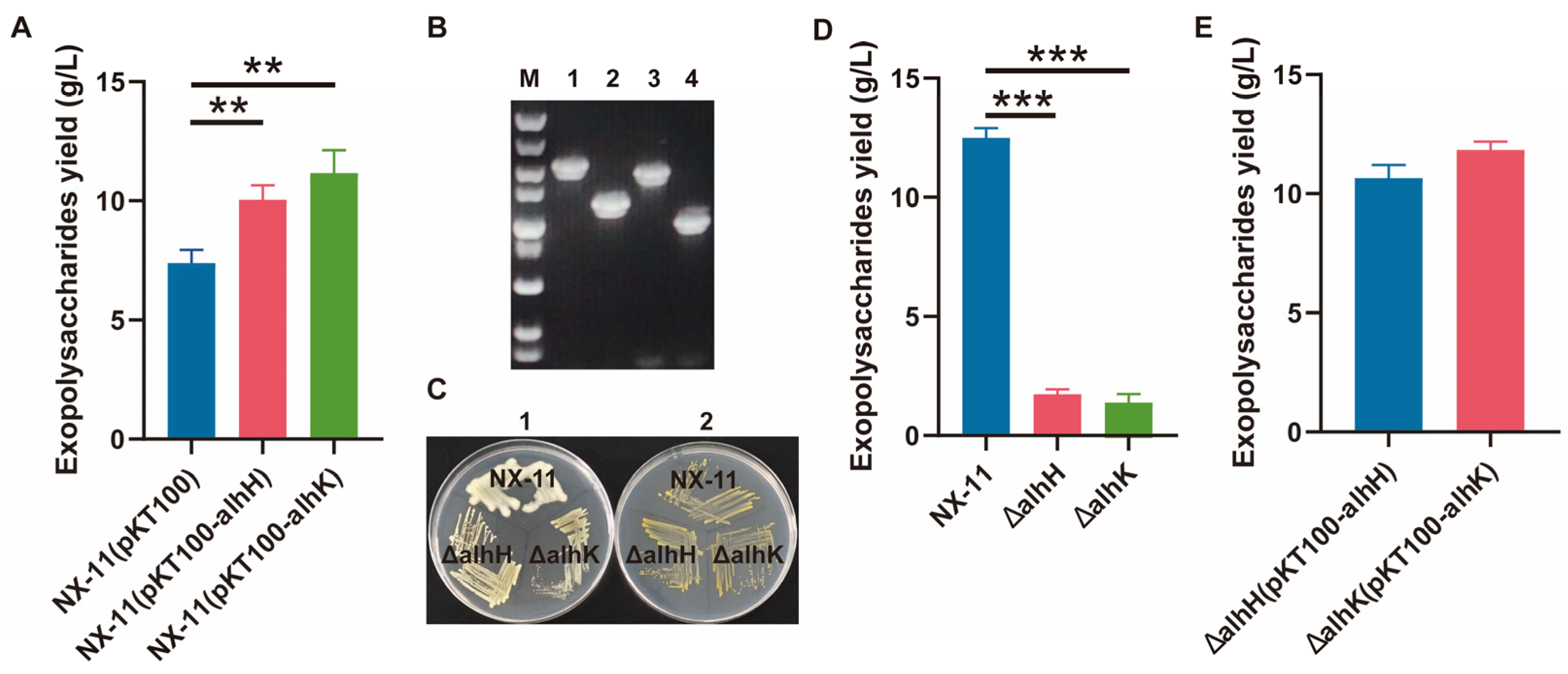
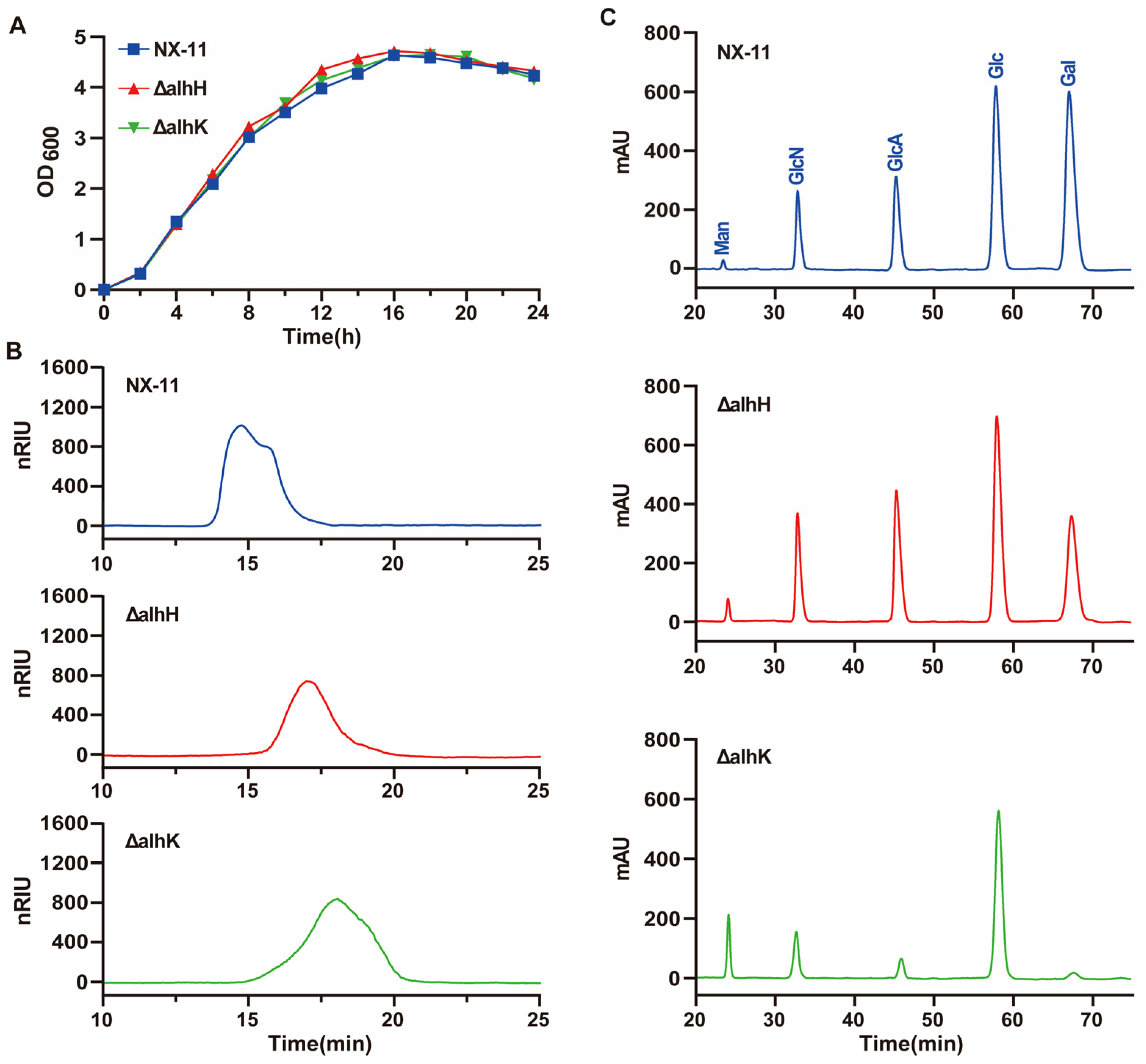
3.6. Characterization of the GT Genes alhH and alhK
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Jiang, F.; Liu, M.; Qu, Y.; Lan, Z.; Dai, X.; Huang, C.; Yue, X.; Zhao, S.; Pan, X. Synthesis, characterization, and bioactivities of polysaccharide metal complexes: A review. J. Agric. Food Chem. 2022, 70, 6922–6942. [Google Scholar] [CrossRef]
- Huo, J.; Wu, Z.; Sun, W.; Wang, Z.; Wu, J.; Huang, M.; Wang, B.; Sun, B. Protective effects of natural polysaccharides on intestinal barrier injury: A review. J. Agric. Food Chem. 2022, 70, 711–735. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, J.; Guo, Z.; Zheng, Y.; Rea, M.C.; Su, H.; Zheng, X.; Zheng, B.; Miao, S. Using polysaccharides for the enhancement of functionality of foods: A review. Trends Food Sci. Technol. 2019, 86, 311–327. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, K.; Zhang, M.; Xu, T.; Du, H.; Pang, B.; Si, C. Sustainable polysaccharide-based materials for intelligent packaging. Carbohydr. Polym. 2023, 313, 120851. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Xing, R.; Liu, S.; Li, P. Chitin and chitosan fragments responsible for plant elicitor and growth stimulator. J. Agric. Food Chem. 2020, 68, 12203–12211. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, R.S. Natural polysaccharides and their interactions with dye molecules: Applications in effluent treatment. Environ. Sci. Technol. 2004, 38, 4905–4909. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J.; Chang, S.; Chung, W. Bacterial biosorbents, an efficient heavy metals green clean-up strategy: Prospects, challenges, and opportunities. Microorganisms 2022, 10, 610. [Google Scholar] [CrossRef]
- Netrusov, A.I.; Liyaskina, E.V.; Kurgaeva, I.V.; Liyaskina, A.U.; Yang, G.; Revin, V.V. Exopolysaccharides producing Bacteria: A review. Microorganisms 2023, 11, 1541. [Google Scholar] [CrossRef]
- Schmid, J.; Sieber, V.; Rehm, B. Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Front. Microbiol. 2015, 6, 147519. [Google Scholar] [CrossRef]
- Schmid, J. Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies. Curr. Opin. Biotechnol. 2018, 53, 130–136. [Google Scholar] [CrossRef]
- Li, H.; Feng, Z.-M.; Sun, Y.-J.; Zhou, W.-L.; Jiao, X.; Zhu, H. Draft genome sequence of Sphingomonas sp. WG, a welan gum-producing strain. Genome Announc. 2016, 4, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Wear, S.S.; Sande, C. Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu. Rev. Microbiol. 2020, 74, 521–543. [Google Scholar] [CrossRef] [PubMed]
- Stephens, Z.; Wilson, L.F.; Zimmer, J. Diverse mechanisms of polysaccharide biosynthesis, assembly and secretion across kingdoms. Curr. Opin. Struct. Biol. 2023, 79, 102564. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Upadhyay, L.S.B. Microbial exopolysaccharides: Synthesis pathways, types and their commercial applications. Int. J. Biol. Macromol. 2020, 157, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, Y.; Lei, P.; Li, S.; Xu, H.; Wang, R.; Qiu, Y.; Zhang, W. Structure characterization, antioxidant and emulsifying capacities of exopolysaccharide derived from Pantoea alhagi NX-11. Carbohydr. Polym. 2021, 261, 117872. [Google Scholar] [CrossRef]
- Sun, L.; Cheng, L.; Ma, Y.; Lei, P.; Wang, R.; Gu, Y.; Li, S.; Zhang, F.; Xu, H. Exopolysaccharides from Pantoea alhagi NX-11 specifically improve its root colonization and rice salt resistance. Int. J. Biol. Macromol. 2022, 209, 396–404. [Google Scholar] [CrossRef]
- Wu, S.; Zheng, R.; Sha, Z.; Sun, C. Genome sequence of Pseudomonas stutzeri 273 and identification of the exopolysaccharide EPS273 biosynthesis locus. Mar. Drugs 2017, 15, 218. [Google Scholar] [CrossRef]
- Padmanabhan, A.; Tong, Y.; Wu, Q.; Zhang, J.; Shah, N.P. Transcriptomic insights into the growth phase-and sugar-associated changes in the exopolysaccharide production of a high EPS-producing Streptococcus thermophilus ASCC 1275. Front. Microbiol. 2018, 9, 1919. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef]
- Reiner, J.; Pisani, L.; Qiao, W.; Singh, R.; Yang, Y.; Shi, L.; Khan, W.A.; Sebra, R.; Cohen, N.; Babu, A. Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet–Biedl Syndrome 9 (BBS9) deletion. NPJ Genom. Med. 2018, 3, 3. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, L.; Cai, L. Transcriptome sequencing: RNA-seq. In Computational Systems Biology: Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 15–27. [Google Scholar]
- Zhang, L.; Li, M.; Li, Q.; Chen, C.; Qu, M.; Li, M.; Wang, Y.; Shen, X. The catabolite repressor/activator Cra is a bridge connecting carbon metabolism and host colonization in the plant drought resistance-promoting bacterium Pantoea alhagi LTYR-11Z. Appl. Environ. Microbiol. 2018, 84, e00054-18. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef]
- Sun, L.; Lei, P.; Wang, Q.; Ma, J.; Zhan, Y.; Jiang, K.; Xu, Z.; Xu, H. The endophyte Pantoea alhagi NX-11 alleviates salt stress damage to rice seedlings by secreting exopolysaccharides. Front. Microbiol. 2020, 10, 3112. [Google Scholar] [CrossRef] [PubMed]
- Cheong, K.-L.; Wu, D.-T.; Zhao, J.; Li, S.-P. A rapid and accurate method for the quantitative estimation of natural polysaccharides and their fractions using high performance size exclusion chromatography coupled with multi-angle laser light scattering and refractive index detector. J. Chromatogr. A 2015, 1400, 98–106. [Google Scholar] [CrossRef]
- Chen, C.; Xin, K.; Liu, H.; Cheng, J.; Shen, X.; Wang, Y.; Zhang, L. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 2017, 7, 41564. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, J. Bacterial exopolysaccharides: Chemical structures, gene clusters and genetic engineering. Int. J. Biol. Macromol. 2021, 173, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Boels, I.C.; Ramos, A.; Kleerebezem, M.; de Vos, W.M. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 2001, 67, 3033–3040. [Google Scholar] [CrossRef]
- Boels, I.C.; van Kranenburg, R.; Hugenholtz, J.; Kleerebezem, M.; De Vos, W.M. Sugar catabolism and its impact on the biosynthesis and engineering of exopolysaccharide production in lactic acid bacteria. Int. Dairy J. 2001, 11, 723–732. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Mainprize, I.L.; Naismith, J.H.; Whitfield, C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 2009, 73, 155–177. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, C.G.; Lee, D.-J. Bacterial diguanylate cyclases: Structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol. Adv. 2015, 33, 124–141. [Google Scholar] [CrossRef] [PubMed]
- Suresh Kumar, A.; Mody, K.; Jha, B. Bacterial exopolysaccharides–a perception. J. Basic Microbiol. 2007, 47, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Das, S. Genetic regulation, biosynthesis and applications of extracellular polysaccharides of the biofilm matrix of bacteria. Carbohydr. Polym. 2022, 291, 119536. [Google Scholar]
- Kang, Y.; Li, P.; Zeng, X.; Chen, X.; Xie, Y.; Zeng, Y.; Zhang, Y.; Xie, T. Biosynthesis, structure and antioxidant activities of xanthan gum from Xanthomonas campestris with additional furfural. Carbohydr. Polym. 2019, 216, 369–375. [Google Scholar] [CrossRef]
- Rehm, B.H. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592. [Google Scholar] [CrossRef]
- Prada-Ramírez, H.A.; Pérez-Mendoza, D.; Felipe, A.; Martínez-Granero, F.; Rivilla, R.; Sanjuán, J.; Gallegos, M.T. AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC 3000. Mol. Microbiol. 2016, 99, 960–977. [Google Scholar] [CrossRef]
- Hay, I.D.; Remminghorst, U.; Rehm, B.H. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 1110–1120. [Google Scholar] [CrossRef]
- He, Y.W.; Ng, A.Y.J.; Xu, M.; Lin, K.; Wang, L.H.; Dong, Y.H.; Zhang, L.H. Xanthomonas campestris cell–cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 2007, 64, 281–292. [Google Scholar] [CrossRef]
- Becker, A. Challenges and perspectives in combinatorial assembly of novel exopolysaccharide biosynthesis pathways. Front. Microbiol. 2015, 6, 144903. [Google Scholar] [CrossRef]
- Van Kranenburg, R.; Vos, H.R.; van Swam, I.I.; Kleerebezem, M.; de Vos, W.M. Functional analysis of glycosyltransferase genes from Lactococcus lactis and other gram-positive cocci: Complementation, expression, and diversity. J. Bacteriol. 1999, 181, 6347–6353. [Google Scholar] [CrossRef] [PubMed]
- Jolly, L.; Stingele, F. Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria. Int. Dairy J. 2001, 11, 733–745. [Google Scholar] [CrossRef]

| Attribute | Values |
|---|---|
| Genome Size (bp) | 4,260,552 |
| GC Content (%) | 53.6 |
| Gene Number | 4192 |
| Gene Length (bp) | 3,711,663 |
| CDS (protein) | 3928 |
| tRNA genes | 82 |
| rRNA genes (5S, 16S, 23S) | 22 |
| Gene ID | Length (aa) | Function | Rename |
|---|---|---|---|
| LQ939_RS12550 | 340 | LPS O-antigen chain length determinant protein | wzzB |
| LQ939_RS12555 | 469 | NADP-dependent phosphogluconate dehydrogenase | gndA |
| LQ939_RS12560 | 183 | dTDP-4-dehydrorhamnose 3,5-epimerase | rfbC |
| LQ939_RS12565 | 193 | DapH/DapD/GlmU-related protein | alhA |
| LQ939_RS12570 | 434 | hypothetical protein | alhB |
| LQ939_RS12575 | 266 | hypothetical protein | alhC |
| LQ939_RS12580 | 363 | EpsG family protein | alhD |
| LQ939_RS12585 | 475 | oligosaccharide flippase family protein | alhE |
| LQ939_RS12590 | 274 | NAD(P)-dependent oxidoreductase | alhF |
| LQ939_RS12595 | 293 | glucose-1-phosphate thymidylyltransferase | rfbA |
| LQ939_RS12600 | 363 | dTDP-glucose 4,6-dehydratase | rfbB |
| LQ939_RS12605 | 349 | UDP-N-acetylglucosamine-undecaprenyl-phosphate N-acetylglucosaminephosphotransferase | wecA |
| LQ939_RS12610 | 338 | UDP-glucose 4-epimerase | galE |
| LQ939_RS12615 | 299 | UTP-glucose-1-phosphate uridylyltransferase | galF |
| LQ939_RS12620 | 612 | hypothetical protein | alhG |
| LQ939_RS12625 | 336 | glycosyltransferase | alhH |
| LQ939_RS12630 | 316 | polysaccharide pyruvyl transferase family protein | alhI |
| LQ939_RS12635 | 494 | MOP flippase family protein | alhJ |
| LQ939_RS12640 | 467 | undecaprenyl-phosphate glucose phosphotransferase | wcaJ |
| LQ939_RS12645 | 361 | glycosyltransferase | alhK |
| LQ939_RS12650 | 360 | hypothetical protein | alhL |
| LQ939_RS12655 | 302 | glycosyltransferase | alhM |
| LQ939_RS12660 | 726 | tyrosine-protein kinase | wzc |
| LQ939_RS12665 | 145 | protein tyrosine phosphatase | wzb |
| LQ939_RS12670 | 377 | polysaccharide export protein | wza |
| LQ939_RS12675 | 525 | TerC family protein | alhN |
| LQ939_RS12680 | 608 | outer membrane assembly protein AsmA | asmA |
| LQ939_RS12685 | 194 | dCTP deaminase | dcd |
| LQ939_RS12690 | 214 | uridine kinase | udk |
| LQ939_RS12695 | 214 | phosphatase PAP2 family protein | alhO |
| LQ939_RS12700 | 992 | diguanylate cyclase | alhP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, K.; Shi, X.; Tao, Z.; Hu, X.; Sun, L.; Wang, R.; Gu, Y.; Xu, H.; Qiu, Y.; Lei, P. Genomic and Transcriptomic Analyses Identify Two Key Glycosyltransferase Genes alhH and alhK of Exopolysaccharide Biosynthesis in Pantoea alhagi NX-11. Microorganisms 2024, 12, 2016. https://doi.org/10.3390/microorganisms12102016
He K, Shi X, Tao Z, Hu X, Sun L, Wang R, Gu Y, Xu H, Qiu Y, Lei P. Genomic and Transcriptomic Analyses Identify Two Key Glycosyltransferase Genes alhH and alhK of Exopolysaccharide Biosynthesis in Pantoea alhagi NX-11. Microorganisms. 2024; 12(10):2016. https://doi.org/10.3390/microorganisms12102016
Chicago/Turabian StyleHe, Kun, Xiaolong Shi, Zhongming Tao, Xing Hu, Liang Sun, Rui Wang, Yian Gu, Hong Xu, Yibin Qiu, and Peng Lei. 2024. "Genomic and Transcriptomic Analyses Identify Two Key Glycosyltransferase Genes alhH and alhK of Exopolysaccharide Biosynthesis in Pantoea alhagi NX-11" Microorganisms 12, no. 10: 2016. https://doi.org/10.3390/microorganisms12102016
APA StyleHe, K., Shi, X., Tao, Z., Hu, X., Sun, L., Wang, R., Gu, Y., Xu, H., Qiu, Y., & Lei, P. (2024). Genomic and Transcriptomic Analyses Identify Two Key Glycosyltransferase Genes alhH and alhK of Exopolysaccharide Biosynthesis in Pantoea alhagi NX-11. Microorganisms, 12(10), 2016. https://doi.org/10.3390/microorganisms12102016






