Abstract
Emerging life-threatening multidrug-resistant (MDR) species such as the C. haemulonii species complex, Clavispora lusitaniae (sin. C. lusitaniae), and other Candida species are considered as an increasing risk for human health in the near future. (1) Background: Many studies have emphasized that the increase in drug resistance can be associated with several virulence factors in Candida and its knowledge is also essential in developing new antifungal strategies. (2) Methods: Hydrophobicity, adherence, biofilm formation, lipase activity, resistance to osmotic stress, and virulence ‘in vivo’ on G. mellonella larvae were studied in isolates of C. haemulonii, C. albicans, and C. lusitaniae with low susceptibility and resistance to fluconazole and amphotericin B. (3) Results: Intra- and interspecies variability were observed. C. haemulonii showed high hydrophobicity and the ability to adhere to and form biofilm. C. lusitaniae was less hydrophobic, was biofilm-formation-strain-dependent, and did not show lipase activity. Larvae inoculated with C. albicans isolates displayed significantly higher mortality rates than those infected with C. haemulonii and C. lusitaniae. (4) Conclusions: The ability to adhere to and form biofilms associated with their hydrophobic capacity, to adapt to stress, and to infect within an in vivo model, observed in these non-wild-type Candida and Clavispora isolates, shows their marked virulence features. Since factors that define virulence are related to the development of the resistance of these fungi to the few antifungals available for clinical use, differences in the physiology of these cells must be considered to develop new antifungal therapies.
1. Introduction
Due to the increase in the number of multidrug-resistant (MDR) microorganisms and the unavailability of new antimicrobials, multidrug resistance (MDR) is recognized as one of the most important public health threats of the 21st century [1]. The World Health Organization (WHO) warns that MDR will constitute one of the leading causes of death in the world, causing approximately 10 million deaths annually in 2050 [2]. Fungi represent a growing clinical threat due to the limited availability of drugs [3]. Acquired antifungal resistance is related to stress response activation such as adaptive mechanisms, drug target modification, or an overexpression of or increase in multidrug transporters [4]. Emerging life-threatening multidrug-resistant Candida species such as C. auris, the C. haemulonii species complex, C. glabrata, Clavispora lusitaniae (sin. C. lusitaniae), Meyerozyma guilliermondii (sin. C. guillermondii), and the C. parapsilosis species complex are considered as an increasing risk for human health in the near future [5,6].
C. haemulonii complex candidiasis is associated with superficial to invasive infections in patients with risk factors like peripheral vascular disease, diabetes mellitus, organ transplants, malignant tumors, and chronic leg ulcers [7]. Knowledge about its biology and virulence factors is scarce. Its adaptive ability which contributes to survival in different host niches was observed. In addition, its capability to evade the action of commonly used antifungal agents, especially azoles and amphotericin B, makes this complex a worrying emerging opportunistic pathogen for either immunocompromised or immunosuppressed humans [8,9,10].
C. lusitaniae was first documented in 1979 and has attracted attention because it exhibits the development of resistance to amphotericin B, 5-fluorocytosine, or fluconazole [11,12,13]. C. lusitaniae was reported as being responsible for approximately 19.3% of fungemia in patients with cancer and 1.7% of genitourinary candidiasis cases in ambulatory patients, and it has also been associated with peritonitis and meningitis [13,14,15].
Candida drug resistance can be associated with several virulence factors [16]. In C. albicans, the main virulence factors are cell wall barriers, adherence, dimorphism, biofilm formation, proteins related to stress tolerance, hydrolytic enzymes (such as proteases, lipases, and hemolysins), and toxin production [17,18]. Adherence contributes to the persistence of the organism within the host, and this virulence factor is thus considered to be essential for the fungus settling and spreading [19]. Regardless of the species, microbial adherence due to hydrophobic interaction depends on the microbe surface hydrophobicity [20]. Likewise, biofilm formation is an important virulence feature that favors fungal pathogenesis. Candida species form a multicellular biofilm which allows yeast cells to anchor to host tissues, catheters, implants, and other devices [21]. It has been highlighted that increasing drug resistance has provided a strong impetus to understand the mechanisms of the increased tolerance of biofilm-associated infections to aid antimicrobial therapy.
Among the main virulence mechanisms of Candida, the ability to secrete hydrolytic enzymes like phospholipase C, caseinase, lipase, and proteinase stands out, to degrade the host cellular components, in order to facilitate penetration and invasion, thus playing a determining role in the pathogenesis [22]. These virulence factors have not been determined for all species and are not well known. Understanding and assessing these virulence factors in emerging drug-resistant yeast pathogens will contribute to the future development of novel target-specific drugs. In this study, we have evaluated hydrophobicity, the ability to adhere to and form biofilm, lipase activity, resistance to osmotic stress, and infection in an in vivo model of isolates of C. albicans, C. haemulonii, and C. lusitaniae with low susceptibility and resistance to fluconazole and amphotericin B.
2. Materials and Methods
2.1. Microorganisms and Growth Conditions
Three vaginal isolates of C. albicans (IMR-M-L 1462, IMR-M-L 1463, and IMR-M-L 1464), three blood isolates of C. haemulonii (IMR-M-L 785, IMR-M-L 1293, and IMR-M-L 1375), and four C. lusitaniae (IMR-M-L 301 and IMR-M-L 1112 isolated from blood and IMR-M-L 522 and IMR-M-L 1384 isolated from urine) were studied. Candida albicans ATCC 24433 was included as the quality control strain. All isolates were identified using the Vitek matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) system (Bruker Daltoniks, Bremen, Germany) and deposited in the culture collection (IMR-M-L) of the Mycology Department, Instituto de Medicina Regional, Universidad Nacional del Nordeste. Prior to studying the virulence factors, all yeast isolates were grown on Sabouraud Dextrose (SD) agar.
2.2. Antifungal Activity of Fluconazole and Amphotericin B
The antimicrobial activity of fluconazole and amphotericin B against all clinical and standard strains were evaluated using the broth microdilution method proposed by the reference CLSI M27-A4 methodology [23] and interpretative breakpoints were used according to the CLSI M27M44S [24].
Amphotericin B and fluconazole clinical breakpoints were not established for C. haemulonii and C. lusitaniae. However, epidemiological cut-off points (ECVs) were established for fluconazole against C. haemulonii and C. lusitaniae and also for amphotericin B against C. albicans and C. lusitaniae [25].
2.3. Hydrophobicity Assay
The cellular surface hydrophobicity (CSH) was determined by a two-phase system following previous reports [26]. In brief, yeasts were grown in SD broth at 28 °C for 24 h. Then, cells were washed with sterile saline buffer and 0.5% Tween 20 was added; they were resuspended in 0.05 M PBS (pH 7.2) at a final concentration of 2 × 106 cells/mL. The cell suspension was transferred to a glass tube containing 500 μL octane (Sigma Aldrich, Saint Louis, MO, USA). The mixture was vortexed for 1 min and maintained at room temperature for phase separation. After the two phases had been separated, the aqueous phase was measured at OD600. The group without the octane overlay was used as the control. Relative CSH was calculated as follows: [(OD600 of the control-OD600 after octane overlay)/OD600 of the control] × 100. The value for each strain was the average of three independent biological replicates.
2.4. Adherence on Plastic Surface by Crystal Violet Assay
The ability of Candida isolates to adhere on polystyrene surface were measured as reported previously [27]. The yeast cells were grown as described above for 24 h at 28 °C, washed twice with sterile PBS, and then resuspended at 37 °C in RPMI 1640 plus 10% FBS at 2.5 × 107 cells. After incubation for 3 h at 37 °C in six-well polystyrene plates (Corning Incorporated, Corning, NY, USA), the medium was aspirated and the non-adherent cells were removed and washed with PBS. The adherent cells were fixed with 99% v/v methanol for 15 min and then 0.02% (v/v) crystal violet was added for 20 min. Cells were washed and 33% (v/v) acetic acid was added again for 30 min. The crystal violet released was measured at 590 nm.
2.5. Biofilm Formation
Following previously described studies, the in vitro biofilm formation assay was carried out [28]. A yeast cell suspension (1.0 × 107 cells/mL) was incubated for 24 and 48 h at 37 °C in 96-well microtiter plates (Corning, NY, USA). After formation of the mature biofilm, the medium was aspirated, and non-adherent cells were removed by washing with sterile PBS. The biofilm development was measured with a 2,3-bis-(2-methoxy- 4-nitro-5- sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction assay, a reaction catalyzed by mitochondrial dehydrogenases. In brief, biofilm cells were washed with PBS and then incubated with 0.5 mg/mL of XTT and 1 μM of menadione in PBS at 37 °C for 2 h. A sample (500 µL) was then transferred from each well into a fresh 12-well plate and the colorimetric change resulting from XTT reduction was measured at 490 nm.
2.6. Lipase Activity Assay
The lipase activity (Lz) was assessed according to Muhsin et al. [29]. Briefly, 10 µL of each strain suspension was cultured in a sterile Petri dish containing a lipid medium (i.e., peptone 1%, sodium chloride 5%, calcium chloride 0.01%, and agar 2%, plus 1% tween 80) and incubated at 32 °C for 20 days. A clear halo zone of precipitation around the colony indicated lipase production. The production of lipase was expressed as a ratio of diameter of a colony to total diameter plus zone of precipitation. Each test was performed in duplicate and the results were expressed as the average of the two obtained values. The ranges of activity according to the Lz index were established as follows: high, Lz ≤ 0.69; moderate, Lz = 0.70–0.89; weak, Lz = 0.90–0.99; none, Lz = 1.
2.7. Sensitivity to Osmotic Stress in Sodium Chloride
With some modifications to determine the sensitivity of Candida isolates to NaCl, the method reported by Chaves and da Silva [30] was applied. Ten microliter volumes of Sabouraud-grown yeast cells were transferred to 100 µL of SD broth, with the addition of 0.03–30% NaCl in 96-well microtiter plates, and incubated at 32 °C for 48 h. Growth was determined when visually perceptible turbidity was observed within each well.
2.8. Infection in Galleria Mellonella Model
For survival analyses, 10 larvae (250–320 mg/each) of G. mellonella per each Candida/Clavispora isolate for testing and two control groups were employed. Larvae were previously incubated at 37 °C. Larvae were selected at random for each group in the procedure and injected with different concentrations of yeast cells (about 5 × 107 cells CFU/mL) into the hemocoel through the last left proleg (Hamilton syringe 701N; volume, 10 μL; needle size, 26 s; cone tip; Sigma-Aldrich, Milan, Italy) [31]. After injection, larvae were incubated in Petri dishes at 37 °C in standard aerobic conditions and survival was recorded at 24 h intervals for five days. Larvae were considered dead when they showed no movement in response to gentle prodding with a pipette tip. As a control, one group that did not receive injection and one group that was injected with phosphate-buffered saline (PBS) plus 0.01% Tween 20 were included. Each experiment was repeated three times.
2.9. Statistical Analysis
Statistical differences among the groups of data were analyzed by two-way ANOVA. In all of the comparisons, a p value of 0.05 or lower was considered to be significant. The analyses were performed using GraphPad Prism Software version 8.0.1.
3. Results
3.1. Susceptibility to Fluconazole and Amphotericin B
Table 1 shows the antifungal activity of fluconazole against the eleven isolates included in this work. The MIC values ranged from 0.25 (μg/mL), obtained for the reference strain, to >64 (μg/mL).

Table 1.
Antifungal activity of fluconazole and amphotericin B.
The values obtained for QC Candida albicans ATCC 24433 were within the ranges established by the reference document. All clinical isolates of C. albicans presented decreased susceptibility to fluconazole (C. albicans IMR-M-L 1462 was resistant and the other was susceptible-dose-dependent).
3.2. Hydrophobicity Assay and Adherence on Plastic Surface
The CSH and the ability to adhere to plastic surfaces are two important virulence factors to initiate infection and biofilm formation. Figure 1A shows the histograms, in percentage, of the CSH obtained for all of the strains of the three species studied. Variability in CSH was observed between the different species. In fact, the statistical analysis of all strains of each species shows significant differences **** (p < 0.0001) between C. albicans and C. lusitaniae, while differences in CSH were not significant for C. haemulonii (Figure S1A). C. lusitaniae was the species with the lowest hydrophobicity in contrast to the other species studied.

Figure 1.
Histograms of hydrophobicity and adherence on plastic surface of C. albicans, C. lusitanieae, and C. haemulonii isolates. (A) Percentage of hydrophobicity. (B) Percentage of adherence on plastic surface.
The ability of C. albicans, C. lusitanieae, and C. haemulonii isolates to adhere to the polystyrene surface is shown in Figure 1B. All strains were highly adherent to the polystyrene surface. Differences between the three species were observed. The statistical analyses of all strains of each species show that C. albicans is the more adherent species with respect to C. haemulonii *** (p < 0.0003) (Figure S1B).
3.3. Biofilm Formation
Histograms of biofilm formation after 24 and 48 h measured using XTT for Candida and Clavispora isolates (IMR-M-L) and the reference strain ATCC 24433 C. albicans are shown in Figure 2. All clinical isolates of the three species were able to produce biofilm with intraspecies variations, as depicted in Figure 2.
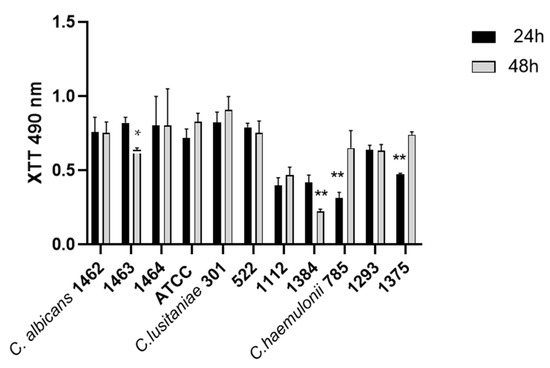
Figure 2.
Histograms of biofilm formation after 24 and 48 h of C. albicans, C. lusitaniae, and C. haemulonii. * p-value < 0.0392, ** p-values < 0.004.
The statistical analysis of all strains of each species indicated significant dissimilarities between C. albicans and C. haemulonii during the initial 24 h of biofilm formation, where C. haemulonii was the lower biofilm producer with *** p-values < 0.0004 (Figure S2). Nonetheless, these variations decreased after 48 h of incubation. A noticeable difference was observed between C. albicans and C. lusitaniae, with a * p-value < 0.0392 (Figure S2).
3.4. Lipase Activity and Osmotic Stress
Table 2 reports the lipase activity expressed as the LZ index in Candida spp. Positive lipase activity (Lz < 1) was measured in 63% of the isolates studied. Lz values ranged from 0.32 to 0.70. Strong lipase production (Lz ≤ 0.69) was observed for five isolates (45.45%), C. albicans (IMR-M-L 1462, 1463, 1464) and C. haemulonii (IMR-M-L 1293 and 1345) and in the reference strain C.albicans ATCC 24433. Only C. lusitaniae IMR-M-L 1384 showed moderate activity (Lz = 0.70 ± 0.00).

Table 2.
Lipase activity in Candida spp.
No enzymatic activity (Lz = 1) was observed for the three isolates of C. lusitaniae (IMR-M-L 301, 522, and 1112) and C. haemulonii IMR-M-L 785.
C. albicans isolates produced higher lipase activity levels (mean LZ of 0.44 ± 0.06) compared to C. haemulonii (mean LZ of 0.73 ± 0.25) or C. lusitaniae (mean LZ of 0.92 ± 0.15). Differences were significant between the three species **** (p-values < 0.0001).
With a gradual increase in NaCl concentrations, yeast cells were inoculated in SD broth to evaluate resistance to osmotic stress. All of the tested isolates were able to grow at a concentration of 15% NaCl, including the reference strain C. albicans ATCC 24433. All of the strains were considered to be resistant to osmotic stress.
3.5. Galleria Mellonella Survival Assay
To determine in vivo infection, G. mellonella larvae were employed as an animal model for a five-day study period (Figure 3). No significant difference in the survival rate of G.mellonella larvae infected with all three Candida species was observed 4 days post infection. Survival of 50% for all three species was four days. Five days after the inoculation, larvae infected with C. albicans strains showed a higher mortality rate compared to those infected with C. lusitaniae and C. haemulonii, with significant differences ** (p < 0.0042). No larval fatalities were observed in the control groups.

Figure 3.
G. mellonella survival assays after infection with C. albicans (IMR-M-L 1462, 1463, 1464, ATCC 24433), C. haemulonii (IMR-M-L 785, 1293, 1375), and C. lusitaniae (IMR-M-L 301, 522, 1112, 1384). Results are expressed as % survival in comparison to uninfected (PBS-treated) larvae. Median values obtained per group (10 larvae) are presented. Larvae infected with C. albicans significantly decreased survival, as assessed using the Mantel–Cox log-rank test (** p < 0.0042).
4. Discussion
AMR is one of the top 10 global public health deteriorates. Antifungal resistance is an increasing problem involving yeast fungi like Candida and allied genera. In recent years, international emphasis has been placed on the emergence of C. auris and its multi-resistance, but less attention has been paid to other yeasts with this same feature. Currently, 20–30% of candidemia cases involve species with intrinsic or cross-resistance to scarce disposal antifungals [32,33]. This work highlights the virulence factors of C. haemulonii and Clavispora lusitaniae, since this knowledge may be associated with their emergence and provide new targets to combat their drug resistance in the near future. Likewise, C. albicans isolates were included to evaluate the behavior of these factors in isolates of this species with low susceptibility.
With the exception of the ATCC quality control strain, according to the CLSI M27M44S clinical breakpoints for C. albicans, all of the isolates presented low susceptibility to fluconazole, one of the most widely used antifungal agents. On the other hand, according to the ECV for antifungal susceptibility testing, high MIC values were obtained for all C. haemulonii isolates but there were variable results for C. lusitaniae, which may reflect the fact that this yeast population is non-wild-type.
The antifungal susceptibility profiles of members of the C. haemulonii complex are of concern. The low susceptibility of C. haemulonii to currently available antifungal drugs was also observed in our results. C. haemulonii complex isolates evaluated in this study showed higher MICs for fluconazole and amphotericin B than C. lusitaniae and C. albicans isolates. For fluconazole, MIC values ranging from 4 to >64 µg/mL were obtained. The fluconazole ECV established by CLSI for C. haemulonii is very high (128 µg/mL) and may be an indication of intrinsic resistance or of limited susceptibility to this agent. The CLSI supplement M57S, 2022, states that MIC values lower than the ECV do not imply that the isolate is susceptible to fluconazole [25]. These results agree with other authors who have reported an increase in the fluconazole resistance of this fungal complex for many years [34,35,36,37,38,39].
With MIC values higher than ECV, 3/4 studied C. lusitaniae isolates can be distinguished as being non-wild-type strains. Heterogeneity in fluconazole resistance largely caused by the presence of at least 12 different alleles of MRR1, which encodes a drug resistance regulator, was explained [40]. The capability of C. lusitaniae to rapidly develop resistance to multiple antifungals during therapy was recognized. Facilitated by the haploid nature of C. lusitaniae, the selection of candins and/or azole-resistant isolates following candins and azole or their combination treatments was reported, but cross-resistance to amphotericin B resistance and candins also occurred without ongoing exposure to amphotericin B [41,42,43]. The M27-A3 document states that Candida species with MICs >1 µg/mL are likely resistant to AMB. Our C. lusitaniae isolates showed an MIC range for amphotericin B from 1 to >16, confirming the low susceptibility reported for this species. C. lusitaniae is not intrinsically resistant to amphotericin B, but some reports describe that phenotype resistance can only be observed using agar gradient strips and cannot be detected using broth microdilution methods. This could explain the ranges of amphotericin MIC values obtained for this species [25].
Many studies have emphasized that an increase in drug resistance can be associated with several virulence factors in Candida [16,44,45,46,47]. In particular, studying the genes that are correlated with drug resistance and virulence factors could represent a way to resolve this problem. The literature on the behavior of these emerging drug-resistant species is scarce for C. haemulonii and C. lusitaniae.
When developing new antifungal strategies, knowledge of CSH is also essential. This biophysical parameter influences cell–cell and also cell–surface interactions, playing a crucial and complex role in the processes of virulence, as well as response to therapies. Differences in CSH may cause variable efficacy of antifungal treatments [48]. Candida isolates with differences in CSH may possess altered lipid metabolism and consequently may differ in their response to treatment [49]. In this study, isolates of C. albicans and C. haemulonii with low MIC values with regard to fluconazole and amphotericin B showed differences in CSH in a range between 20% and 80% and 35% and 80%, respectively. Hydrophobic C. albicans strains showed more resistance to fluconazole due to ergosterol overproduction and ERG11 gene overexpression, as well as overproduction and higher activity of the Cdr1 transporter [49]. While C. albicans and C. haemulonii showed higher CHS values, low values ranging between 5 and 15% were obtained for C. lusitaniae. Our isolates were obtained from patients who had had several previous treatments. The CSH could be reduced by antifungal therapies [48]. Further exploration of the mechanisms through which multiple drugs reduce CSH could explain additional pathways involved in pathogenesis and lead to the development of novel antifungal therapeutic strategies.
Adhesion is considered the first step in the development of biofilm formation, a significant virulence factor described for Candida and yeast-like fungus with medical inference. Moreover, Candida is adept at adhering and this process is favored, in part, by the hydrophobic interactions between yeast cells and surfaces [50,51]. Despite this low CSH values were obtained for C. lusitaniae in this study, high adherence performance was observed for all of the strains studied. Our results contrast those of Muadcheingka et al., 2015, who reported that it was more hydrophobic but less adherent [52]. It is well known that other factors, different to CHS, are related to adherence [13]. Furthermore, and related to this ability to adhere, all three Candida/Clavispora species tested demonstrated their capability to produce biofilm; in particular, with C. haemulonii and C. albicans, the latter is already cited as being one of the most adherent species. The ability of the C. haemulonii species complex to form biofilm on different types of surfaces was reported [5,38,43,53]. Although all C. lusitaniae isolates were adherent, they showed more interspecies variability in terms of biofilm formation. These low properties correlate with the lower virulence reported for C. lusitaniae than the other species [13]. In a biofilm, microorganisms are in a stable environment where they can tolerate high concentrations of antimicrobials. Moreover, biofilms’ exposure to antimicrobial drugs usually results in the induction of resistance genes. For these reasons, biofilm is a recognized virulence trait directly associated with enhanced antimicrobial resistance [54,55,56].
Lipases are enzymes of pharmacological interest because they can act as virulence factors in several infectious diseases. The role of lipases in the virulence of Candida has been shown to favor morphological transition, colonization, cytotoxicity, and penetration, thus enhancing their survival within the host [57]. Although most of the isolates studied in this work showed lipase activity, no uniformity in the expression of this activity for any of the three species was observed. In contrast, all of the isolates were considered to be resistant to osmotic stress. Exposure to NaCl or KCl produces rapid and effective adaptation to nutrients and stress, which benefits the virulence of C. albicans [58,59]. The ability to rapidly adapt to host-imposed stresses in varied environments contributes significantly to the survival of the fungus in host niches and its ability to cause infection [60].
The larvae of Galleria mellonella are now broadly accepted as being a model system for assessing the virulence of microbial pathogens. According to other reports, the virulence of non-C. albicans species in G. mellonella infection models is dependent on the fungal species. In our study, C. albicans showed higher virulence and produced lower survival rates than C. haemulonii and C. lusitaniae. C. lusitaniae was shown to be more virulent than the C. haemulonii species complex [9].
The emergence of multidrug-resistant Candida species, like the C. haemulonii species complex and C. lusitaniae, and the growing number of resistant isolates of the cosmopolitan C. albicans pose a threat and we are likely to witness the rise of new multidrug-resistant pathogenic yeast species in the near future. Candida and Clavispora may use different mechanisms for the evasion of the host immune system but also exhibit different virulence-related phenotypes to resist antifungal effects. Mutations were found in cell adhesion genes and biofilm formation, indicating that resistance is co-evolving with virulence [32].
The ability to adhere to and form biofilms associated with their hydrophobic capacity, to adapt to stress, and to infect in an in vivo model, observed in non-wild-type Candida and Clavispora isolates, shows their marked virulence features. The observed behavior is not general for an entire species; furthermore, virulence factors can vary depending on the strain. Since the factors that define virulence are related to the development of resistance of these fungi to the few antifungals available for clinical use, differences in the physiology of these cells must be considered to develop new antifungal therapies. Likewise, the patient’s history and previous treatments are also a point of analysis, since previous antifungal treatments alter the virulence factors of these yeasts, either decreasing or exacerbating them.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms12010212/s1: Figure S1. Hydrophobicity and adherence on plastic surface of C. albicans, C. lusitanieae, and C. haemulonii isolates. Median values obtained for species are presented (* p < 0.05; **** p< 0.0001). Figure S2. Biofilm formation after 24 and 48 h of C. albicans, C. lusitanieae, and C. haemulonii isolates. Median values obtained for species are presented (* p < 0.05; **** p< 0.0001).
Author Contributions
Conceptualization, L.A. and G.G.; methodology, L.A., F.R., A.G., N.B., and G.G.; software, L.A., A.G., and G.G.; validation, L.A. and G.G.; formal analysis, L.A. and G.G.; investigation, F.R., A.G., and N.B.; resources, L.A., F.R., and G.G.; data curation, L.A., F.R., A.G., N.B., and G.G.; writing—original draft preparation, L.A. and G.G.; writing—review and editing, L.A. and G.G.; supervision, L.A. and G.G.; project administration, L.A.; funding acquisition, L.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Bando Ateneo Sapienza 2022, grant number RP12218147BEEF07.
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors want to acknowledge ‘Cooperativa gruppo Comunale Logistica Industriale Integrata. Soc. Coop.’ for its donations in kind (e.g., the materials used for experiments).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Temkin, E.; Adler, A.; Lerner, A.; Carmeli, Y. Carbapenem-resistant Enterobacteriaceae: Biology, epidemiology, and management. Ann. N. Y. Acad. Sci. 2014, 1323, 22–42. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, G.; Fleck, F. United Nations meeting on antimicrobial resistance. Bull. World Health Organ. 2016, 94, 638–639. [Google Scholar]
- Stevenson, E.M.; Gaze, W.H.; Gow, N.A.R.; Hart, A. Antifungal Exposure and Resistance Development: Defining Minimal Selective Antifungal Concentrations and Testing Methodologies. Front. Fungal Biol. 2022, 3, 918717. [Google Scholar] [CrossRef] [PubMed]
- Hossain, C.M.; Ryan, L.K.; Gera, M.; Choudhuri, S.; Lyle, N.; Ali, K.A.; Diamond, G. Antifungal and drug resistance. Encyclopedia 2022, 2, 1722–1737. [Google Scholar] [CrossRef]
- Ramos, L.S.; Figueiredo-Carvalho, M.H.G.; Silva, L.N.; Siqueira, N.L.; Lima, J.C.; Oliveira, S.S.; Almeida-Paes, R.; Zancopé-Oliveira, R.M.; Azevedo, F.S.; Ferreira, A.L.; et al. The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes. J. Fungi 2022, 8, 574. [Google Scholar] [CrossRef] [PubMed]
- Cendejas-Bueno, E.; Kolecka, A.; Alastruey-Izquierdo, A.; Theelen, B.; Groenewald, M.; Kostrzewa, M.; Cuenca-Estrella, M.; Gomez-Lopez, A.; Boekhout, T. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: Three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 2012, 50, 3641–3651. [Google Scholar] [PubMed]
- Ben-Ami, R.B.; Berman, J.; Novikov, A. Multi-drug resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef]
- Lima, S.L.; Rossato, L.; Salles de Azevedo Melo, A. Evaluation of the potential virulence of Candida haemulonii species complex and Candida auris isolates in Caenorhabditis elegans as an in vivo model and correlation to their biofilm production capacity. Microbial Pathogenesis. 2020, 148, 104461. [Google Scholar] [CrossRef]
- Silva, L.N.; Oliveira, S.S.; Magalhães, L.B.; Andrade Neto, V.V.; Torres-Santos, E.C.; Carvalho, M.D.; Pereira, M.D.; Branquinha, M.H.; Santos, A.L. Unmasking the Amphotericin B Resistance Mechanisms in Candida haemulonii Species Complex. ACS Infect. Dis. 2020, 6, 1273–1282. [Google Scholar] [CrossRef]
- Deng, Y.; Li, S.; Bing, J.; Liao, W.; Tao, L. Phenotypic Switching and Filamentation in Candida haemulonii, an Emerging Opportunistic Pathogen of Humans. Microbiol. Spectrum 2021, 9, e00779-21. [Google Scholar] [CrossRef]
- Pappagianis, D.; Collins, M.S.; Hector, R.; Remington, J. Development of resistance to amphotericin B in Candida lusitaniae infecting a human. Antimicrob. Agents Chemother. 1979, 16, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Fakhim, H.; Vaezi, A.; Dannaoui, E.; Chowdhary, A.; Nasiry, D.; Faeli, L.; Meis, J.F.; Badali, H. Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses 2018, 61, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Reyes, D.; Gómez-Gaviria, M.; Mora-Montes, H. Candida lusitaniae: Biology, Pathogenicity, Virulence Factors, Diagnosis, and Treatment. Infect. Drug Resist. 2022, 15, 5121–5135. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.S.; Farmakiotis, D.; Jiang, Y.; Tarrand, J.J.; Kontoyiannis, D.P. Uncommon Candida species fungemia among cancer patients, Houston, Texas, USA. Emerg. Infect. Dis. 2015, 21, 1942–1950. [Google Scholar] [CrossRef]
- Obisesan, O.J.; Olowe, O.A.; Taiwo, S.S. Phenotypic detection of genitourinary candidiasis among sexually transmitted disease clinic attendees in Ladoke Akintola University Teaching Hospital, Osogbo, Nigeria. J. Environ. Public. Health. 2015, 2015, 401340. [Google Scholar] [CrossRef] [PubMed]
- Angiolella, L.; Stringaro, A.R.; De Bernardis, F.; Posteraro, B.; Bonito, M.; Toccacieli, L.; Torosantucci, A.; Colone, M.; Sanguinetti, M.; Cassone, A.; et al. Increase of Virulence and Its Phenotypic Traits in Drug-Resistant Strains of Candida albicans. Antimicrob. Agents Chemother. 2008, 52, 927–936. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rajendran, R.; Sherry, L.; Nile, C.J.; Sherriff, A.; Johnson, E.M.; Hanson, M.F.; Williams, C.; Munro, C.A.; Jones, B.J.; Ramage, G. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection—Scotland, 2012–2013. Clin. Microbiol. Infect. 2016, 22, 87–93. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, G.; Dong, J.; Xing, X.H.; Dai, J.; Zhang, C. An efficient machine-learning workflow in conjunction with the Yeas tFab assembly strategy for combinatorial optimization of heterologous metabolic pathways in Saccharomyces cerevisiae. Metab. Eng. 2018, 47, 294–302. [Google Scholar] [CrossRef]
- Ghannoum, M.A.; Abu-Elteen, K.; Ibrahim, A.; Stretton, R. Protection against Candida albicans gastrointestinal colonization and dissemination by saccharides in experimental animals. Microbios 1991, 67, 95–105. [Google Scholar]
- Yoshijima, Y.; Murakami, K.; Kayama, S.; Liu, D.; Hirota, K.; Ichikawa, T.; Miyake, Y. Effect of substrate surface hydrophobicity on the adherence of yeast and hyphal Candida. Mycoses 2010, 53, 221–226. [Google Scholar] [CrossRef]
- Silva, S.; Henriques, M.; Martins, A.; Oliverira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.G.; Latha, R.; Vedhagiri, K.; Sathiamoorthi, T.; Javarani, G.; Sakala, R.; Selvin, J.; Natarajaseenivasan, K. Phospholipase C, proteinase and hemolytic activities of Candida spp. isolated from pulmonary tuberculosis patients. J. Mycol. Med. 2009, 19, 3–10. [Google Scholar] [CrossRef]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, CLSI M27 A4, 4th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- CLSI. Performance Standard for Antifungal Susceptibility Testing of Yeasts, CLSI M27M44S, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- CLSI. Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 4th ed.; CLSI Supplement M57S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- de Groot, P.W.; Kraneveld, E.A.; Yin, Q.Y.; Dekker, H.L.; Groß, U.; Crielaard, W.; de Koster, C.G.; Bader, O.; Klis, F.M.; Weig, M. The cell wall of the human pathogen Candida glabrata: Differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 2008, 7, 1951–1964. [Google Scholar] [CrossRef] [PubMed]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016, 2016, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Finkel, J.S.; Mitchell, A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 2010, 9, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Muhsin, T.M.; Salih, T.H. Exocellular enzyme activity of dermatophytes and other fungi isolated from ruminants in Southern Iraq. Mycopathologia 2001, 150, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Chaves, G.M.; da Silva, W.P. Superoxide dismutases and glutaredoxins have a distinct role in the response of Candida albicans to oxidative stress generated by the chemical compounds menadione and diamide. Mem. Inst. Oswaldo Cruz 2012, 107, 998–1005. [Google Scholar] [CrossRef]
- Daniel, S.F.Q.; Casadevall, A. Fungal immunity and pathogenesis in mammals versus the invertebrate model organism Galleria mellonella. Pathog. Dis. 2021, 79, ftab013. [Google Scholar]
- Ksiezopolska, E.; Gabaldón, T. Evolutionary Emergence of Drug Resistance in Candida Opportunistic Pathogens. Genes 2018, 9, 461. [Google Scholar] [CrossRef]
- Pappas, P.; Lionakis, M.; Arendrup, M.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.S.; Mello, T.P.; Branquinha, M.H.; Santos, A.L.S. Biofilm Formed by Candida haemulonii Species Complex: Structural Analysis and Extracellular Matrix Composition. J. Fungi 2020, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Xiao, M.; Chen, S.C.A.; Wang, H.; Cheng, J.W.; Chen, X.X.; Xu, Z.P.; Fan, X.; Kong, F.; Xu, Y.C. Identification and Antifungal Susceptibility Profiles of Candida haemulonii Species Complex Clinical Isolates from a Multicenter Study in China. J. Clin. Microbiol. 2016, 54, 2676–2680. [Google Scholar] [CrossRef]
- Ruan, S.Y.; Kuo, Y.W.; Huang, C.T.; Hsiue, H.C.; Hsueh, P.R. Infections Due to Candida haemulonii: Species Identification, Antifungal Susceptibility and Outcomes. Int. J. Antimicrob. Agents 2010, 35, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Rodero, L.; Cuenca-Estrella, M.; Córdoba, S.; Cahn, P.; Davel, G.; Kaufman, S.; Guelfand, L.; Rodríguez-Tudela, J.L. Transient Fungemia Caused by an Amphotericin B-Resistant Isolate of Candida haemulonii. J. Clin. Microbiol. 2002, 40, 2266–2269. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.U.; Al-Sweih, N.A.; Ahmad, S.; Al-Kazemi, N.; Khan, S.; Joseph, L.; Chandy, R. Outbreak of Fungemia Among Neonates Caused by Candida haemulonii Resistant to Amphotericin B, Itraconazole, and Fluconazole. J. Clin. Microbiol. 2007, 45, 2025–2027. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Results from the Artemis Disk Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-Year Analysis of Susceptibilities of Candida Species to Fluconazole and Voriconazole as Determined by CLSI Standardized Disk Diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Demers, E.G.; Biermann, A.R.; Masonjones, S.; Hogan, D.A. Evolution of drug resistance in an antifungal-naive chronic Candida lusitaniae infection. Proc. Natl. Acad. Sci. USA 2018, 115, 12040–12045. [Google Scholar] [CrossRef]
- Chibabhai, V. Incidence of candidemia and prevalence of azole-resistant candidemia at a tertiary South African hospital—A retrospective laboratory analysis 2016–2020. S. Afr. J. Infect. Dis. 2022, 37, 326. [Google Scholar] [CrossRef]
- Asner, S.A.; Giulieri, S.; Diezi, M.; Marchetti, O.; Sanglard, D. Acquired Multidrug Antifungal Resistance in Candida lusitaniae during Therapy. Antimicrob. Agents Chemother. 2015, 59, 7715–7722. [Google Scholar] [CrossRef]
- Kannan, A.; Asner, S.A.; Trachsel, E.; Kelly, S.; Parker, J.; Sanglard, D. Comparative Genomics for the Elucidation of Multidrug Resistance in Candida lusitaniae. mBio 2019, 10, e02512-19. [Google Scholar] [CrossRef]
- Zuza-Alves, D.L.; De Medeiros, S.S.; De Souza, L.B.; Silva-Rocha, W.P.; Francisco, E.C.; De Araujo, M.C.; Lima-Neto, R.G.; Neves, R.P.; Melo, A.S.D.A.; Chaves, G.M. Evaluation of Virulence Factors in vitro, Resistance to Osmotic Stress and Antifungal Susceptibility of Candida tropicalis Isolated from the Coastal Environment of Northeast Brazil. Front. Microbiol. 2016, 7, 1783. [Google Scholar] [CrossRef] [PubMed]
- Maras, B.; Maggiore, A.; Mignogna, G.; D’Erme, M.; Angiolella, L. Hyperexpression of CDRs and HWP1 genes negatively impacts on Candida albicans virulence. PLoS ONE 2021, 16, e0252555. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Zambrano, L.J.; Puig-Asensio, M.; Pérez-García, F.; Escribano, P.; Sánchez-Carrillo, C.; Zaragoza, O.; Padilla, B.; Cuenca-Estrella, M.; Almirante, B.; Martín-Gómez, M.T.; et al. CANDIPOP study. Candida guilliermondii complex is characterized by high antifungal resistance but low mortality in 22 cases of candidemia. Antimicrob. Agents Chemother. 2017, 61, e00099-17. [Google Scholar] [PubMed]
- Schikora-Tamarit, M.A.; Gabaldón, T. Using genomics to understand the mechanisms of virulence and drug resistance in fungal pathogens. Biochem. Soc. Trans. 2022, 50, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Danchik, C.; Casadevall, A. Role of Cell Surface Hydrophobicity in the Pathogenesis of Medically-Significant Fungi. Front. Cell Infect. Microbiol. 2021, 10, 594973. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.; Muraszko, J.; Korba, A.; Bernat, P.; Krasowka, A. Lipid composition and cell surface hydrophobicity of Candida albicans influence the efficacy of fluconazole-gentamicin treatment. Yeast 2020, 37, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Tronchin, G.; Pihet, M.; Lopes-Bezerra, L.; Bouchara, J.P. Adherence mechanisms in human pathogenic fungi. Med. Mycol. 2008, 46, 749–772. [Google Scholar] [CrossRef]
- Tsui, C.; Kong, E.F.; Jabra-Rizk, M.A. Pathogenesis of Candida albicans biofilm. Pathog. Dis. 2016, 74, ftw018. [Google Scholar] [CrossRef] [PubMed]
- Muadcheingka, T.; Tantivitayakul, P. Distribution of Candida albicans and non-albicans Candida species in oral candidiasis patients: Correlation between cell surface hydrophobicity and biofilm forming activities. Arch. Oral Biol. 2015, 60, 894–901. [Google Scholar] [CrossRef]
- Oh, B.J.; Shin, J.H.; Kim, M.N.; Sung, H.; Lee, K.; Joo, M.Y.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med. Mycol. 2011, 49, 98–102. [Google Scholar] [CrossRef]
- Santos, A.L.S.; Mello, T.P.; Ramos, L.S.; Branquinha, M.H. Biofilm: A robust and efficient barrier to antifungal chemotherapy. J. Antimicrob. 2015, 1, e101. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Chandra, J. Candida biofilms: Development, architecture, and resistance. Microbiol. Spectr. 2015, 3, 115–134. [Google Scholar]
- Kean, R.; Delaney, C.; Sherry, L.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R.; Williams, C.; Ramage, G. Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm-mediated resistance. mSphere 2018, 3, e00334-18. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Do, E.; Jung, W.H. Lipolytic enzymes involved in the virulence of human pathogenic fungi. Mycobiology 2013, 41, 67–72. [Google Scholar] [CrossRef]
- Brown, A.J.P.; Brown, G.D.; Netea, M.G.; Gow, N.A.R. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014, 22, 614–622. [Google Scholar] [CrossRef]
- Brown, A.J.; Budge, S.; Kaloriti, D.; Tillmann, A.; Jacobsen, M.D.; Yin, Z.; Ene, I.V.; Bohovych, I.; Sandai, D.; Kastora, S.; et al. Stress adaptation in a pathogenic fungus. J. Exp. Biol. 2013, 217, 144–155. [Google Scholar] [CrossRef]
- Brown, A.J.P.; Haynes, K.; Gow, N.A.R.; Quinn, J. Stress Responses in Candida. In Candida and Candidiasis; Calderone, R.A., Clancy, C.J., Eds.; ASM Press: Washington, DC, USA, 2012; pp. 225–242. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).