Salmonella Typhimurium with Eight Tandem Copies of blaNDM-1 on a HI2 Plasmid
Abstract
1. Introduction
2. Materials and Methods
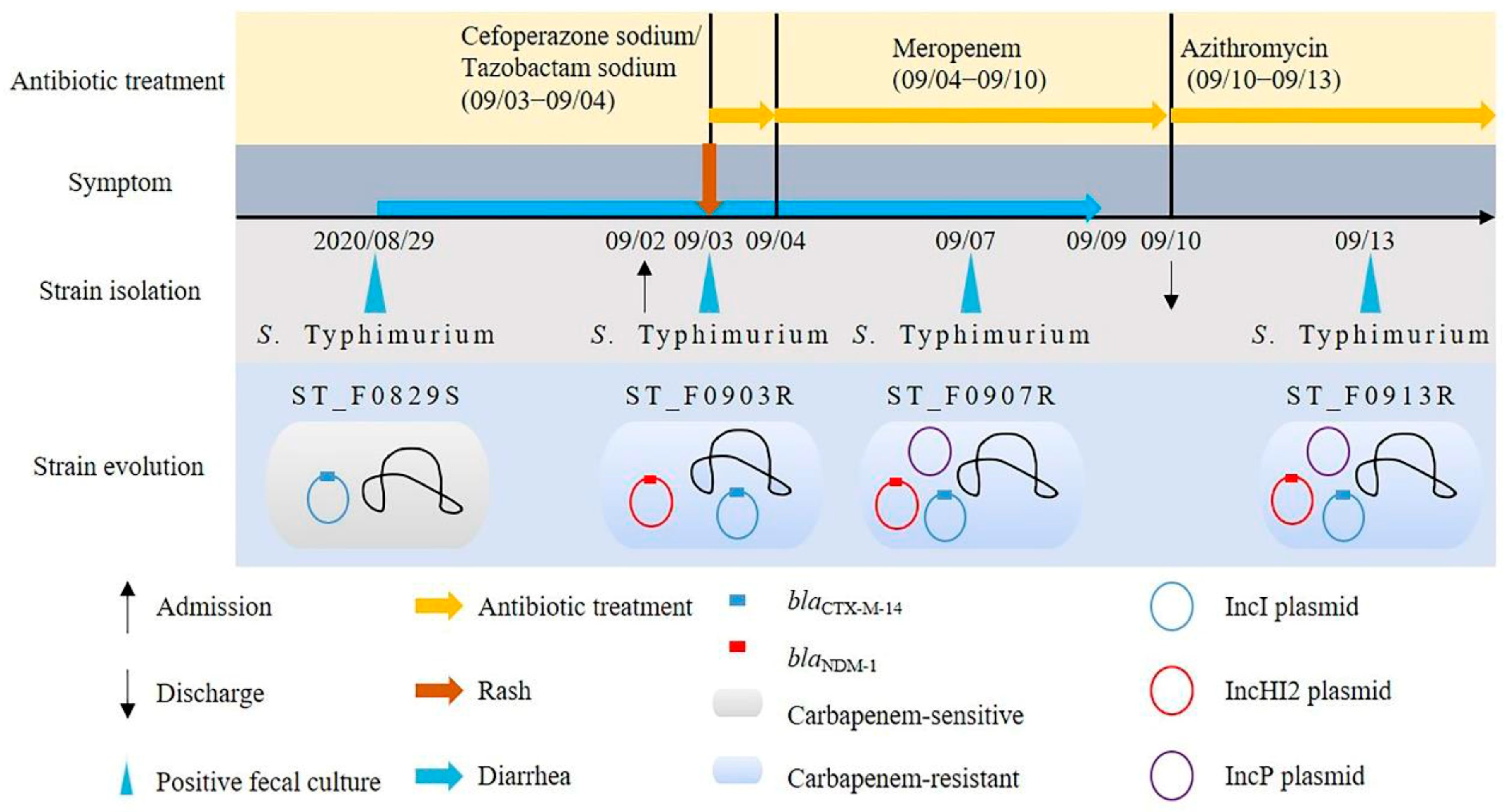
2.1. Bacterial Isolates and Case Information
2.2. Strain Identification and Sequence Typing
2.3. Antibiotic Susceptibility Testing
2.4. Screening for Carbapenemases
2.5. Conjugation Assay and PCR-Based Replicon Typing (PBRT)
2.6. Pulsed-Field Gel Electrophoresis (PFGE)
2.7. Illumina Short Reads Sequencing and Assembly
2.8. Pacific Biosciences (PacBio) High-Accuracy Long-Read (HiFi) Sequencing and Assembly
2.9. Genome Annotation and Comparative Genomics Analysis
2.10. Real-Time Quantitative PCR (qRT-PCR)
3. Results
3.1. Strain Identification
3.2. Phenotypic and Genotypic Characteristics of the Strain
3.3. Identification of Conjugative Resistance Plasmid
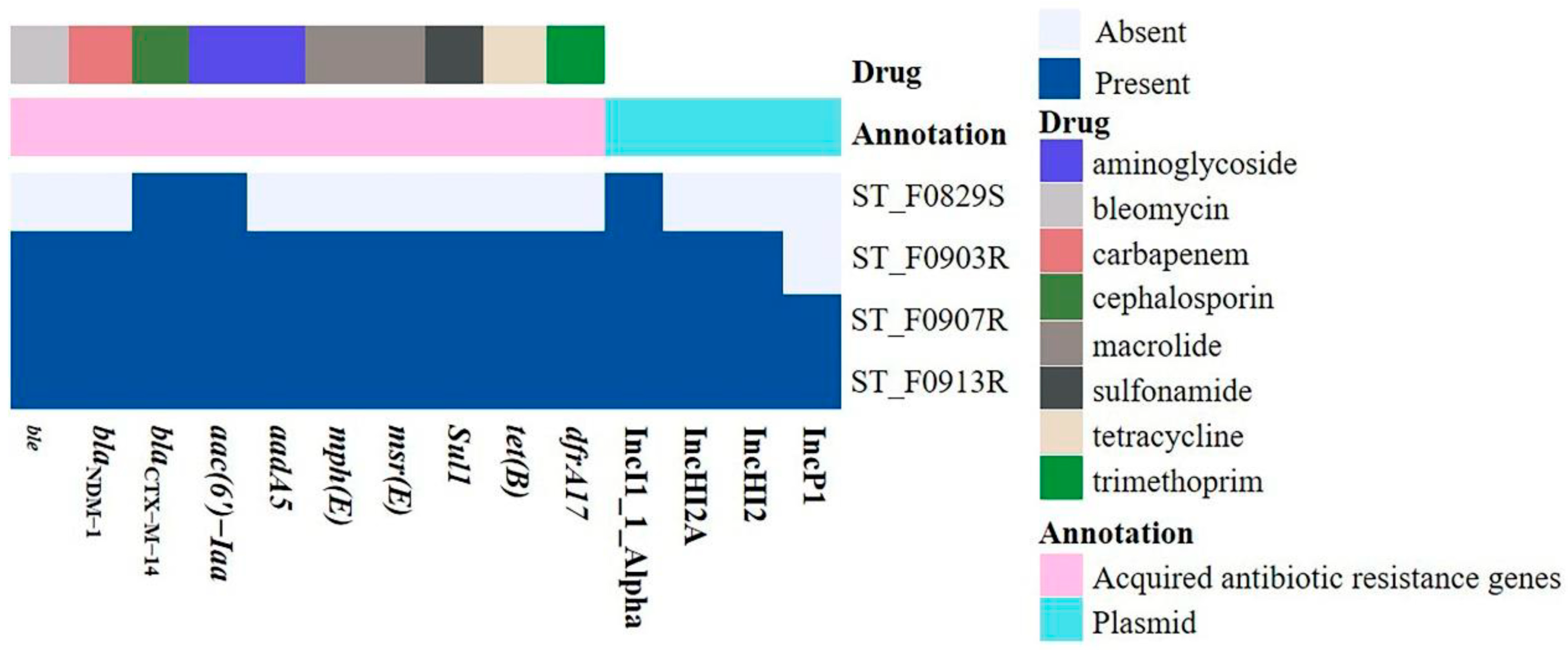
3.4. The ARGs and Plasmid Replicons of the Four S. Typhimurium Isolates
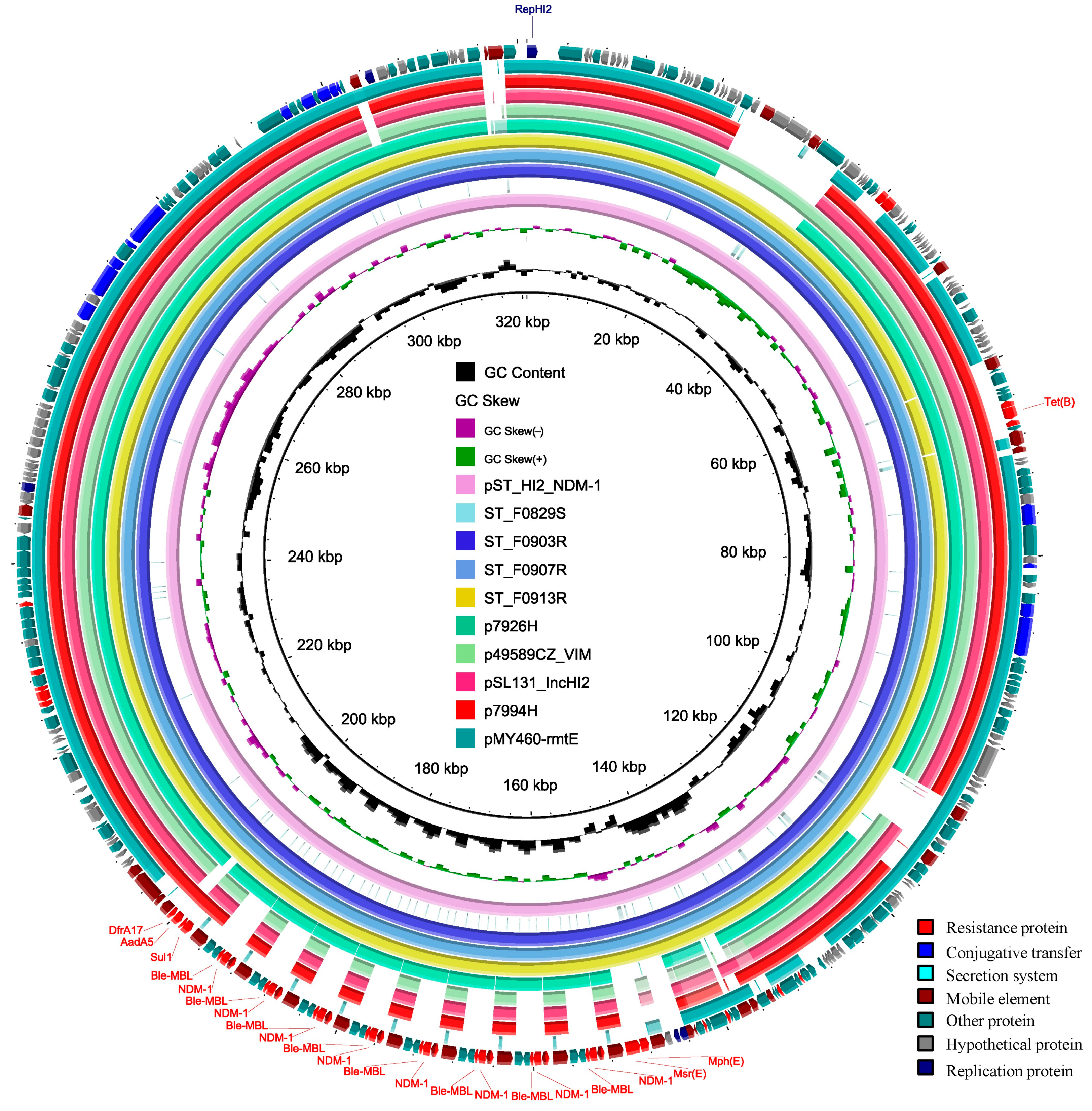
3.5. The Features of the Chromosome and Plasmids of the ST_F0903R Isolate
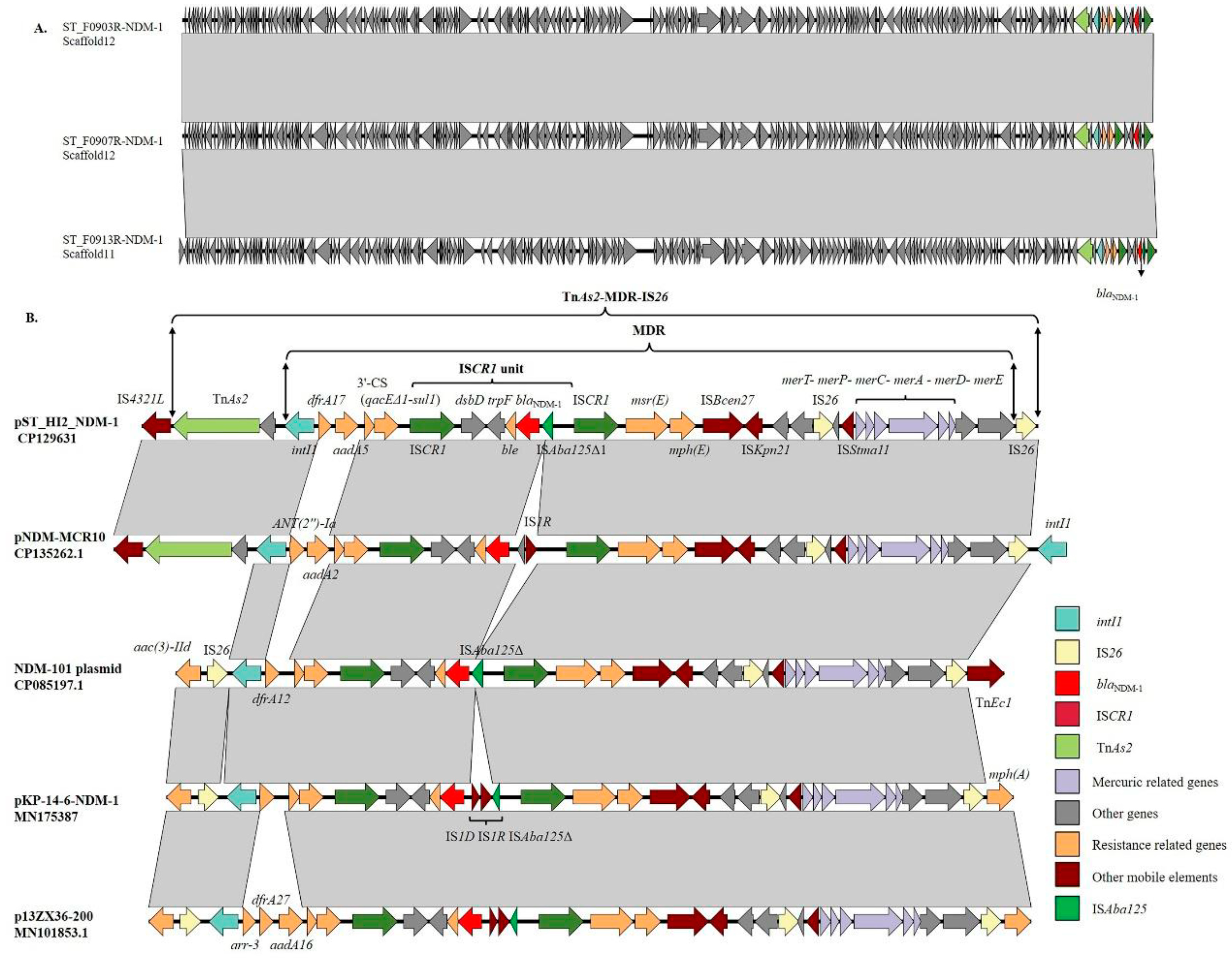
3.6. Genetic Context of the blaNDM-1 Gene
3.7. Amplification of ISCR1 Unit Carrying blaNDM-1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Knodler, L.A.; Elfenbein, J.R. Salmonella enterica. Trends Microbiol. 2019, 27, 964–965. [Google Scholar] [CrossRef] [PubMed]
- Shane, A.L.; Mody, R.K.; Crump, J.A.; Tarr, P.I.; Steiner, T.S.; Kotloff, K.; Langley, J.M.; Wanke, C.; Warren, C.A.; Cheng, A.C.; et al. 2017 Infectious Diseases Society of America Clinical Practice Guidelines for the Diagnosis and Management of Infectious Diarrhea. Clin. Infect. Dis. 2017, 65, e45–e80. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.S.; Hong, Y.P.; Wang, Y.W.; Chen, B.H.; Teng, R.H.; Song, H.Y.; Liao, Y.S. Antimicrobial Resistance and Mechanisms of Azithromycin Resistance in Nontyphoidal Salmonella Isolates in Taiwan, 2017 to 2018. Microbiol. Spectr. 2023, 11, e0336422. [Google Scholar] [CrossRef] [PubMed]
- Phu, D.H.; Wongtawan, T.; Truong, D.B.; Van Cuong, N.; Carrique-Mas, J.; Thomrongsuwannakij, T. A systematic review and meta-analysis of integrated studies on antimicrobial resistance in Vietnam, with a focus on Enterobacteriaceae, from a One Health perspective. One Health 2022, 15, 100465. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Guerra, B.; Rodicio, M.R. Resistance to Carbapenems in Non-Typhoidal Salmonella enterica Serovars from Humans, Animals and Food. Vet. Sci. 2018, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, E.; Bautista, A.; Barrero, L. First report of a Salmonella enterica serovar typhimurium isolate with carbapenemase (KPC-2) in Colombia. Antimicrob. Agents Chemother. 2014, 58, 1263–1264. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, M.; Ding, H.; Ye, M.; Hu, F.; Guo, Q.; Xu, X.; Wang, M. New Delhi metallo-β-lactamase-1 in carbapenem-resistant Salmonella strain, China. Emerg. Infect. Dis. 2013, 19, 2049–2051. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Mak, J.K.; White, P.A.; McIver, C.J.; Taylor, P. Multidrug-resistant Salmonella strains expressing emerging antibiotic resistance determinants. Clin. Infect. Dis. 2008, 46, 324–325. [Google Scholar] [CrossRef]
- Fischer, J.; Rodríguez, I.; Schmoger, S.; Friese, A.; Roesler, U.; Helmuth, R.; Guerra, B. Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J. Antimicrob. Chemother. 2013, 68, 478–480. [Google Scholar] [CrossRef]
- Seiffert, S.N.; Perreten, V.; Johannes, S.; Droz, S.; Bodmer, T.; Endimiani, A. OXA-48 carbapenemase-producing Salmonella enterica serovar Kentucky isolate of sequence type 198 in a patient transferred from Libya to Switzerland. Antimicrob. Agents Chemother. 2014, 58, 2446–2449. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Jin, S.; Ge, H.; Xu, Z.; Jiao, X.; Chen, X. First Detection of bla(NDM-1)-Haboring IncHI2 Plasmid in Escherichia coli Strain Isolated from Goose in China. Foodborne Pathog. Dis. 2023, 20, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hang, X.; Xiao, X.; Chu, W.; Li, X.; Liu, Y.; Li, X.; Zhou, Q.; Li, J. Co-occurrence of bla(NDM-1) and mcr-9 in a Conjugative IncHI2/HI2A Plasmid From a Bloodstream Infection-Causing Carbapenem-Resistant Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 756201. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.C.; da Silva, M.J.; dos Santos, P.R.; Barros, E.M.; Pereira Mde, O.; Seco, B.M.; Magagnin, C.M.; Leiroz, L.K.; de Oliveira, T.G.; de Faria-Júnior, C.; et al. Characterization of Tn3000, a Transposon Responsible for blaNDM-1 Dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob. Agents Chemother. 2015, 59, 7387–7395. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.W.; Song, Y.; Mu, Y.; Zhang, G.; Fu, S.; Wang, C.; Li, J.; Feng, J. Amplification of bla(NDM-1) mediated by ISCR1 confers hyperresistance to carbapenem. J. Glob. Antimicrob. Resist. 2022, 30, 180–182. [Google Scholar] [CrossRef]
- Zhao, Q.Y.; Zhu, J.H.; Cai, R.M.; Zheng, X.R.; Zhang, L.J.; Chang, M.X.; Lu, Y.W.; Fang, L.X.; Sun, J.; Jiang, H.X. IS26 Is Responsible for the Evolution and Transmission of bla(NDM)-Harboring Plasmids in Escherichia coli of Poultry Origin in China. mSystems 2021, 6, e0064621. [Google Scholar] [CrossRef]
- Shen, P.; Yi, M.; Fu, Y.; Ruan, Z.; Du, X.; Yu, Y.; Xie, X. Detection of an Escherichia coli Sequence Type 167 Strain with Two Tandem Copies of blaNDM-1 in the Chromosome. J. Clin. Microbiol. 2017, 55, 199–205. [Google Scholar] [CrossRef]
- Banerjee, K.; Sekar, P.; Krishnan, P.; Wattam, A.R.; Roy, S.; Hays, J.P.; Menezes, G.A. Whole genome sequence analysis of NDM-1, CMY-4, and SHV-12 coproducing Salmonella enterica serovar Typhimurium isolated from a case of fatal burn wound infection. Infect. Drug Resist. 2018, 11, 2491–2495. [Google Scholar] [CrossRef]
- Woh, P.Y.; Yeung, M.P.S.; Goggins, W.B., 3rd; Lo, N.; Wong, K.T.; Chow, V.; Chau, K.Y.; Fung, K.; Chen, Z.; Ip, M. Genomic Epidemiology of Multidrug-Resistant Nontyphoidal Salmonella in Young Children Hospitalized for Gastroenteritis. Microbiol. Spectr. 2021, 9, e0024821. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. EUCAST Clinical Breakpoint Table. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 20 March 2022).
- Clinical and Laboratory Standards Institute. Performance Standard for Antimicrobial Susceptibility Testing, 31st ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2021. [Google Scholar]
- Pierce, V.M.; Simner, P.J.; Lonsway, D.R.; Roe-Carpenter, D.E.; Johnson, J.K.; Brasso, W.B.; Bobenchik, A.M.; Lockett, Z.C.; Charnot-Katsikas, A.; Ferraro, M.J.; et al. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wang, Y.; Yu, J.; Li, S.; Zhang, Y.; Wang, H.; Lai, X.; Liu, D.; Mao, L.; Luo, Y.; et al. Bacterial characteristics of carbapenem-resistant Enterobacteriaceae (CRE) colonized strains and their correlation with subsequent infection. BMC Infect. Dis. 2021, 21, 638. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Lee, E.H.; Yoon, Y.; Chua, B.; Son, A. Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 2016, 120, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Hon, T.; Mars, K.; Young, G.; Tsai, Y.C.; Karalius, J.W.; Landolin, J.M.; Maurer, N.; Kudrna, D.; Hardigan, M.A.; Steiner, C.C.; et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data 2020, 7, 399. [Google Scholar] [CrossRef] [PubMed]
- Nakandala, U.; Masouleh, A.K.; Smith, M.W.; Furtado, A.; Mason, P.; Constantin, L.; Henry, R.J. Haplotype resolved chromosome level genome assembly of Citrus australis reveals disease resistance and other citrus specific genes. Hortic. Res. 2023, 10, uhad058. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Liu, S.; Liu, P.; Luo, X.; Zhao, J.; Yan, F.; Pan, Y.; Liu, J.; Zhai, Y.; Hu, G. Synergistic antibacterial activity of tetrandrine combined with colistin against MCR-mediated colistin-resistant Salmonella. Biomed. Pharmacother. 2022, 149, 112873. [Google Scholar] [CrossRef]
- Bitar, I.; Papagiannitsis, C.C.; Kraftova, L.; Marchetti, V.M.; Petinaki, E.; Finianos, M.; Chudejova, K.; Zemlickova, H.; Hrabak, J. Implication of different replicons in the spread of the VIM-1-encoding integron, In110, in Enterobacterales from Czech hospitals. Front. Microbiol. 2022, 13, 993240. [Google Scholar] [CrossRef]
- Izdebski, R.; Biedrzycka, M.; Urbanowicz, P.; Papierowska-Kozdój, W.; Dominiak, M.; Żabicka, D.; Gniadkowski, M. Multiple secondary outbreaks of NDM-producing Enterobacter hormaechei in the context of endemic NDM-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2022, 77, 1561–1569. [Google Scholar] [CrossRef]
- Hepner, S.; Kuleshov, K.; Tooming-Kunderud, A.; Alig, N.; Gofton, A.; Casjens, S.; Rollins, R.E.; Dangel, A.; Mourkas, E.; Sheppard, S.K.; et al. A high fidelity approach to assembling the complex Borrelia genome. BMC Genom. 2023, 24, 401. [Google Scholar] [CrossRef]
- Yan, S.; Zhang, W.; Li, C.; Liu, X.; Zhu, L.; Chen, L.; Yang, B. Serotyping, MLST, and Core Genome MLST Analysis of Salmonella enterica From Different Sources in China During 2004–2019. Front. Microbiol. 2021, 12, 688614. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; Liao, S.; Zhou, X.; Jia, C.; Feng, M.; Pan, H.; Ma, Z.; Yue, M. Prevalence and Genomic Investigation of Multidrug-Resistant Salmonella Isolates from Companion Animals in Hangzhou, China. Antibiotics 2022, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zeng, Z.; Liu, J.; Liu, C.; Pan, Y.; Zhang, Y.; Li, Y. Emergence of IncHI2 Plasmid-Harboring blaNDM-5 from Porcine Escherichia coli Isolates in Guangdong, China. Pathogens 2021, 10, 954. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.; Piedra-Carrasco, N.; Bartolomé, R.; Quintero-Zarate, J.N.; Larrosa, N.; Cornejo-Sánchez, T.; Prats, G.; Garcillán-Barcia, M.P.; de la Cruz, F.; González-Lopéz, J.J. Role of IncHI2 plasmids harbouring blaVIM-1, blaCTX-M-9, aac(6′)-Ib and qnrA genes in the spread of multiresistant Enterobacter cloacae and Klebsiella pneumoniae strains in different units at Hospital Vall d’Hebron, Barcelona, Spain. Int. J. Antimicrob. Agents 2012, 39, 514–517. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, J.; Li, X.P.; Fang, L.X.; Sun, R.Y.; He, Y.Z.; Lin, J.; Liao, X.P.; Feng, Y.; Liu, Y.H. Co-occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: Persistence of colistin resistance in Escherichia coli. Int. J. Antimicrob. Agents 2018, 51, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Jovcić, B.; Lepsanović, Z.; Begović, J.; Rakonjac, B.; Perovanović, J.; Topisirović, L.; Kojić, M. The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob. Agents Chemother. 2013, 57, 3405–3407. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Shen, W.; Zhang, Z.; Zhang, J.; Wang, G.; Xiang, L.; She, J.; Hu, X.; Zou, G.; Zhu, B.; et al. Whole-Genome Analysis of Two Copies of bla (NDM-1) Gene Carrying Acinetobacter johnsonii Strain Acsw19 Isolated from Sichuan, China. Infect. Drug Resist. 2020, 13, 855–865. [Google Scholar] [CrossRef]
- Chen, Y.T.; Liao, T.L.; Liu, Y.M.; Lauderdale, T.L.; Yan, J.J.; Tsai, S.F. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob. Agents Chemother. 2009, 53, 1235–1237. [Google Scholar] [CrossRef]
- Gaibani, P.; Bianco, G.; Amadesi, S.; Boattini, M.; Ambretti, S.; Costa, C. Increased bla(KPC) Copy Number and OmpK35 and OmpK36 Porins Disruption Mediated Resistance to Imipenem/Relebactam and Meropenem/Vaborbactam in a KPC-Producing Klebsiella pneumoniae Clinical Isolate. Antimicrob. Agents Chemother. 2022, 66, e0019122. [Google Scholar] [CrossRef]
- Gaibani, P.; Re, M.C.; Campoli, C.; Viale, P.L.; Ambretti, S. Bloodstream infection caused by KPC-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: Epidemiology and genomic characterization. Clin. Microbiol. Infect. 2020, 26, 516.e1–516.e4. [Google Scholar] [CrossRef]
- San Millan, A.; Toll-Riera, M.; Escudero, J.A.; Cantón, R.; Coque, T.M.; MacLean, R.C. Sequencing of plasmids pAMBL1 and pAMBL2 from Pseudomonas aeruginosa reveals a blaVIM-1 amplification causing high-level carbapenem resistance. J. Antimicrob. Chemother. 2015, 70, 3000–3003. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Mostafa, H.H.; Bergman, Y.; Ante, M.; Tekle, T.; Adebayo, A.; Beisken, S.; Dzintars, K.; Tamma, P.D. Progressive Development of Cefiderocol Resistance in Escherichia coli During Therapy is Associated With an Increase in blaNDM-5 Copy Number and Gene Expression. Clin. Infect. Dis. 2022, 75, 47–54. [Google Scholar] [CrossRef] [PubMed]

| Drug | MIC (mg/L) | ||||||
|---|---|---|---|---|---|---|---|
| ST_F0829S | ST_F0903R | ST_F0907R | ST_F0913R | ST_F0903Rtrans | ST_F0907Rtrans | ST_F0913Rtrans | |
| IPM | 0.125 | 16 | 16 | 16 | 16 | 16 | 16 |
| IMR | 0.25 | 16 | 16 | 16 | 16 | 16 | 16 |
| MEM | ≤0.06 | 64 | 128 | 64 | 64 | 128 | 64 |
| MEV | ≤0.06 | 128 | 128 | 64 | 128 | 128 | 64 |
| CAZ | 4 | >128 | >128 | >128 | >128 | >128 | >128 |
| CZA | 0.5 | >128 | >128 | >128 | >128 | >128 | >128 |
| COL | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| POL | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| AMK | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| FEP | 16 | >128 | >128 | >128 | >128 | >128 | >128 |
| ATM | 16 | 16 | 16 | 16 | 16 | 16 | 16 |
| CIP | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 |
| AZA | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 |
| ERA | 1 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 |
| SXT | 0.125 | >128 | >128 | >128 | >128 | >128 | >128 |
| TGC | 2 | 2 | 2 | 2 | 0.25 | 0.25 | 0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Zou, S.; Huang, Y.; Jian, C.; Liu, W.; Tian, L.; Gong, L.; Chen, Z.; Sun, Z.; Wang, Y. Salmonella Typhimurium with Eight Tandem Copies of blaNDM-1 on a HI2 Plasmid. Microorganisms 2024, 12, 20. https://doi.org/10.3390/microorganisms12010020
Song H, Zou S, Huang Y, Jian C, Liu W, Tian L, Gong L, Chen Z, Sun Z, Wang Y. Salmonella Typhimurium with Eight Tandem Copies of blaNDM-1 on a HI2 Plasmid. Microorganisms. 2024; 12(1):20. https://doi.org/10.3390/microorganisms12010020
Chicago/Turabian StyleSong, Huijuan, Siyu Zou, Yi Huang, Cui Jian, Weiyong Liu, Lei Tian, Lu Gong, Zhongju Chen, Ziyong Sun, and Yue Wang. 2024. "Salmonella Typhimurium with Eight Tandem Copies of blaNDM-1 on a HI2 Plasmid" Microorganisms 12, no. 1: 20. https://doi.org/10.3390/microorganisms12010020
APA StyleSong, H., Zou, S., Huang, Y., Jian, C., Liu, W., Tian, L., Gong, L., Chen, Z., Sun, Z., & Wang, Y. (2024). Salmonella Typhimurium with Eight Tandem Copies of blaNDM-1 on a HI2 Plasmid. Microorganisms, 12(1), 20. https://doi.org/10.3390/microorganisms12010020







