Abstract
The Vibrio genus includes bacteria widely distributed in aquatic habitats and the infections caused by these bacteria can affect a wide range of hosts. They are able to adhere to numerous surfaces, which can result in biofilm formation that helps maintain them in the environment. The involvement of the biofilm lifestyle in the virulence of Vibrio pathogens of aquatic organisms remains to be investigated. Vibrio harveyi ORM4 is a pathogen responsible for an outbreak in European abalone Haliotis tuberculata populations. In the present study, we used a dynamic biofilm culture technique coupled with laser scanning microscopy to characterize the biofilm formed by V. harveyi ORM4. We furthermore used RNA-seq analysis to examine the global changes in gene expression in biofilm cells compared to planktonic bacteria, and to identify biofilm- and virulence-related genes showing altered expression. A total of 1565 genes were differentially expressed, including genes associated with motility, polysaccharide synthesis, and quorum sensing. The up-regulation of 18 genes associated with the synthesis of the type III secretion system suggests that this virulence factor is induced in V. harveyi ORM4 biofilms, providing indirect evidence of a relationship between biofilm and virulence.
1. Introduction
Bacteria belonging to the Vibrio genus are ubiquitous in aquatic environments, being found in estuaries, marine coastal waters and sediments, and aquaculture settings. Some of these bacteria can form symbiotic or pathogenic relationships with a wide range of hosts. Thus, they alternate between survival and growth in aquatic environments and hosts [1]. One of the strategies adopted by Vibrio species to persist in changing environments is the conversion to a viable but non-culturable state (VBNC) [2]. Another strategy is the formation of biofilm which increases resistance to various stresses [3,4].
Biofilms are communities of microorganisms, usually attached to a surface, embedded in a self-produced extracellular matrix mainly composed of extracellular polysaccharides, DNA, and proteins. Changes in gene expression during biofilm formation allow the production of molecules and structures responsible for adhesion, aggregation, and community growth [5,6,7]. Some of the genes involved in biofilm formation are the same as those associated with host colonization. For Vibrio species, the presence of flagella and type IV pilus (T4P) is associated with bacterial attachment to surfaces, whether for biofilm formation or host colonization [8,9,10]. After adhesion onto a surface, bacteria begin the biofilm maturation process by producing an extracellular matrix. Different polysaccharides can be produced depending on the Vibrio species. The vps gene cluster (for Vibrio polysaccharide synthesis) was initially discovered in Vibrio cholerae but has since been identified in Vibrio fischeri and Vibrio tapetis [6,11,12]. VPS polysaccharides can constitute up to 50% of the total matrix of biofilms formed by V. cholerae and are mainly composed of glucose and galactose [13,14]. Vibrio parahaemolyticus possesses a cps (for capsular polysaccharides) gene locus and produces capsular polysaccharides (CPS) [15,16] involved in biofilm formation and containing various sugars such as glucose, galactose, or N-acetylglucosamine [15,17]. Polysaccharides synthesized by the products of the syp cluster (for symbiotic polysaccharides) in V. fischeri are necessary for the initiation of symbiotic relationships between V. fischeri and the squid Euprymna scolopes [18,19,20]. In a context of pathogenicity, the biofilm formation appears important for infections caused by the waterborne pathogen V. cholerae. There is evidence that V. cholerae can form aggregates similar to biofilms during infection, which may be crucial for pathogenesis and disease spread [21,22]. Indeed, biofilm growth promotes a hyperinfectious phenotype in V. cholerae through the up-regulation of several virulence genes [23,24]. Recently, a study demonstrated the production of biofilm around human immune cells as an aggressive cell-killing strategy [25].
In contrast to human pathogens, the role of the biofilm lifestyle in the virulence of Vibrio pathogens of marine organisms is still unclear. Vibrio harveyi strains are marine pathogens that affect a large range of marine vertebrates and invertebrates, inducing gastroenteritis, inflammation of the circulatory system, and skin lesions [26,27]. The V. harveyi ORM4 strain was the causative agent of a severe outbreak of the European abalone Haliotis tuberculata which occurred in natural populations and farmed stocks in the late 1990s [28]. This bacterium colonizes abalone gills, despite the presence of mucus, and was observed in aggregates on these organs, suggesting the formation of biofilm [29]. After a few hours of contact, the bacteria are found in the hemolymph of their host, where they are able to proliferate [29]. The immune depression generally associated with the abalone summer spawning period facilitates V. harveyi ORM4 multiplication into the hemolymph [30,31]. The mechanisms behind the bacterium’s pathogenicity against the European abalone are not well understood; a recent work demonstrated the involvement of quorum sensing (QS) and type III secretion system (T3SS) in the virulence of V. harveyi ORM4 but also showed that the presence of abalone serum, i.e., cell-free hemolymph, induces the production of biofilm by the bacteria [32]. These observations suggest that biofilm formation could be involved in the pathogenicity of V. harveyi ORM4 against European abalone. Unfortunately, no study described the molecular mechanisms involved in biofilm formation by V. harveyi.
Bacterial biofilm formation is generally quantified in polystyrene microplates, but this technique does not allow observation of the biofilm architecture [33]. Several other devices have been set up to grow and observe biofilms. Among these, the development of flow cell culture techniques coupled with the use of confocal laser scanning microscopy (CLSM) enables a more complete characterization, such as visualization of the three-dimensional structure, labeling of the various biofilm constituents (bacteria, matrix components), and quantification of various parameters (maximum and average thicknesses, biovolume), and allows monitoring of the biofilm formation over time [34]. The objective of the present study was to provide a comprehensive view of biofilm formation by the virulent V. harveyi ORM4 strain using microscopic observations coupled to a transcriptomic analysis allowing the identification of differentially expressed genes in biofilm compared to planktonic bacteria.
2. Materials and Methods
2.1. Strains and Culture Medium
V. harveyi ORM4 [28] was used for RNA-seq experiments and V. harveyi ORM4-GFP, carrying the pFD086 plasmid (TrimR) [32], was only used for biofilm microscopic observations. V. harveyi ORM4 was grown in LBS (LB containing 20 g/L NaCl) and trimethoprim (10 µg/mL) was added to maintain pFD086 inside V. harveyi ORM4-GFP cells.
2.2. Planktonic Growth
One colony of V. harveyi ORM4 was used to inoculate 5 mL of LBS which was incubated overnight at 20 °C with shaking. Next, 100 µL of this pre-culture was used to inoculate 5 mL LBS in triplicate and the tubes were incubated for 8 h at 20 °C with shaking. Then, 50 µL of each triplicate was used to inoculate in triplicate 20 mL LBS contained in a 100 mL Erlenmeyer flask, which was then incubated at 20 °C with shaking for 15 h (bacteria were in stationary phase). Finally, 1 mL aliquots of planktonic cells were harvested by centrifugation (6000× g, 6 min, 4 °C), and pellets from each culture were used for RNA extraction.
2.3. Biofilm Culture
V. harveyi ORM4 and V. harveyi ORM4-GFP biofilms were grown at 20 °C under hydrodynamic conditions in a three-channel flow cell (1 mm × 40 mm × 44 mm, Biocentrum, DTU, Lyngby, Denmark) [34], on a glass coverslip (24 × 50 st1 [KnittelGlasser, Braunschweig, Germany]). The system was assembled as described previously [35] and sterilized for 2 h with bleach before being washed with a flow of autoclaved distilled water overnight (running volume: 800 mL). The next day, the system was filled with LBS. A quantity of 300 µL of an overnight culture of V. harveyi ORM4 or V. harveyi ORM4-GFP diluted to an OD600nm of 0.1 in artificial seawater (30 g/L sea salts [Sigma-Aldrich, Saint-Louis, MO, USA]) was injected in each channel with a 1 mL syringe and a needle (25G, 0.5 × 16 mm [Terumo, Tokyo, Japan]). An attachment step of the bacteria onto the glass surface was performed for 2 h without medium flow. After this step, LBS was applied at a flow rate of 2.5 mL/h for 24 h during which the system was incubated at 20 °C. The resulting biofilms were observed by CLSM (strain V. harveyi ORM4-GFP) or used to extract RNAs (strain V. harveyi ORM4) as described below.
2.4. Confocal Laser Scanning Microscopy (CLSM)
Biofilms formed by V. harveyi ORM4-GFP were observed by monitoring the GFP fluorescence. Different dyes were used to observe the matrix components after biofilm growth. Polysaccharides and proteins were, respectively, stained using 100 µM Calcofluor White (Sigma Aldrich, Saint-Louis, MO, USA) [36] and 1X FilmTracer SYPRO Ruby (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). Extracellular DNA (eDNA) was stained with 1 µM of 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO) (Invitrogen, CarIsbad, CA, USA) [37]. All dyes were prepared in artificial seawater (ASW: 30 g/L sea salts [Sigma Aldrich, Saint-Louis, MO, USA]) and staining was done by injecting in each channel 300 µL of appropriate dye(s) after biofilm growth. Biofilms were stained for 15 min in the dark in the absence of flow, before washing with an LBS flow for 10 min (2.5 mL/h). Stained biofilms were observed with a confocal laser scanning microscope (LSM 710, Zeiss, Oberkochen, Germany). GFP was excited at 488 nm and fluorescence emission was detected between 500 and 550 nm. FilmTracer SYPRO Ruby and DDAO were excited at 488 and 633 nm, and detected between 604–714 nm and 637–716 nm, respectively. These two dyes have close emission spectra that do not allow differentiating them; they could thus not be used on the same biofilms. Calcofluor White was excited at 400 nm and fluorescence emission was detected between 498 and 501 nm. Images were acquired at intervals of 1 µm throughout the whole depth of the biofilm. At least three image stacks from each eight of independent experiments (24 stacks in total) were analyzed using the COMSTAT v1 2000 software [38].
2.5. RNA Extraction
Total RNA was isolated for each condition (planktonic or biofilm) from three independent experiments, using the MasterPure Complete RNA purification kit (Lucigen, Biosearch Technologies, LGC, Kidlington, UK). For V. harveyi ORM4 planktonic cells, RNA extraction was performed from cell pellets obtained as described in Section 2.2, following the supplier’s protocol. For V. harveyi ORM4 biofilm cells, we followed the procedure described by Rodrigues et al., 2018 [6] to ensure that RNA was only extracted from biofilm-forming cells. Briefly, directly after microscopic observations of V. harveyi ORM4 biofilms, the flow was progressively increased from 2.5 mL/h to 25 mL/h for 5 min. The tubing at the end of the flow cell was subsequently cut and the content was aseptically recovered by manual flushing with 1 mL cold LBS in a syringe and placed on ice until RNA extraction. The glass slide of each flow cell was then observed to ensure the absence of residual cells. The cellular suspension was centrifuged (6000× g, 6 min, 4 °C) and the pellet was used for RNA extraction. The amount and quality of total RNA were assessed using a NanoPhotometer N60 (Implen, Munich, Germany) spectrophotometer.
2.6. RNA-Sequencing and Data Analysis
Ribosomal RNA depletion, cDNA library preparation, and Illumina sequencing were performed by GATC laboratory (Eurofins, GATC Biotech, Konstanz, Germany).
Raw data analysis was performed using the Galaxy France platform (Galaxy France, https://usegalaxy.fr, accessed on September 2022). Raw data quality was analyzed with the FastQC tool (version 0.73) [39] and sequence cleaning was performed with the Trimmomatic tool (version 0.38.1) [40]. The sequences were then mapped onto the V. harveyi ORM4 genome (GenBank assembly accession number GCA_963920535) using the Boowtie2 software (Version 2.4.5) [41]. The abundance of each transcript was calculated using the union model with the HTseq-count tool (Version 0.9.1) [42]. Subsequently, statistical analysis of the differentially expressed genes was performed with the R packages SARTools and DESeq2 [43]. These packages allow data normalization and differential expression tests for each feature between conditions and raw p-value adjustment. Genes were considered significantly differentially expressed if the p-value adjusted (padj) by False Discovery Rate (FDR) correction [44] was less than 0.05. The raw data obtained in this study have been deposited in the SRA database and are accessible under PRJNA1010657 accession number. The RNA-seq data were plotted on a volcano plot with the negative log of the adjusted p-value on the y axis and the log2 of the fold change between the biofilm and planktonic condition on the x axis using the ggplot2 v3.4.4 package [45]. R version 4.2.0 was used for all analysis.
2.7. mRNA Quantification by Reverse Transcription Followed by Quantitative PCR (RT-qPCR)
The RNA-seq results were validated by RT-qPCR assays. Total RNA was converted to cDNA using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems, Foster City, CA, USA). Eight genes were chosen for which primers (Table 1) were designed using the Primer Express v3 software (Applied Biosystems, Foster City, CA, USA). qPCR reactions were then performed with a 7300 RT PCR System (Applied Biosystems), using Master Mix PCR SYBR Green (Applied Biosystems). Gene level expression was obtained by the comparative CT (2−∆∆CT) method [46], using 16S ribosomal RNA (rRNA) as endogenous control.

Table 1.
Primers used in RT-qPCR. Primers are oriented 5′ to 3′.
3. Results and Discussion
3.1. Biofilm Formation by V. harveyi
3.1.1. V. harveyi ORM4-GFP Biofilm Presents a Non-Uniform Structure
In order to characterize the structure and composition of the biofilm of a pathogenic V. harveyi strain, the ORM4-GFP strain was grown in a flow cell system. The first microscopic observations showed that V. harveyi ORM4-GFP was able to produce a biofilm on a glass slide in the presence of a constant LBS flow (2.5 mL/h) during 24 h at 20 °C (Figure 1). Our observations highlighted a curved structure (convex shape) of the biofilm: the highest biofilm thickness was observed in the center of the glass slide, in the same orientation as the LBS flow (Figure 1B). On both sides, the biofilm thickness was decreased (Figure 1B) and bacteria were found in monolayer until their total absence on the edges of the glass slide [47].
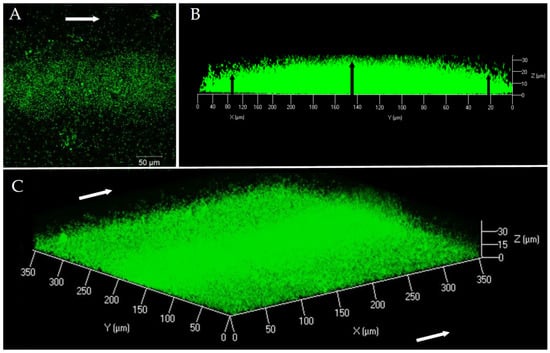
Figure 1.
V. harveyi ORM4-GFP biofilm observation at 40× magnification by CLSM after 24 h of culture at 20 °C under a constant flow of LBS (2.5 mL/h). Top view (A), side view (B), and three-dimensional view (C). White arrows represent the LBS flow direction and black arrows indicate the difference in biofilm thickness between the center and the ends.
This biofilm structure was found on the whole length of the flow cell channel. Biofilms of V. harveyi ORM4-GFP reached a biovolume of 5.4 ± 0.7 µm3/µm2 and a maximal thickness of 27.5 ± 1.7 µm. However, due to this curved structure, the average thickness was only 9.5 ± 1 µm. Such a structure is unusual since we could not find any other example in the literature. Actually, CLSM observations of biofilms formed under the same conditions by other Vibrio species revealed biofilms that uniformly covered the surfaces [48,49].
3.1.2. Matrix Component Distribution
In addition to the observation of V. harveyi ORM4-GFP biofilm-forming cells, the exploration of the biofilm matrix produced by V. harveyi ORM4-GFP cells was performed with specific dyes allowing the visualization of matrix components. Biofilms were stained with Calcofluor White (which binds to β1–3 and β1–4 polysaccharides), DDAO (which binds to eDNA), and FilmTracer SYPRO Ruby (which binds to proteins). CLSM observations enabled the visualization of the distribution of these different components surrounding the bacterial cells. β-polysaccharides and proteins were observed across the entire biofilm thickness (Figure 2B,C), while eDNA were mostly localized in the lower half of the biofilm (Figure 2A). The presence of these different components was previously reported in V. cholerae, V. parahaemolyticus, and V. tapetis biofilms [11,48,49,50,51].
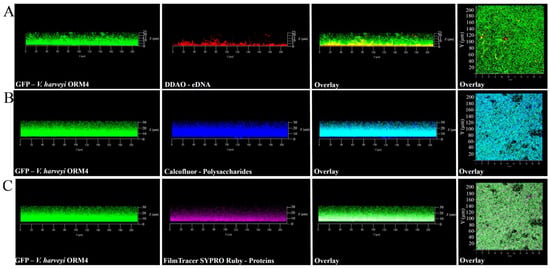
Figure 2.
Observations with the CLSM at 40× magnification of matrix components of 24 h V. harveyi ORM4-GFP biofilms. The matrix components were stained using the following dyes: eDNA with DDAO (A), polysaccharides with Calcofluor (B), and proteins with FilmTracer SYPRO Ruby (C). The right panels show a top view of the biofilm.
Although V. harveyi ORM4-GFP forms biofilms with a curved structure, this does not appear to be induced by matrix structuring since the matrix components were observed across the entire width of the biofilm (Figure 2). Several hypotheses can be put forward to explain this particular biofilm structure, such as the nature of the substrate, the availability of nutrients, and the pH [52,53,54]. However, V. harveyi ORM4-GFP cells grown under static conditions in glass-bottom microplates and in the same medium yielded uniformly thick biofilms which evenly covered the glass surface at the bottom of the wells [47]. Altogether, this suggests that the flow may induce the curved structure of the biofilm and that this particular biofilm structure did not result from a non-homogeneous accumulation of the matrix.
3.2. Transcriptomic Comparison between Planktonic and Biofilm Cells
In order to obtain an insight into the molecular mechanisms behind biofilm formation by V. harveyi ORM4, an RNA-seq transcriptome analysis was performed using V. harveyi ORM4 planktonic cells as the control group and V. harveyi ORM4 biofilm cells as the experimental group. Analysis of 12 million sequences obtained for each condition revealed that 1565 genes (29% of the total V. harveyi ORM4 genes) were significantly differentially expressed (adjusted p-value < 0.05), out of which 868 and 697 were, respectively, down- and up-regulated in biofilm cells (Figure 3A).
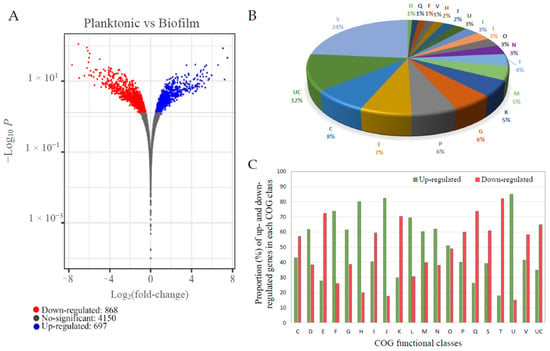
Figure 3.
Summary of the V. harveyi ORM4 RNA-seq analysis results. Volcano plot representation of differentially expressed genes (DEGs) in V. harveyi ORM4 biofilm cells versus planktonic cells. The blue and red dots represent significantly up- and down-regulated genes (adjusted p-value < 0.05), respectively, and the grey dots represent the non-differentially expressed genes (adjusted p-value > 0.05) (A). Distribution of DEGs in V. harveyi ORM4 biofilm cells in functional classes (COG) (B). Percentage of up- and down-regulated genes for each COG class in biofilm cells versus planktonic cells (C). COG classes: C: Energy production and conversion. D: Cell cycle control, cell division, and chromosome partitioning. E: Amino acid transport and metabolism. F: Nucleotide transport and metabolism. G: Carbohydrate transport and metabolism. H: Coenzyme transport and metabolism. I: Lipid transport and metabolism. J: Translation, ribosomal structure, and biogenesis. K: Transcription. L: Replication, recombination, and repair. M: Cell wall/membrane/envelope biogenesis. N: Cell motility. O: Posttranslational modification, protein turnover, and chaperones. P: Inorganic ion transport and metabolism. Q: Secondary metabolite biosynthesis, transport, and catabolism. S: Function unknown. T: Signal transduction mechanisms. U: Intracellular trafficking, secretion, and vesicular transport. V: Defense mechanisms. UC: Unclassified.
In order to validate the results obtained by RNA-seq analysis, we selected eight genes involved in biofilm formation (flp, luxR, and sypH) [18,55,56], virulence (ompV, vscN, and vpa0450) [57,58,59], or encoding unknown proteins (HORM4_920008, HORM4_1130010) and their relative expression levels (Figure 4) were evaluated using RT-qPCR. These genes showed a wide range of fold-change values in RNA-seq, and were up-regulated (vscN, vpa0450 and ompV), down-regulated (luxR, sypH and HORM4_920008), or not differentially expressed (flp and HORM4_1130010). These eight genes were found to have high Pearson correlation coefficients (0.93) between their fold-change levels assessed by RT-qPCR and by RNA-seq, thereby validating the RNA-seq findings (Figure 4B).
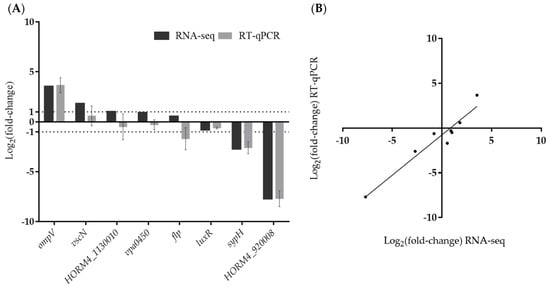
Figure 4.
Validation of RNA-seq data by RT-qPCR. Comparison of log2(fold-change) (log2FC) gene expression values obtained in RT-qPCR and RNA-seq (A). Correlation of log2FC values determined by RNA-seq and RT-qPCR in biofilm-forming V. harveyi ORM4 versus planktonic cells (B).
3.3. Differential Expression Analysis of Biofilm- and Virulence-Related Genes
To obtain further insights into biofilm formation by a pathogenic strain of V. harveyi, and into the possible links between biofilm and virulence, we then focused our analysis of the RNA-seq data on genes encoding proteins which could be associated with these two phenotypes.
3.3.1. DEGs Involved in Motility and Attachment
A necessary step prior to successful biofilm formation or infection is the ability to swim and adhere to surfaces. Bacteria switch from a planktonic swimming state, associated with the presence of a polar flagellum, to attachment via filamentous appendages such as lateral flagella or pili. A total of 3% of the DEGs were associated with motility (COG class N), among which 62% were up-regulated in biofilm-forming cells (Figure 3B,C). In biofilm cells, genes encoding protein forming key parts of the polar flagellum (hook, basal body, and motor) were significantly down-regulated, while lateral flagellum genes were up-regulated (Figure 5 and Table S1). The lack of transcriptional activity linked with swimming motility involving the polar flagellum in mature biofilm cells has been previously described for Pseudomonas aeruginosa [60,61], which is in agreement with our observations. In parallel, bacteria attach to surfaces by the production of proteinaceous appendages such as pili. In Vibrio species, several types of pili are involved in early attachment to abiotic surfaces [12]. In the genome of V. harveyi ORM4, we found two sets of genes encoding type IV pili. The first one is a gene locus (HORM4_370011 to HORM4_370026) homologous to the gene cluster for the mannose-sensitive haemagglutinin pilus (MSHA pilus) of V. cholerae O1 El Tor [62], and the second one includes genes similar to the genes encoding the PapC porin and the PapD chaperone of the P pilus of V. parahaemolyticus RIMD 2210633 (HORM4_920006 and HORM4_920007, respectively).
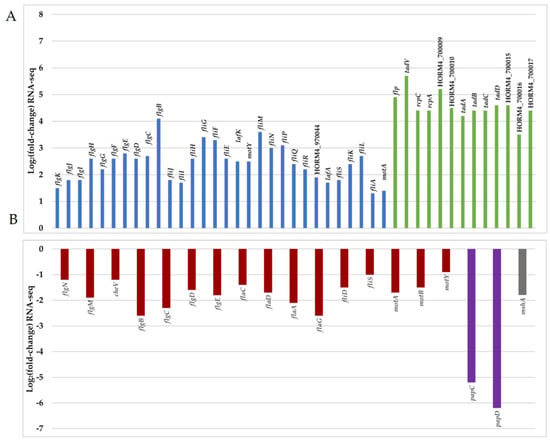
Figure 5.
Representation of Log2FC values of 64 DEGs (adjusted p-value < 0.05) related to motility and attachment in V. harveyi ORM4 biofilm cells versus planktonic cells. Positive (A) and negative (B) values of Log2FC are represented in two separated graphics. Red bar represents DEGs encoding the polar flagellum, blue bar represents DEGs encoding lateral flagella, green bar represents DEGs encoding a putative Tad pilus (gene cluster HORM4_700005 to HORM4_700017), and purple and grey bars represent, respectively, DEGs encoding the P pilus (HORM4_920006 and HORM4_920007) and the MSHA pilin protein (HORM4_370020).
In V. cholerae, the MSHA pilus plays important roles in the persistence of the bacteria by mediating their attachment to nutritive substrates, in biofilm formation, and in chitin utilization [63,64]. V. parahaemolyticus also produces an MSHA pilus, which is used to anchor the bacteria to chitinous surfaces or produce biofilms, whereas the P pilus is associated with host adhesion [8,65]. In V. harveyi ORM4 biofilm cells, the HORM4_370020 gene encoding the pilin protein MshA is 3.8-fold down-regulated and the genes encoding PapC and PapD were 46 to 100-fold down-regulated (log2FC between −6.2 and −5.2) (Figure 5 and Table S1). Although the encoding genes are down-regulated in mature biofilms, the MSHA and P pili could participate in the V. harveyi ORM4 anchoring to the glass slide during an earlier stage of biofilm formation, since stained proteins are localized at the biofilm basis (Figure 2). This would be consistent with previous reports showing that Vibrio bacteria defective in the production of these pili failed to adhere or to progress from the monolayer formation to three-dimensional biofilms [17,66,67].
An expert annotation of the V. harveyi ORM4 genome was done using the MaGe tool of the Microscope platform [68], leading to the identification of three distinct loci putatively encoding a tight adherence (Tad) pilus (Table S2). One of these loci (HORM4_700005 to HORM4_700017) is fully induced in V. harveyi ORM4 biofilm cells, with up to 53-fold (log2FC between 3.5 and 5.7) induction levels (Figure 5 and Table S1). Genes homologous to Tad pilus biogenesis genes are widespread in Vibrionaceae [69]. In V. vulnificus, the Tad pilus has been shown to be involved both in adhesion and in biofilm maturation since cells lacking the Tad pilin required more time to attach to a surface and to form a biofilm of a substantial biomass [55,70]. Concerning V. harveyi ORM4, our transcriptomic results suggested the involvement of the Tad pilus in the process of biofilm maturation, possibly in its structuration as is the case for the type IV pili in P. aeruginosa biofilm or fimbriae in E. coli and Salmonella enterica serovar Typhimurium biofilms [71,72].
3.3.2. DEGs Involved in Polysaccharide Production
Staining of the biofilm matrix revealed the presence of β1–3 and β1–4 polysaccharides on the whole thickness of V. harveyi ORM4 biofilm (Figure 2). Several studies have reported the involvement of vps and syp gene clusters in Vibrio biofilm formation [11,12,20]. Within the V. harveyi ORM4 genome, we identified a gene cluster (HORM4_940043 to HORM4_940054) which encodes proteins sharing over 67% amino acid identities with the proteins produced by the V. parahaemolyticus cps cluster (vpa1403 to vpa1413) [15] (Table S2). However, there was no significant difference in their expression between biofilm and planktonic conditions (adjusted p-value ≥ 0.05) (Table S3). Nevertheless, we furthermore found in the V. harveyi ORM4 genome a gene cluster homologous to the syp cluster, initially identified in V. fischeri but conserved in many pathogenic Vibrio species such as V. parahaemolyticus, V. tapetis, and V. vulnificus (Table 2) [6,12]. In the V. harveyi ORM4 genome, all the syp genes were found (HORM4v2_640011 to HORM4_640026), except the genes encoding SypE, SypF, and SypM. SypE and SypF are both regulatory proteins [73,74,75], while the role of SypM remains to be defined. Among the 15 identified syp genes, 5 were down-regulated in V. harveyi ORM4 biofilm cells (Table 2).

Table 2.
syp gene expression in V. harveyi ORM4 biofilm cells (adjusted p-value < 0.05).
Our transcriptomic analysis revealed that, despite the presence of polysaccharide synthesis genes, these genes are either not more expressed or down-regulated in V. harveyi ORM4 after 24 h of biofilm formation compared to the planktonic condition. These results suggest that if an over-production of polysaccharides occurs in the process of biofilm formation by V. harveyi ORM4, it likely occurs earlier. This would be consistent with previous reports showing that the synthesis of polysaccharides starts during the adhesion step [76,77].
3.3.3. Genes Involved in Quorum Sensing
In bacteria, the control of gene expression in response to cell population density is known as quorum sensing (QS) [78,79]. In the V. harveyi ORM4 genome, we were able to identify genes encoding the three QS systems previously described in Vibrio campbellii: luxM/luxN, luxS/luxPQ, and cqsA/cqsS [56]. Among these genes, cqsS was weakly down-regulated (log2FC: −0.7) in V. harveyi ORM4 biofilm cells, whereas the other genes were not differentially expressed (Table 3). In V. campbellii, the different QS systems converge to the transcriptional regulator proteins LuxO and LuxR [56]. V. harveyi ORM4 possesses both a luxO [32] and a luxR gene. Our data showed that both luxO and luxR were weakly down-regulated (log2FC: −0.8 for both genes) in V. harveyi ORM4 biofilm cells (Table 3). In V. tapetis or V. parahaemolyticus biofilms, transcriptomic studies described the induction of QS-related genes [6,7]. In contrast to these species, the expression of V. harveyi ORM4 QS-related genes was found to be weakly down-regulated or unchanged in comparison with planktonic cells (Table 3), suggesting a low QS activity in V. harveyi ORM4 biofilm cells. It remains possible that an up-regulation of these genes occurs earlier in the process of biofilm formation.

Table 3.
Differential expression of genes involved in quorum sensing when V. harveyi ORM4 produced biofilm in comparison with planktonic cells.
3.3.4. Type III Secretion System (T3SS)-Associated Genes
In a previous study [32], we identified a 33-kb genetic region homologous to the region encoding the T3SS1 in V. parahaemolyticus [79,80]. The deletion of exsA, encoding the major regulator of the T3SS1 genes, abolished the bacterial virulence against European abalones [32]. Our RNA-seq analysis revealed an up-regulation (two- to four-fold) of 18 of the 49 T3SS genes in biofilm cells (Table 4). Proteins encoded by these genes are part of the structure of the T3SS or are involved in its functioning. This includes genes encoding the VscC, VscD, VscI, VscJ, VscT, VscR, and VscQ proteins, which compose the basal body of the T3SS and are located on the bacterial inner and outer membranes [58,81]. In addition, vscN encoding an ATPase and the vscF gene enabling synthesis of the needle found in the extracellular space were also over-expressed (Table 4) [58,82]. On the other hand, genes encoding the ExsA, ExsC, ExsD, and ExsE proteins, which compose the cascade of the T3SS regulation, were not differentially expressed. Genes encoding the VopB and VopD proteins, required for translocation of T3SS effectors across the host cell membrane, showed no difference in expression between the two culture conditions [83]. These results suggest that the T3SS is assembled and that the biofilm lifestyle induces the T3SS in V. harveyi ORM4. To our knowledge, this is the first time that the potential involvement of T3SS in bacterial biofilm formation has been suggested. This secretion system is mainly associated with bacterial virulence [10,32,58]. It is therefore surprising to observe an induction of T3SS genes in a monospecies V. harveyi biofilm. Induced genes encode structural proteins that make up the basal body and the needle found in the extracellular environment [58,82]. If the T3SS needle is produced, it is possible that the apparatus is stained by the FilmTracer SYPRO Ruby dye we used to stain the V. harveyi ORM4 biofilm matrix (Figure 2).

Table 4.
Differentially expressed genes belonging to the T3SS in V. harveyi ORM4 biofilm cells.
Among the secretion systems, the type VI secretion system (T6SS) was the only one described as being involved in both biofilm production and bacterial virulence. Many bacterial pathogens, including P. aeruginosa and V. cholerae, are known to use their T6SS during infection [84]. Some studies have shown an induction of the expression of T6SS genes in biofilms of P. aeruginosa or V. tapetis, including genes encoding T6SS effectors [6,85]. Gallique et al. hypothesized that secretion of T6SS proteins in Pseudomonas fluorescens biofilm could participate in cell communication [86]. This hypothesis cannot be made in the case of T3SS since, to our knowledge, only eukaryotic cells are targeted by effectors [84,87,88,89]. Even if its role remains unknown in biofilm formation, it is important to note that T3SS genes are induced by this lifestyle in V. harveyi ORM4 cells. The infection of European abalone by V. harveyi ORM4 starts by the colonization of the abalone gills on which the bacteria were observed in aggregates [29]. Here, the induction of T3SS genes in V. harveyi ORM4 biofilm may suggest that this lifestyle could be the first step of the infection.
4. Conclusions
In summary, this is the first report describing a V. harveyi biofilm and the molecular changes that occurred during the biofilm production. In mature V. harveyi biofilms, the gene expression pattern was largely modified compared to planktonic stationary phase bacteria since 29% of the genes were differentially expressed, and these DEGs were assigned to various functional classes. Among the functions related to biofilm formation, the impairment of swimming motility was indicated by the down-regulation of polar flagellum-related genes. Our transcriptomic analysis revealed a 53-fold induction of the gene Tad pilus in V. harveyi ORM4 cell biofilm, a type IV pilus involved in both biofilm formation and host colonization in V. vulnificus [55,70]. Finally, we also found that genes for T3SS are up-regulated while earlier work demonstrated its requirement for V. harveyi virulence [32], suggesting that the biofilm formation might be involved in the infection of European abalone by V. harveyi. Future work will be undertaken to study the role of T3SS in biofilm formation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12010186/s1, Table S1: Expression of genes involved in motility and attachment in V. harveyi ORM4 biofilm cells (adjusted p-value < 0.05); Table S2: V. harveyi ORM4 Tad proteins and amino acidic percentage of identity with Tad proteins of V. vulnificus CMCP6. Table S3: V. harveyi ORM4 CPS proteins and amino acidic percentage of identity with CPS proteins of V. parahaemolyticus RIMD2210633; Table S4: Expression of genes associated to the cps cluster in V. harveyi ORM4 biofilm-forming cells.
Author Contributions
Conceptualization, S.R., A.D., A.M., F.D. and C.P.; methodology, S.R., A.D., A.B. and A.M.; validation, S.R. and A.D.; data analysis and curation, A.M. and S.R.; investigation, A.M. and S.R.; writing—original draft preparation, A.M.; writing—review and editing, S.R., A.D., A.B., A.M., F.D. and C.P.; supervision, S.R., A.B. and A.D.; project administration, A.D. and C.P.; funding acquisition, A.D. and C.P. All authors have read and agreed to the published version of the manuscript.
Funding
AM was the recipient of a doctoral fellowship (PhD COMPARO project) co-funded by the Université de Bretagne Sud (UBS) and the Région Bretagne. This work was supported by the ISblue project, Interdisciplinary graduate school for the blue planet (ANR-17-EURE-0015) and co-funded by a grant from the Région Bretagne under the project GIS EUROPOLE MER 2009. The LBCM is supported by European FEDER.
Data Availability Statement
The RNA-seq raw data obtained in this study are available at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA1010657 (sample accession numbers: SAMN37193765, SAMN37193766, SAMN37193767, SAMN37193768, SAMN37193769 and SAMN37193770).
Acknowledgments
The LABGeM (CEA/Genoscope and CNRS UMR8030) and the France Génomique and French Bioinformatics Institute national infrastructures (funded as part of Investissement d’Avenir program managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) are acknowledged for support within the MicroScope annotation platform.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
Capsular polysaccharides (cps), cluster of orthologous genes (COG), confocal laser scanning microscopy (CLSM), 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO), differentially expressed genes (DEGs), extracellular DNA (eDNA), green fluorescent protein (GFP), lysogeny broth (LB), mannose-sensitive haemagglutinin pilus (MSHA), p-value adjusted (padj), quorum sensing (QS), ribosomal RNA (rRNA), symbiotic polysaccharides (syp), tight adherence (Tad), trimethoprim resistance (TrimR), type IV pilus (T4P), type III secretion system (T3SS), type VI secretion system (T6SS), viable but non-culturable state (VBNC), Vibrio polysaccharide synthesis (vps).
References
- Thompson, F.L.; Iida, T.; Swings, J. Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 2004, 68, 403–431. [Google Scholar] [CrossRef]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The Importance of the Viable but Non-Culturable State in Human Bacterial Pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef]
- Hung, D.T.; Zhu, J.; Sturtevant, D.; Mekalanos, J.J. Bile Acids Stimulate Biofilm Formation in Vibrio cholerae. Mol. Microbiol. 2006, 59, 193–201. [Google Scholar] [CrossRef]
- He, H.; Cooper, J.N.; Mishra, A.; Raskin, D.M. Stringent Response Regulation of Biofilm Formation in Vibrio cholerae. J. Bacteriol. 2012, 194, 2962–2972. [Google Scholar] [CrossRef]
- Beloin, C.; Ghigo, J.-M. Finding Gene-Expression Patterns in Bacterial Biofilms. Trends Microbiol. 2005, 13, 16–19. [Google Scholar] [CrossRef]
- Rodrigues, S.; Paillard, C.; Van Dillen, S.; Tahrioui, A.; Berjeaud, J.-M.; Dufour, A.; Bazire, A. Relation between Biofilm and Virulence in Vibrio tapetis: A Transcriptomic Study. Pathogens 2018, 7, 92. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, P.; Liu, P.; Ou, J. Comparative Transcriptome Analysis Reveals Regulatory Factors Involved in Vibrio parahaemolyticus Biofilm Formation. Front. Cell. Infect. Microbiol. 2022, 12, 917131. [Google Scholar] [CrossRef]
- Frischkorn, K.R.; Stojanovski, A.; Paranjpye, R. Vibrio Parahaemolyticus Type IV Pili Mediate Interactions with Diatom-Derived Chitin and Point to an Unexplored Mechanism of Environmental Persistence: V. parahaemolyticus and Chitin. Environ. Microbiol. 2013, 15, 1416–1427. [Google Scholar] [CrossRef]
- Luo, G.; Huang, L.; Su, Y.; Qin, Y.; Xu, X.; Zhao, L.; Yan, Q. flrA, flrB and flrC Regulate Adhesion by Controlling the Expression of Critical Virulence Genes in Vibrio alginolyticus. Emerg. Microbes Infect. 2016, 5, e85. [Google Scholar] [CrossRef]
- Aagesen, A.M.; Phuvasate, S.; Su, Y.-C.; Häse, C.C. Persistence of Vibrio parahaemolyticus in the Pacific Oyster, Crassostrea gigas, Is a Multifactorial Process Involving Pili and Flagella but Not Type III Secretion Systems or Phase Variation. Appl. Environ. Microbiol. 2013, 79, 3303–3305. [Google Scholar] [CrossRef]
- Yildiz, F.H.; Schoolnik, G.K. Vibrio cholerae O1 El Tor: Identification of a Gene Cluster Required for the Rugose Colony Type, Exopolysaccharide Production, Chlorine Resistance, and Biofilm Formation. Proc. Natl. Acad. Sci. USA 1999, 96, 4028–4033. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, F.H.; Visick, K.L. Vibrio Biofilms: So Much the Same yet so Different. Trends Microbiol. 2009, 17, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2017, 3, 223–247. [Google Scholar]
- Ruhal, R.; Kataria, R. Biofilm Patterns in Gram-Positive and Gram-Negative Bacteria. Microbiol. Res. 2021, 251, 126829. [Google Scholar] [CrossRef]
- Güvener, Z.T.; McCarter, L.L. Multiple Regulators Control Capsular Polysaccharide Production in Vibrio parahaemolyticus. J. Bacteriol. 2003, 185, 5431–5441. [Google Scholar] [CrossRef]
- Enos-Berlage, J.L.; McCarter, L.L. Relation of Capsular Polysaccharide Production and Colonial Cell Organization to Colony Morphology in Vibrio parahaemolyticus. J. Bacteriol. 2000, 182, 5513–5520. [Google Scholar] [CrossRef]
- Enos-Berlage, J.L.; Guvener, Z.T.; Keenan, C.E.; McCarter, L.L. Genetic Determinants of Biofilm Development of Opaque and Translucent Vibrio parahaemolyticus: V. parahaemolyticus Biofilm Development. Mol. Microbiol. 2004, 55, 1160–1182. [Google Scholar] [CrossRef]
- Yip, E.S.; Grublesky, B.T.; Hussa, E.A.; Visick, K.L. A Novel, Conserved Cluster of Genes Promotes Symbiotic Colonization and σ54-Dependent Biofilm Formation by Vibrio fischeri: Syp, a Novel Symbiosis Locus in Vibrio fischeri. Mol. Microbiol. 2005, 57, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Yip, E.S.; Geszvain, K.; DeLoney-Marino, C.R.; Visick, K.L. The Symbiosis Regulator RscS Controls the Syp Gene Locus, Biofilm Formation and Symbiotic Aggregation by Vibrio fischeri. Mol. Microbiol. 2006, 62, 1586–1600. [Google Scholar] [CrossRef]
- Shibata, S.; Yip, E.S.; Quirke, K.P.; Ondrey, J.M.; Visick, K.L. Roles of the Structural Symbiosis Polysaccharide (syp) Genes in Host Colonization, Biofilm Formation, and Polysaccharide Biosynthesis in Vibrio fischeri. J. Bacteriol. 2012, 194, 6736–6747. [Google Scholar] [CrossRef]
- Millet, Y.A.; Alvarez, D.; Ringgaard, S.; Von Andrian, U.H.; Davis, B.M.; Waldor, M.K. Insights into Vibrio cholerae Intestinal Colonization from Monitoring Fluorescently Labeled Bacteria. PLoS Pathog. 2014, 10, e1004405. [Google Scholar] [CrossRef]
- Faruque, S.M.; Biswas, K.; Udden, S.M.N.; Ahmad, Q.S.; Sack, D.A.; Nair, G.B.; Mekalanos, J.J. Transmissibility of Cholera: In vivo-Formed Biofilms and Their Relationship to Infectivity and Persistence in the Environment. Proc. Natl. Acad. Sci. USA 2006, 103, 6350–6355. [Google Scholar] [CrossRef] [PubMed]
- Tamayo, R.; Patimalla, B.; Camilli, A. Growth in a Biofilm Induces a Hyperinfectious Phenotype in Vibrio cholerae. Infect. Immun. 2010, 78, 3560–3569. [Google Scholar] [CrossRef]
- Gallego-Hernandez, A.L.; DePas, W.H.; Park, J.H.; Teschler, J.K.; Hartmann, R.; Jeckel, H.; Drescher, K.; Beyhan, S.; Newman, D.K.; Yildiz, F.H. Upregulation of Virulence Genes Promotes Vibrio cholerae Biofilm Hyperinfectivity. Proc. Natl. Acad. Sci. USA 2020, 117, 11010–11017. [Google Scholar] [CrossRef] [PubMed]
- Vidakovic, L.; Mikhaleva, S.; Jeckel, H.; Nisnevich, V.; Strenger, K.; Neuhaus, K.; Raveendran, K.; Ben-Moshe, N.B.; Aznaourova, M.; Nosho, K.; et al. Biofilm Formation on Human Immune Cells Is a Multicellular Predation Strategy of Vibrio cholerae. Cell 2023, 186, 2690–2704. [Google Scholar] [CrossRef] [PubMed]
- Austin, B.; Zhang, X.-H. Vibrio harveyi: A Significant Pathogen of Marine Vertebrates and Invertebrates. Lett. Appl. Microbiol. 2006, 43, 119–124. [Google Scholar] [CrossRef]
- Darshanee Ruwandeepika, H.A.; Sanjeewa Prasad Jayaweera, T.; Paban Bhowmick, P.; Karunasagar, I.; Bossier, P.; Defoirdt, T. Pathogenesis, Virulence Factors and Virulence Regulation of Vibrios Belonging to the Harveyi Clade: Virulence and Pathogenesis of Vibrios. Rev. Aquac. 2012, 4, 59–74. [Google Scholar] [CrossRef]
- Nicolas, J.; Basuyaux, O.; Mazurié, J.; Thébault, A. Vibrio carchariae, a Pathogen of the Abalone Haliotis tuberculata. Dis. Aquat. Org. 2002, 50, 35–43. [Google Scholar] [CrossRef]
- Cardinaud, M.; Barbou, A.; Capitaine, C.; Bidault, A.; Dujon, A.M.; Moraga, D.; Paillard, C. Vibrio harveyi Adheres to and Penetrates Tissues of the European Abalone Haliotis tuberculata within the First Hours of Contact. Appl. Environ. Microbiol. 2014, 80, 6328–6333. [Google Scholar] [CrossRef]
- Travers, M.-A.; Le Goïc, N.; Huchette, S.; Koken, M.; Paillard, C. Summer Immune Depression Associated with Increased Susceptibility of the European Abalone, Haliotis tuberculata to Vibrio harveyi Infection. Fish Shellfish. Immunol. 2008, 25, 800–808. [Google Scholar] [CrossRef]
- Cardinaud, M.; Dheilly, N.M.; Huchette, S.; Moraga, D.; Paillard, C. The Early Stages of the Immune Response of the European Abalone Haliotis tuberculata to a Vibrio harveyi Infection. Dev. Comp. Immunol. 2015, 51, 287–297. [Google Scholar] [CrossRef]
- Morot, A.; El Fekih, S.; Bidault, A.; Le Ferrand, A.; Jouault, A.; Kavousi, J.; Bazire, A.; Pichereau, V.; Dufour, A.; Paillard, C.; et al. Virulence of Vibrio harveyi ORM4 towards the European Abalone Haliotis tuberculata Involves Both Quorum Sensing and a Type III Secretion System. Environ. Microbiol. 2021, 23, 5273–5288. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hébraud, M.; Jaglic, Z.; et al. Critical Review on Biofilm Methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed]
- Tolker-Nielsen, T.; Sternberg, C. Methods for Studying Biofilm Formation: Flow Cells and Confocal Laser Scanning Microscopy. In Pseudomonas Methods and Protocols; Filloux, A., Ramos, J.-L., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2014; Volume 1149, pp. 615–629. ISBN 978-1-4939-0472-3. [Google Scholar]
- Weiss Nielsen, M.; Sternberg, C.; Molin, S.; Regenberg, B. Pseudomonas aeruginosa and Saccharomyces cerevisiae Biofilm in Flow Cells. J. Vis. Exp. 2011, 15, 2383. [Google Scholar]
- Chen, M.-Y.; Lee, D.-J.; Tay, J.-H.; Show, K.-Y. Staining of Extracellular Polymeric Substances and Cells in Bioaggregates. Appl. Microbiol. Biotechnol. 2007, 75, 467–474. [Google Scholar] [CrossRef]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A Characterization of DNA Release in Pseudomonas aeruginosa Cultures and Biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of Biofilm Structures by the Novel Computer Program Comstat. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on September 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Morot, A. Elucidation des Mécanismes Moléculaires de Colonisation D’organismes Marins par le Pathogène Vibrio harveyi. Ph.D. Thesis, Université Bretagne Sud, Lorient, France, 2023. [Google Scholar]
- Bilecen, K.; Yildiz, F.H. Identification of a Calcium-Controlled Negative Regulatory System Affecting Vibrio cholerae Biofilm Formation. Environl. Microbiol. 2009, 11, 2015–2029. [Google Scholar] [CrossRef]
- Rodrigues, S.; Paillard, C.; Le Pennec, G.; Dufour, A.; Bazire, A. Vibrio tapetis, the Causative Agent of Brown Ring Disease, Forms Biofilms with Spherical Components. Front. Microbiol. 2015, 6, 1384. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, J.J.; Qian, H.; Tan, L.; Zhang, Z.; Liu, H.; Pan, Y.; Zhao, Y. Insights into the Role of Extracellular DNA and Extracellular Proteins in Biofilm Formation of Vibrio parahaemolyticus. Front. Microbiol. 2020, 11, 813. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.C.N.; Karplus, K.; Schoolnik, G.K.; Yildiz, F.H. Identification and Characterization of RbmA, a Novel Protein Required for the Development of Rugose Colony Morphology and Biofilm Structure in Vibrio cholerae. J. Bacteriol. 2006, 188, 1049–1059. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Henares, B.; Xu, Y.; Boon, E. A Nitric Oxide-Responsive Quorum Sensing Circuit in Vibrio harveyi Regulates Flagella Production and Biofilm Formation. Int. J. Mol. Sci. 2013, 14, 16473–16484. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Duong-Nu, T.-M.; Jeong, K.; Hong, S.H.; Puth, S.; Kim, S.Y.; Tan, W.; Lee, K.H.; Lee, S.E.; Rhee, J.H. A Stealth Adhesion Factor Contributes to Vibrio vulnificus Pathogenicity: Flp Pili Play Roles in Host Invasion, Survival in the Blood Stream and Resistance to Complement Activation. PLoS Pathog. 2019, 15, e1007767. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum Sensing Signal–Response Systems in Gram-Negative Bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Kaur, D.; Mukhopadhaya, A. Outer Membrane Protein OmpV Mediates Salmonella enterica Serovar Typhimurium Adhesion to Intestinal Epithelial Cells via Fibronectin and α1β1 Integrin. Cell. Microbiol. 2020, 22, e13172. [Google Scholar] [CrossRef]
- Izoré, T.; Job, V.; Dessen, A. Biogenesis, Regulation, and Targeting of the Type III Secretion System. Structure 2011, 19, 603–612. [Google Scholar] [CrossRef]
- Broberg, C.A.; Zhang, L.; Gonzalez, H.; Laskowski-Arce, M.A.; Orth, K. A Vibrio Effector Protein Is an Inositol Phosphatase and Disrupts Host Cell Membrane Integrity. Science 2010, 329, 1660–1662. [Google Scholar] [CrossRef]
- Whiteley, M.; Bangera, M.G.; Bumgarner, R.E.; Parsek, M.R.; Teitzel, G.M.; Lory, S.; Greenberg, E.P. Gene Expression in Pseudomonas aeruginosa Biofilms. Nature 2001, 413, 860–864. [Google Scholar] [CrossRef]
- Guttenplan, S.B.; Kearns, D.B. Regulation of Flagellar Motility during Biofilm Formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef]
- Marsh, J.W.; Taylor, R.K. Genetic and Transcriptional Analyses of the Vibrio cholerae Mannose-Sensitive Hemagglutinin Type 4 Pilus Gene Locus. J. Bacteriol. 1999, 181, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Meibom, K.L.; Li, X.B.; Nielsen, A.T.; Wu, C.-Y.; Roseman, S.; Schoolnik, G.K. The Vibrio cholerae Chitin Utilization Program. Proc. Natl. Acad. Sci. USA 2004, 101, 2524–2529. [Google Scholar] [CrossRef] [PubMed]
- Watnick, P.I.; Fullner, K.J.; Kolter, R. A Role for the Mannose-Sensitive Hemagglutinin in Biofilm Formation by Vibrio cholerae El Tor. J. Bacteriol. 1999, 181, 3606–3609. [Google Scholar] [CrossRef]
- Hospenthal, M.K.; Costa, T.R.D.; Waksman, G. A Comprehensive Guide to Pilus Biogenesis in Gram-Negative Bacteria. Nat. Rev. Microbiol. 2017, 15, 365–379. [Google Scholar] [CrossRef]
- Watnick, P.I.; Lauriano, C.M.; Klose, K.E.; Croal, L.; Kolter, R. The Absence of a Flagellum Leads to Altered Colony Morphology, Biofilm Development and Virulence in Vibrio cholerae O139. Mol. Microbiol. 2001, 39, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Paranjpye, R.N.; Strom, M.S. A Vibrio vulnificus Type IV Pilin Contributes to Biofilm Formation, Adherence to Epithelial Cells, and Virulence. Infect. Immun. 2005, 73, 1411–1422. [Google Scholar] [CrossRef]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An Integrated Platform for the Annotation and Exploration of Microbial Gene Functions through Genomic, Pangenomic and Metabolic Comparative Analysis. Nucleic Acids Res. 2020, 48, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Tomich, M.; Planet, P.J.; Figurski, D.H. The Tad Locus: Postcards from the Widespread Colonization Island. Nat. Rev. Microbiol. 2007, 5, 363–375. [Google Scholar] [CrossRef]
- Pu, M.; Rowe-Magnus, D.A. A Tad Pilus Promotes the Establishment and Resistance of Vibrio vulnificus Biofilms to Mechanical Clearance. NPJ Biofilms Microbiomes 2018, 4, 10. [Google Scholar] [CrossRef]
- Van Schaik, E.J.; Giltner, C.L.; Audette, G.F.; Keizer, D.W.; Bautista, D.L.; Slupsky, C.M.; Sykes, B.D.; Irvin, R.T. DNA Binding: A Novel Function of Pseudomonas aeruginosa Type IV Pili. J. Bacteriol. 2005, 187, 1455–1464. [Google Scholar] [CrossRef]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Romling, U. The Multicellular Morphotypes of Salmonella Typhimurium and Escherichia coli Produce Cellulose as the Second Component of the Extracellular Matrix. Mol. Microbiol. 2001, 39, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.R.; Visick, K.L. Control of Biofilm Formation and Colonization in Vibrio fischeri: A Role for Partner Switching?: Partner Switching in Vibrio fischeri. Environ. Microbiol. 2010, 12, 2051–2059. [Google Scholar] [CrossRef]
- Morris, A.R.; Visick, K.L. Inhibition of SypG-Induced Biofilms and Host Colonization by the Negative Regulator SypE in Vibrio fischeri. PLoS ONE 2013, 8, e60076. [Google Scholar] [CrossRef]
- Ludvik, D.A.; Bultman, K.M.; Mandel, M.J. Hybrid Histidine Kinase BinK Represses Vibrio Fischeri Biofilm Signaling at Multiple Developmental Stages. J. Bacteriol. 2021, 203, e0015521. [Google Scholar] [CrossRef]
- Tsuneda, S.; Aikawa, H.; Hayashi, H.; Yuasa, A.; Hirata, A. Extracellular Polymeric Substances Responsible for Bacterial Adhesion onto Solid Surface. FEMS Microbiol. Lett. 2003, 223, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Sharma, P.K.; Busscher, H.J.; Van Der Mei, H.C.; Krom, B.P. Role of Extracellular DNA in Initial Bacterial Adhesion and Surface Aggregation. Appl. Environ. Microbiol. 2010, 76, 3405–3408. [Google Scholar] [CrossRef]
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular Control of the Synthesis and Activity of the Bacterial Luminescent System. J. Bacteriol. 1970, 104, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Zhou, X.; Konkel, M.E.; Call, D.R. Regulation of Type III Secretion System 1 Gene Expression in Vibrio parahaemolyticus Is Dependent on Interactions between ExsA, ExsC, and ExsD. Virulence 2010, 1, 260–272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Deng, Y.; Feng, J.; Guo, Z.; Chen, H.; Wang, B.; Hu, J.; Lin, Z.; Su, Y. Functional Characterization of VscCD, an Important Component of the Type III Secretion System of Vibrio harveyi. Microb. Pathog. 2021, 157, 104965. [Google Scholar] [CrossRef]
- Casselli, T.; Lynch, T.; Southward, C.M.; Jones, B.W.; DeVinney, R. Vibrio parahaemolyticus Inhibition of Rho Family GTPase Activation Requires a Functional Chromosome I Type III Secretion System. Infect. Immun. 2008, 76, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Park, K.-S.; Ueta, M.; Iida, T.; Honda, T. Identification of Proteins Secreted via Vibrio parahaemolyticus Type III Secretion System 1. Infect. Immun. 2006, 74, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Virul. Mech. Bact. Pathog. 2016, 4, 213–239. [Google Scholar]
- Chen, L.; Zou, Y.; Kronfl, A.A.; Wu, Y. Type VI Secretion System of Pseudomonas aeruginosa Is Associated with Biofilm Formation but Not Environmental Adaptation. MicrobiologyOpen 2020, 9, e991. [Google Scholar] [CrossRef] [PubMed]
- Gallique, M.; Decoin, V.; Barbey, C.; Rosay, T.; Feuilloley, M.G.J.; Orange, N.; Merieau, A. Contribution of the Pseudomonas fluorescens MFE01 Type VI Secretion System to Biofilm Formation. PLoS ONE 2017, 12, e0170770. [Google Scholar] [CrossRef]
- Salomon, D.; Guo, Y.; Kinch, L.N.; Grishin, N.V.; Gardner, K.H.; Orth, K. Effectors of Animal and Plant Pathogens Use a Common Domain to Bind Host Phosphoinositides. Nat. Commun. 2013, 4, 2973. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Krachler, A.M.; Broberg, C.A.; Li, Y.; Mirzaei, H.; Gilpin, C.J.; Orth, K. Type III Effector VopC Mediates Invasion for Vibrio Species. Cell Rep. 2012, 1, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, J.; Deng, Y.; Huang, W.; Ren, C.; Call, D.R.; Hu, C. The Vibrio alginolyticus T3SS Effectors, Val1686 and Val1680, Induce Cell Rounding, Apoptosis and Lysis of Fish Epithelial Cells. Virulence 2018, 9, 318–330. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).