Abstract
A relevant aspect in the epidemiology of Tegumentary Leishmaniasis (TL) are the Leishmania parasites carrying a viral endosymbiont, Leishmania RNA Virus 1 (LRV1), a dsRNA virus. Leishmania parasites carrying LRV1 are prone to causing more severe TL symptoms, increasing the likelihood of unfavorable clinical outcomes. LRV1 has been observed in the cultured strains of five L. (Viannia) species, and host specificity was suggested when studying the LRV1 from L. braziliensis and L. guyanensis strains. The coevolution hypothesis of LRV1 and Leishmania was based on phylogenetic analyses, implying an association between LRV1 genotypes, Leishmania species, and their geographic origins. This study aimed to investigate LRV1 specificity relative to Leishmania (Viannia) species hosts by analyzing LRV1 from L. (Viannia) species. To this end, LRV1 was screened in L. (Viannia) species other than L. braziliensis or L. guyanensis, and it was detected in 11 out of 15 L. naiffi and two out of four L. shawi. Phylogenetic analyses based on partial LRV1 genomic sequencing supported the hypothesis of host specificity, as LRV1 clustered according to their respective Leishmania species’ hosts. These findings underscore the importance of investigating Leishmania and LRV1 coevolution and its impact on Leishmania (Viannia) species dispersion and pathogenesis in the American Continent.
1. Introduction
Endosymbiotic viruses are found in different Leishmania species. Although some Leishmania species present an association with viruses from the families Leishbunyaviridae [1] and Narnaviridae [2], several Leishmania species human pathogens bear viruses from the Totiviridae family, and these are named Leishmaniavirus; it is a double-strand RNA virus classified into two species, Leishmaniavirus 1 (LRV1) and Leishmaniavirus 2 (LRV2), and the two species are associated with Leishmania (Viannia) and Leishmania (Leishmania) species, respectively [3]. Similar genetic relationships observed for Leishmania species are observed for LRV1 and LRV2 [4].
Cutaneous leishmaniasis (CL) is a neglected disease that primarily affects impoverished populations in developing countries [5]. In the American Continent, Brazil holds the highest incidence of CL. The disease involves several Leishmania species: L. (Leishmania) amazonensis and others within the subgenus Viannia—L. (Viannia) guyanensis, L. (V.) braziliensis, L. (V.) shawi, L. (V.) lainsoni, L. (V.) naiffi, and L. (V.) lindenbergi [6]. A significant aspect of CL epidemiology in Brazil, and possibly beyond, is the presence of LRV1 [7,8]. This viral endosymbiont may influence the outcome and severity of CL [8,9,10,11,12,13,14]. Previously, in an experimental model, it was demonstrated that Leishmania infections harboring high burdens of LRV1 exhibit a metastatic infection profile. This is instigated by the recognition of the virus via the host’s Toll-like Receptor 3 pathway, coupled with the ability to undermine the host’s immune response via the same pathway. Consequently, this leads to the sustained presence of LRV1+ parasites in cutaneous lesions [13].
LRV1 is associated with exacerbating infections, driving the mucosal form of leishmaniasis. This viral role relies on the production of Type I interferons (Type I IFNs) by macrophages [15,16]. LRV1 subverts innate immunity, negatively regulating the NLRP3 inflammasome and thereby promoting parasite survival and chronic disease [16]. Notably, the presence of LRV1 increases the risk of mucosal development by nearly threefold compared to CL cases without the virus [8]. However, while some studies establish a link between LRV1 and mucosal manifestations, others show associations with therapeutic failure [17,18,19]. Differences in the parasites circulating in the different regions were evaluated and/or in the LRV1 infecting these parasites, which are possible explanations for the observed differences among some studies.
Previous studies have highlighted host-specific interactions between LRV and Leishmania species. A monophyletic group was observed for LRV2 from L. major and L. aethiopica, raising the idea of the co-evolution of LRV2 and Leishmania (Leishmania) species [20]. The co-evolution of LRV1 and L. (Viannia) species was also proposed based on evidence of host specificity between L. (Viannia) species and LRV1 genotypes [21].
The genomic sequences of LRV1 from various Leishmania species suggest specificity in L. (Viannia) species–LRV1 interactions [21,22]. For instance, LRV1 from L. (V.) shawi was akin to LRV1 from L. guyanensis, corroborating the similarity observed between these two Leishmania species [21,23]. Although LRV1 was detected already in the cultivated strains of L. naiffi and L. panamensis [18,24,25,26], there is no information on the phylogenetic relationship of viruses from these species compared to LRV1 from other species. LRV1 was already associated with human infections caused by L. lainsoni [8] and L. peruviana [27], but not in the cultivated strains from these species, thus limiting the comparisons with LRV1 from other species.
Given the importance of Leishmania-LRV1 symbiosis in the epidemiology of cutaneous and mucosal leishmaniasis, a comprehensive understanding of the virus’s diversity and spread within parasite populations is crucial. Thus, our study aimed to examine the presence of LRV1 in various strains of Brazilian L. (Viannia) species, excluding L. braziliensis and L. guyanensis, since many LRV1 sequences are available for these two species. To enhance our understanding of LRV1’s interactions across Leishmania species, a comparative analysis was performed between the available sequences of L. braziliensis and L. guyanensis, along with newly acquired LRV1 data from L. naiffi and L. shawi.
2. Materials and Methods
2.1. Leishmania Culture
Strains from L. naiffi (n = 18), L. lainsoni (n = 4), L. shawi (n = 4), L. lindenbergi (n = 3), and L. utingensis (n = 1)—from different geographic regions and available at the Leishmania Collection of Fiocruz (CLIOC)—were screened (Table S1). Parasites were grown in NNN (Novy–MacNeal–Nicolle), and Schneider medium supplemented with 20% fetal bovine serum and incubated in a BOD (biochemical oxygen demand) incubator at 25 °C until reaching the average amount of 5 × 106 parasites.
Cultures were centrifuged at 1400× g for 10 min at 4 °C, resuspended in DNA/RNA Shield™ (Zymo Research Corporation—Irvine, CA, USA), and stored at −20 °C until RNA extraction.
2.2. RNA Extraction and cDNA Synthesis
The RNA of the strains was extracted using the TRIzol® reagent (Invitrogen—Carlsbad, CA, USA). RNA concentration and purity were determined using a NanoDrop® 2000 spectrophotometer (Thermo Scientific™—Wilmington, CA, USA). Reverse transcription was performed using 2 μg of RNA and the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™—Foster City, CA, USA) following the manufacturer’s recommendations.
2.3. LRV1 Detection
An LRV1 screening protocol was performed using primers LRV F–5′-ATGCCTAAGAGTTTGGATTCG-3′ and LRV R–5′-ACAACCAGACGATTGCTGTG-3′ [8] (Figure 1). L. guyanensis (MHOM/BR/1975/M4147) and L. braziliensis (MHOM/BR/1975/M2903) strains were used as positive and negative controls for the experiments, respectively [28,29]. For Leishmania HSP70 fragment amplification, which was used as an endogenous control of all RT-PCR reactions, primers Hsp70cF 5-GGACGAGATCGAGCGCATGGT-3′ and Hsp70cR 5′-TCCTTCGACGCCTCCTGGTTG-3′ were used [30]. For both reactions, a final volume of 50 μL was used: 10X Buffer (1X), MgCl2 (1.5 mM), dNTPs (0.2 mM), Primer F (0.2 μM), Primer R (0.2 mM), Taq Platinum (1.0 U/μL), and 1 μL of cDNA. RT-PCR was performed at 94 °C for 2 min, 35 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 45 s, with a final extension phase at 72 °C for 5 min.
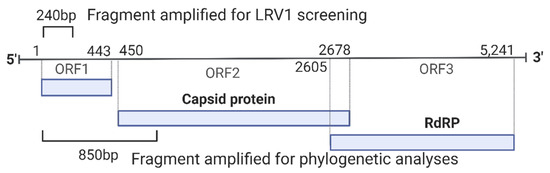
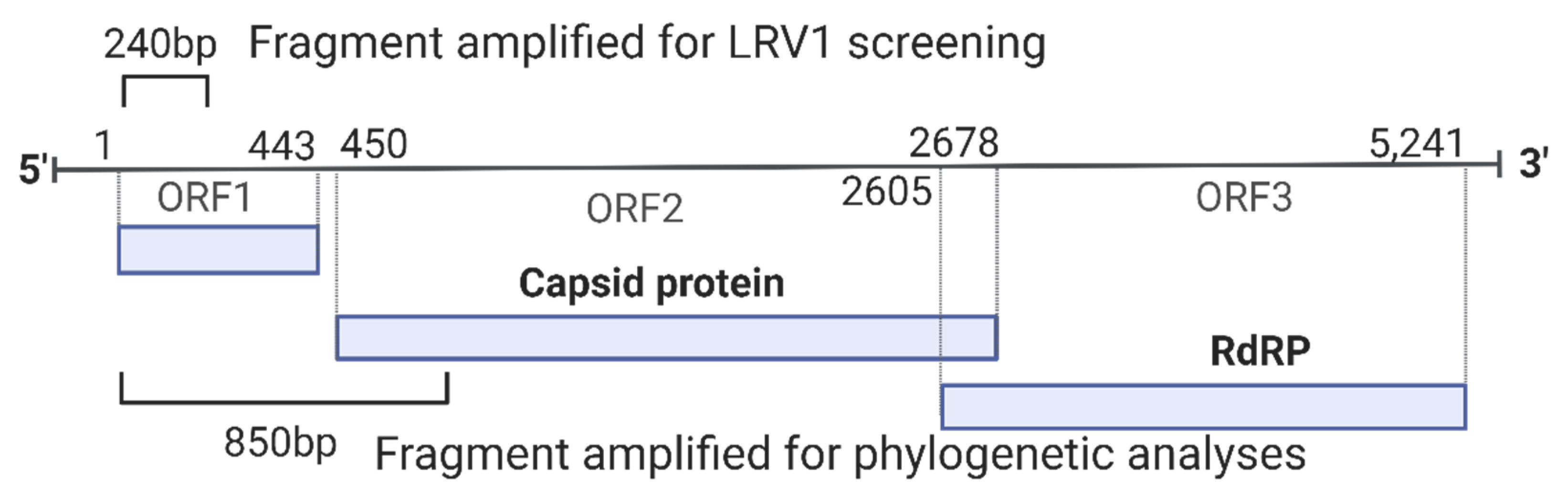
Figure 1.
Schematic representation of the genome of LRV1 showing the region amplified for each primer used for screening (240 bp) and phylogenetic analyses (850 bp). RdRP: RNA-dependent RNA polymerase. Genome length based on NCBI RefSeq NC_003601.
Positive strains for LRV1 (n = 12; 11 L. naiffi and 1 L. shawi) were submitted to another PCR reaction that aimed to amplify a fragment of approximately 850 base pairs for LRV1, which is a phylogenetically informative fragment. For this, primers LRV1 F orf1 5′-ATGCCTAAGAGTTTGGATTCG-3′ and LRV1 R orf2 5′-AATCAATTTTCCCAGTCATGC-3′ [21] were used, amplifying a fragment corresponding to a part of the orf1 region and the beginning of the orf2 region, including the portion responsible for encoding the viral capsid protein (Figure 1). A final volume of 50 μL was used: 10X Buffer (1X), MgCl2 (1.5 mM), dNTPs (0.2 mM), Primer F (0.2 μM), Primer Rg (0.2 mM), Taq Platinum (1.0 U/µL), and 3 µL of cDNA. RT-PCR was performed at 95 °C for 2 min, followed by 35 cycles at 95 °C for 30 s, 57 °C for 45 s, and 72 °C for 45 s, with a final extension phase at 72 °C for 5 min. All RT-PCR products were stained using GelRed® (Biotium—Fremont, CA, USA) and visualized in a 2% agarose gel.
2.4. Sequencing
For sequencing, 45 μL of RT-PCR products (850 bp) was purified using the Wizard® SV Gel and RT-PCR Clean-Up System kit (Promega—Madison, WI, USA) following the manufacturer’s recommendations. Sanger sequencing was performed on the Fiocruz DNA Sequencing Platform—Rio de Janeiro (RPT01A).
2.5. Analyses of LRV1 Sequences
The consensus sequences were created using the BioEdit program [31]. Then, the inference of the best model to build the phylogenetic tree was performed in MEGA X [32] software, where the Tamura 3-parameter model with gamma distribution parameter (G) and invariable sites (I) presented the lowest BIC (Bayesian information criterion) score. The tree was constructed using the maximum likelihood method, employing 10,000 replicates (bootstrap).
In addition, to increase the chances of finding the most parsimonious connections, networks using NeighborNet were built using the SplitsTree program [33]. For that, we used the MEGAX function to exclude sites with missing/ambiguous data and those with sequence gaps, which resulted in sequences that were 509-nucleotide-long and were common across the groups of each species.
The analyzed sequences correspond to the LRV1 detected in L. naiffi and L. shawi strains that were isolated from sandflies and humans from Amazonian regions, and these were already deposited in GenBank (Table 1). The analysis was conducted by comparing the new LRV1 sequences to those that are already available (Table S1).

Table 1.
Leishmania (Viannia) strains from different species screened for the presence of the viral LRV1 endosymbiont and information on accession for LRV1 sequences obtained for the positive strains.
3. Results
3.1. LRV1 Was Not Detected in All L. (Viannia) Species Analyzed but Was Frequent in L. naiffi Strains
All RT-PCR reactions were performed with the promastigotes of available strains identified as L. naiffi (n = 18), L. shawi (n = 4), L. lainsoni (n = 4), L. lindenbergi (n = 3), and L. utingensis (n = 1). Of these, seven L. naiffi, three L. shawi, and all L. lainsoni, L. lindenbergi and L. utingensis strains were negative for the virus. LRV1 was detected in 11 L. naiffi strains (61%) and one analyzed L. shawi. The geographical distributions of all analyzed strains that are negative or positive for LRV1 are demonstrated in Figure 2. After quality checking, sequences ranging from 632 nt to 799 nt (Table 1) were employed in the analyses described below.
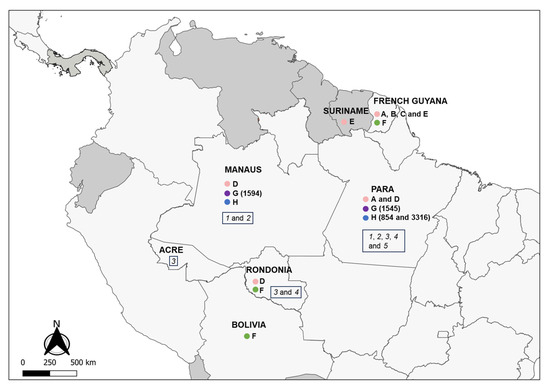
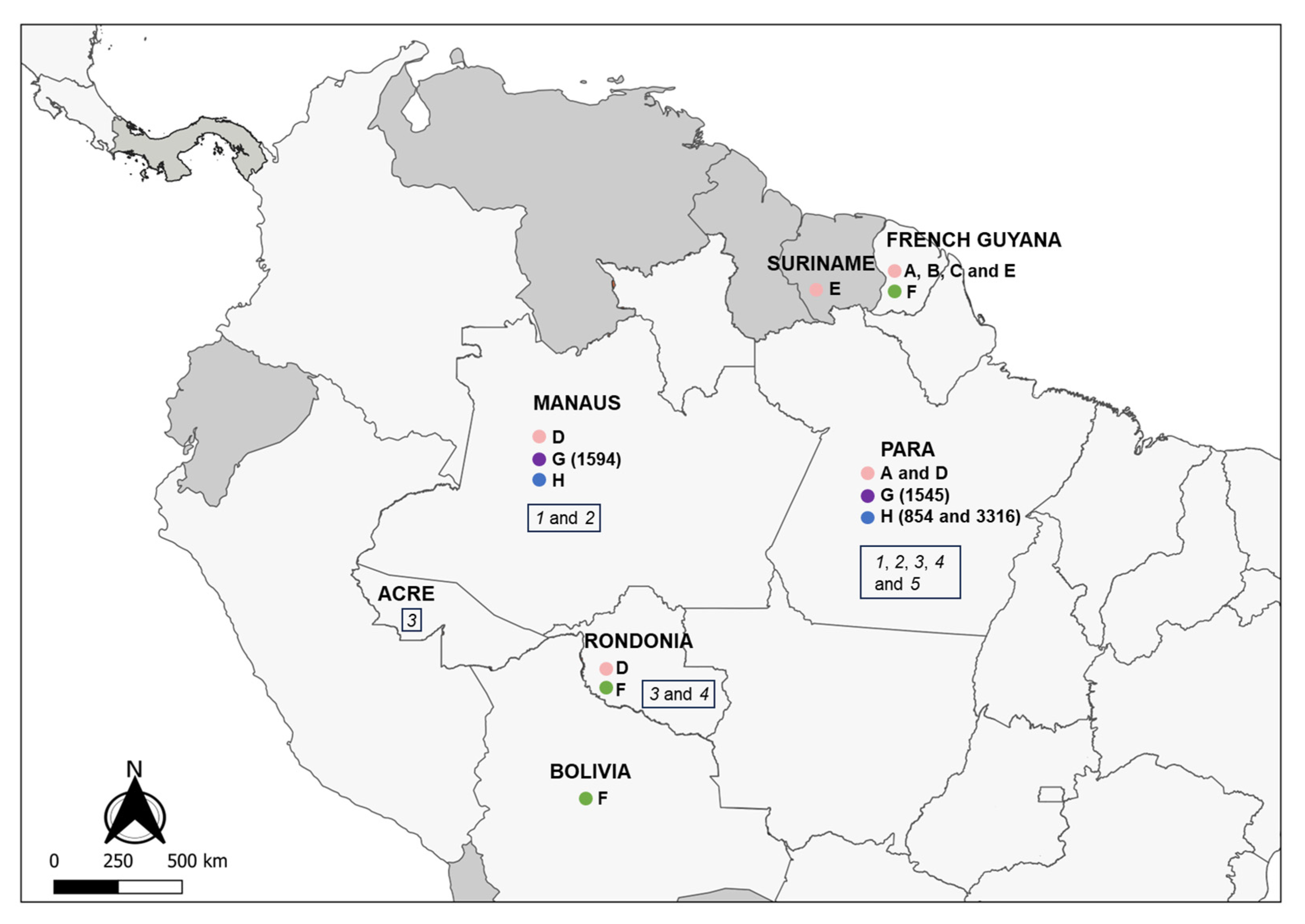
Figure 2.
Partial map of South and Central America indicating the countries and Brazilian states where LRV1 has been detected in the Leishmania (Viannia) spp. analyzed in this study. Colored circles refer to species presenting LRV1: pink = L. guyanensis; green = L. braziliensis; purple = L. shawi; blue = L. naiffi. Letters (A to H) indicate groups that are defined according to the phylogenetic analyses (Figure 3); numbers indicate the IOCL for specific strains (Table 1) in such groups; italic numbers inside the rectangles indicate the negative strains analyzed: 1 = L. naiffi; 2 = L. shawi; 3 = L. lainsoni; 4 = L. lindenbergi; 5 = L. utingensis.
3.2. Variability of LRV1 Diversity across Leishmania Host Species
In addition to the sequences obtained in the present study, another 47 LRV1 sequences from L. guyanensis (n = 35), L. braziliensis (n = 11), and L. shawi (n = 1) strains available on GenBank were included in the analyses (Table S1), corresponding to the sequences reported from studies in French Guiana [22], Bolivia [34,35], and Brazil [21,33,35]. Considering the LRV1 sequences analyzed, the same level of diversity was observed within LRV1 from L. guyanensis and LRV1 from L. braziliensis, and LRV1 sequences from L. naiffi were highly similar, such as those for L. shawi (Table 2). The level of similarity observed among sequences from L. guyanensis clustering in Groups A, B, D, and E was similar to those observed for L. naiffi and L. shawi. However, the similarities for LRV1 sequences from L. braziliensis were lower and similar to those observed for L. guyanensis when considering Groups A, B, and C as one group, based on the phylogenetic tree (Table S2; Figure 3).
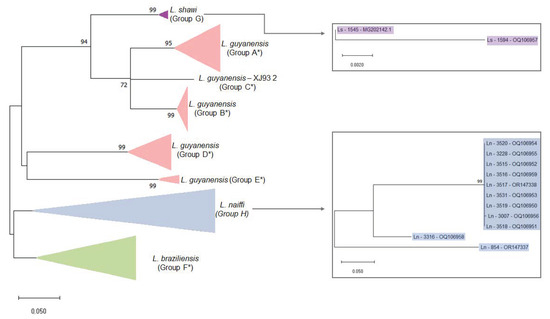
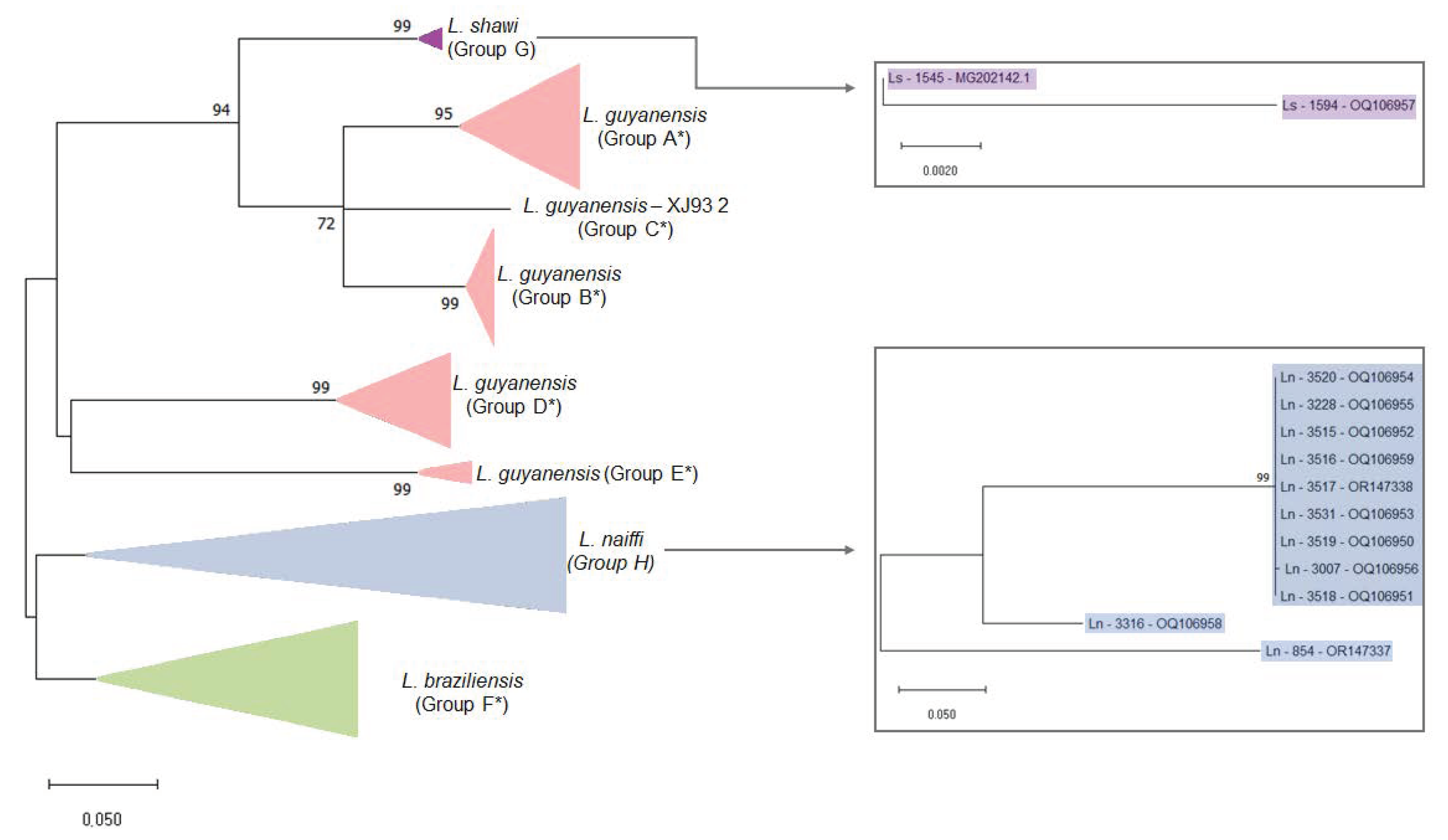

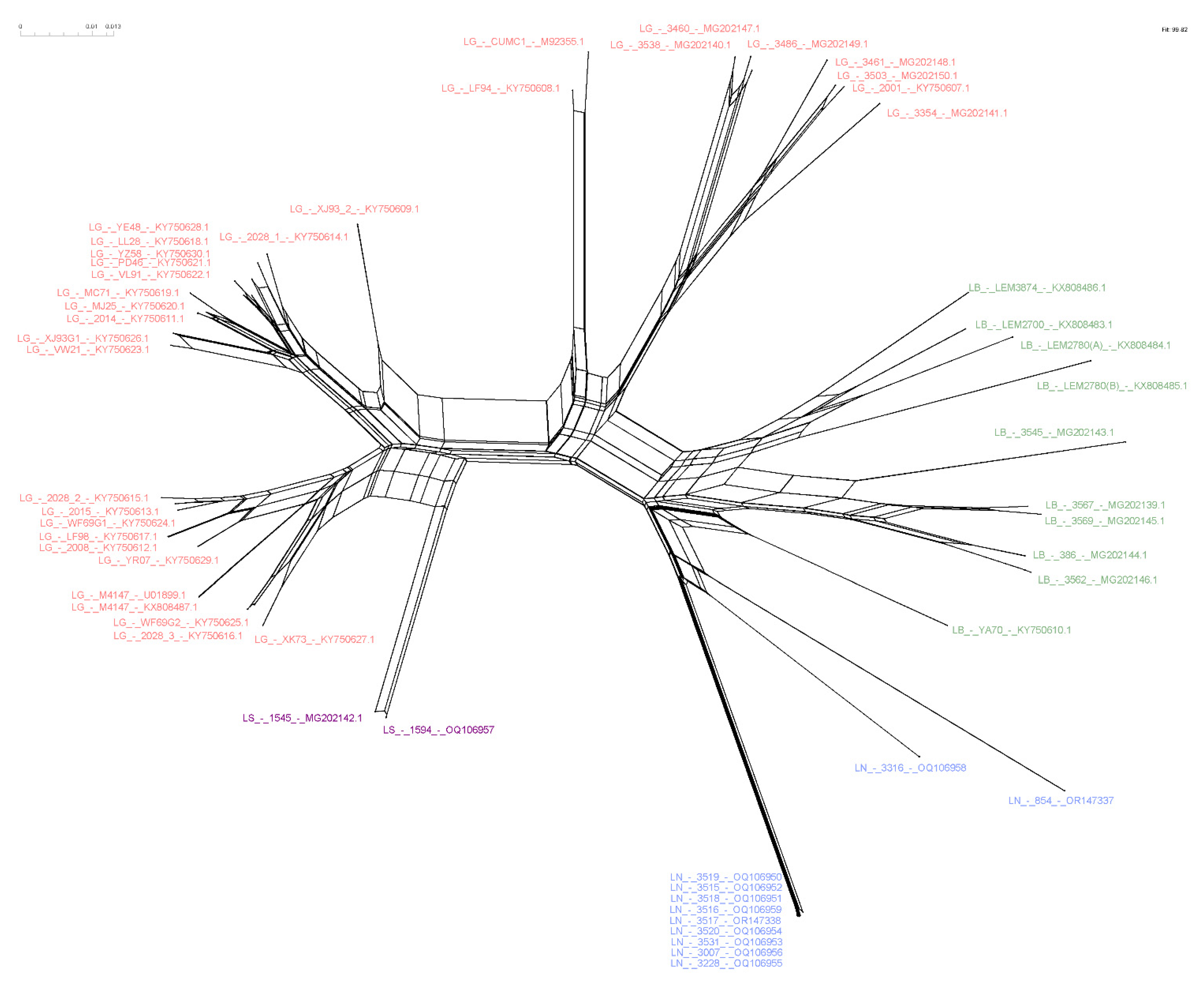
Figure 3.
Maximum likelihood phylogenetic tree of Leishmania RNA Virus 1 found in Leishmania (Viannia) species. The boxes detail the new LRV1 sequences presented in this study that are from L. shawi (in purple) and L. naiffi (in blue). The evolutionary history was inferred by using the maximum likelihood method and the Tamura 3-parameter model [36]. The tree with the highest log likelihood (−4303.97) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances that were estimated using the Tamura 3-parameter model and then selecting the topology with the superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.8242). The rate variation model allowed for some sites to be evolutionarily invariable (+I, 28.96% sites). The tree is drawn to scale, with branch lengths measured in terms of the number of substitutions per site. This analysis involved 59 nucleotide sequences. There were a total of 442 positions in the final dataset. Ls = L. shawi; Ln = L. naiffi. For details of each strain, see Table 1 and Table S1. * Groups are defined following the proposal of Tirera et al. [22]. Groups G and H are defined in the present study.
Figure 3.
Maximum likelihood phylogenetic tree of Leishmania RNA Virus 1 found in Leishmania (Viannia) species. The boxes detail the new LRV1 sequences presented in this study that are from L. shawi (in purple) and L. naiffi (in blue). The evolutionary history was inferred by using the maximum likelihood method and the Tamura 3-parameter model [36]. The tree with the highest log likelihood (−4303.97) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances that were estimated using the Tamura 3-parameter model and then selecting the topology with the superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.8242). The rate variation model allowed for some sites to be evolutionarily invariable (+I, 28.96% sites). The tree is drawn to scale, with branch lengths measured in terms of the number of substitutions per site. This analysis involved 59 nucleotide sequences. There were a total of 442 positions in the final dataset. Ls = L. shawi; Ln = L. naiffi. For details of each strain, see Table 1 and Table S1. * Groups are defined following the proposal of Tirera et al. [22]. Groups G and H are defined in the present study.


Table 2.
Estimates of average evolutionary divergence, based on the number of differences and Tamura 3-parameter, over sequence pairs within mean groups.
3.3. Higher Similarity Is Observed for LRV1 Sequences among Closely Related Leishmania Species
Putting together the results obtained in this study and the sequences of LRV1 that are publicly available, it was possible to compare LRV1 from four different species: L. guyanensis, L. braziliensis, L. naiffi, and L. shawi. The analyses between the group mean distance reveals that L. shawi and L. guyanensis exhibit lower distances, particularly within Groups A, B, and C (Tables S3 and S4). However, there is considerable diversity among LRV1 from L. guyanensis, with L. braziliensis and L. naiffi showing a significant distance between them (Table 3).

Table 3.
Estimates of evolutionary divergence over sequence pairs between groups based on the Tamura 3-parameter model (below the diagonal) and the number of differences (above the diagonal).
3.4. Host-Specificity Is Clearly Observed in the LRV1-L. (Viannia) Species Relationship
To investigate phylogenetic relationships between LRV1 from L. naiffi and L. shawi, along with the other 47 LRV1 sequences available (from L. guyanensis and L. braziliensis and only one L. shawi), a maximum likelihood (ML)-based tree (Figure 3) and phylogenetic network (Figure 4) were constructed. The maximum likelihood tree shows several groups with strong support (higher than 70%), and the results are in agreement with previous studies [21,22], indicating that LRV1 from the L. guyanensis grouping in a cluster comprised different subclusters (named accordingly to Tirera et al. [22] here) and only one cluster for LRV1 was observed to be from L. braziliensis, despite the variability observed within this group (Figure 3 and Table 2). The new LRV1 sequence obtained for L. shawi clustered together with the other sequence available, keeping a close relationship with the L. guyanensis cluster (Figure 3). Although a clear group is observed for LRV1 sequences from L. naiffi, two strains presented very divergent LRV1 sequences and were grouped with other LRV1s from L. naiffi with low bootstrap support, but the most parsimonious connections for these two sequences were observed with the LRV1 from L. naiffi (Figure 4). These two strains are from Pará, and all the others are from another but the same endemic region. Although there is a clear clustering of LRV1 sequences based on their Leishmania host species, there is no association with the geographic distribution of the analyzed sptrain. For example, strains from cluster D (LRV1 from L. guyanensis) were observed in Manaus, Rondônia, and Pará, and strains from cluster F (LRV1 from L. braziliensis) were observed in Bolivia, Rondônia, and French Guyana (Figure 2).
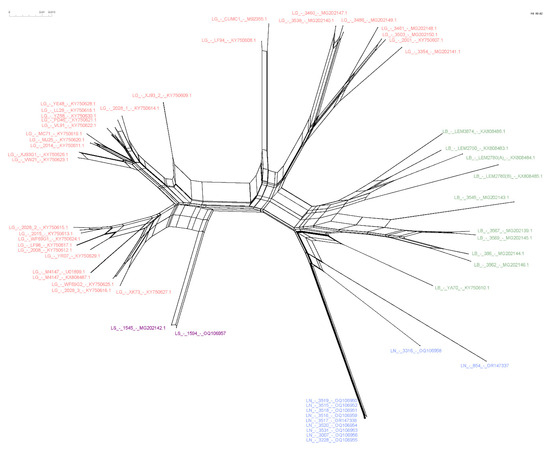
Figure 4.
NeighborNet showing the relationship among LRV1 sequences from different Leishmania (Viannia) species. The network was computed using SplitsTree software. Text colors refer to each analyzed group. Pink = L. guyanensis; purple = L. shawi; blue = L. naiffi; green = L. braziliensis. Strains 854 and 3316, belonging to the species of L. naiffi, presented a divergent profile compared to the other strains of the same group.
The phylogenetic network shows sequences of clustering similar to those observed with the ML tree. The network, however, suggests a common ancestor relative to the structure for all LRV1s (Figure 4). The structure could be the result of the low amount of recombination between the sequences of the network, representing a bottleneck, or it may be simply a result of divergent phylogenetic signals.
4. Discussion
Virus-like particles were demonstrated in Leishmania parasites in the late 1970s [37] but the first molecular description of Leishmania RNA Virus was in 1988 for viruses found in the cytoplasm of an L. (V.) guyanensis strain [28]. Ever since LRV1 was detected in clinical isolates from Peru [38], Brazil [8,18], Colombia [7], Bolivia [19], Costa Rica [39], French Guiana [40], and more recently in Panamá [24], these viruses were observed not only in L. guyanensis but also in strains identified as L. braziliensis, L. shawi, L. naiffi, and L. panamensis [8,21,24,25,26,41,42], indicating an old relationship between LRV1 and the L. (Viannia) subgenus. Furthermore, LRV1 was detected in clinical samples collected from patients infected by L. lainsoni [8] and L. peruviana [27], but no cultivated strains of these two strains that are positive for LRV1 are available yet, limiting our analyses. Considering the epidemiological and medical importance of the symbiosis between Leishmania and LRV, there is a distinct requirement to comprehend the variety and spread of the virus within parasite populations.
The coevolution hypothesis for the LRV-Leishmania species emerged in 1995 when Widmer and Dooley carried out a phylogenetic analysis and found that the genetic distances between LRV types that mirrored the heterogeneity observed for Leishmania species based on random amplified polymorphic DNA (RAPD) fingerprints [4]. More than ten years later, a study presenting a genetic characterization and phylogenetic analysis of LRV1 sequences from 27 L. guyanensis strains and two L. braziliensis was published, and host specificity for LRV1 began to be revealed [22]. A year later, a robust phylogenetic analysis was presented, including 35 LRV1 sequences from L. guyanensis, 11 from L. braziliensis, and for the first time, a sequence of LRV1 was found in an L. shawi strain [21]. Both studies presented evidence corroborating the hypothesis of the coevolution of LRV1 and L. (Viannia) parasites, grouping LRV1 sequences according to their host–parasite species. Host specificity was also demonstrated for LRV2-infecting L. major and L. aethiopica [20].
The abovementioned studies, which suggest a specific relationship between LRV1 and L. (Viannia) species, combined with the observation of LRV1 in other species, motivate the present study in screening for LRV1 in strains representing L. (Viannia) species that have not been analyzed so far and that are available in the Leishmania Collection from Fiocruz. To this end, we analyzed all available strains for L. lainsoni, L. lindenbergi, L. naiffi, L. shawi, and L. utingensis. As previously demonstrated, LRV1 was detected in L. naiffi and L. shawi strains [21,43] but not in L. lainsoni, despite the fact that LRV1 was previously detected in clinical samples that were collected from a patient presenting cutaneous leishmaniasis caused by this species [8]. Of note, Leishmania parasites were isolated from this patient, and the identified strain was included in our analyses (IOCL 3398), but it was negative for LRV1. There are some possibilities for explaining these results, including the possibility of mixed infection due to two or more Leishmania species but with isolation and growth in the culture medium of L. lainsoni to the detriment of another L. (Viannia) species that does not grow very well in a culture medium such as L. lainsoni [44]. The loss of LRV1 during the process of cultivation is also another possibility [45]. Thus, herein, we were able to screen LRV1s in different strains from different L. (Viannia) species, and we were able to analyze the nucleotide sequences of LRV1 from L. naiffi and L. shawi. Interestingly, more than 50% of analyzed L. naiffi strains were positive for LRV1, but we do not know yet if this symbiotic relationship confers any advances to L. naiffi parasites, such as the capacity of interacting with different sandfly species and/or dispersion in different geographic regions [46]. It was demonstrated that a L. naiffi infection cannot have a self-healing nature, as described years ago [47,48]. Patients could experience a poor response to antimonial or pentamidine therapy [43]. A case of a patient who was infected by L. naiffi carrying LRV1 was only first reported in 2019, raising the possibility that the presence of this virus could increase Leishmania spp. virulence and thereby influence therapeutic failure [18], which are aspects already observed for L. braziliensis and L. guyanensis but need further investigation. Similarly, the first L. shawi infection in mucosal secretion was recently observed in Brazil, and it represents a warning for the possible association between L. (V.) shawi and mucosal lesions [49].
Although it is still important to investigate the specificity of LRV1 relative to other Leishmania species, such as L. panamensis, since LRV1 is already detected in cultivated strains of this species [24,25,26], our results strongly support this kind of relationship, keeping L. shawi in a separate cluster that is closely related to L. guyanensis. Here, we assumed the groups suggested by Tirera et al. 2017 [22], where L. guyanensis was divided into five subclusters (A–E). The divergence within L. guyanensis is higher than that observed between L. guyanensis and L. shawi, corroborating the assumption that L. guyanensis comprises complex species [23]. Following this, LRV1 sequences from L. shawi formed another subcluster, named here as Group G, and this subcluster is closely related to the L. guyanensis subclusters A, B, and C. Although LRV1 sequences were obtained for only two L. shawi strains so far, the fact that they clustered together despite the strains being from different geographic regions is also an important aspect that supports host specificity for LRV1. The diversity of LRV1 from L. guyanensis, which forms several subclusters, must be better explored, but the number of LRV1 sequences from the analyzed L. guyanensis and the geographic dispersion of these parasites in the Amazon region might contribute to this observation.
Host specificity was also observed for LRV1 genotypes from L. naiffi. The phylogenetic tree and NeighborNet (Figure 3 and Figure 4) show that LRV1 sequences from L. naiffi were clustered in a well-supported monophyletic clade. Of note, most LRV1 sequences from the analyzed L. naiffi were very similar; in contrast, few differences were observed between the two LRV1 sequences from the analyzed L. shawi, despite being isolated from different hosts in different regions. The diversity observed within LRV1 sequences from the same species must be further investigated, but it is important to consider that all but one L. naiffi strain, presenting highly similar LRV1s, were obtained from patients who were infected in the same endemic region and who were included in the same study, suggesting possible problems during laboratory manipulation. However, this very similar group contained one strain that was previously isolated before the mentioned study, and it was not manipulated together with the other strains, suggesting a homogeneity for the L. naiffi population circulating within this area and causing human disease, which can represent an epidemic clone. The high similarity among LRV1 sequences from L. naiffi is also interesting, since only LRV1 sequences obtained for two L. naiffi strains, IOCL 3316 and IOCL 854, which are both isolated in Pará, showed a different phylogenetic pattern compared with all other L. naiffi strains (n = 9) isolated from Manaus (Amazonas—Brazil). The IOCL 854 strain was obtained from a sandfly species, Lutzomyia squamiventris, and the LRV1 from this strain reflected a basal position relative to the L. naiffi clade, despite the close relationship of this strain to other L. naiffi, including IOCL 3007, as previously demonstrated [23,50]. Considering the Leishmania (Viannia) species was depicted by microsatellite analyses, it is expected that parasites from populations that circulate the Amazon basin (POP1 and POP3 after Kuhls et al. [50]) carry LRV1, and each subpopulation has an association with specific LRV1 genotypes.
By analyzing LRV1 sequences from several strains that represent different L. (Viannia) species, we demonstrated LRV1 genotypes form distinct clusters corresponding to their Leishmania species’ host, suggesting that the transfer of viral particles between strains from different species does not occur frequently. Altogether, our results reinforce the concordance between the phylogenetic patterns of LRV1 and Leishmania (Viannia) species, providing support for the prevailing hypothesis that LRV1 is an ancient virus that has undergone co-evolution with their hosts [3,4]. Recently, it was shown that parasite hybridization might explain the high occurrence of the symbiotic interaction between LRV1 and L. braziliensis in Peru and Bolivia [51]. It is possible that this also explains the high frequency of LRV1 in parasites from the Brazilian Amazon Region, since many possible hybrids were described in the region [52], and analyses of microsatellite markers have shown extensive diversity in the subgenus L. (Viannia), with an indication of both clonality and recombination as a strategy of reproduction [50].
5. Conclusions
Our study adds to the growing body of evidence supporting a specific relationship between Leishmania RNA Virus 1 (LRV1) and species within the L. (Viannia) subgenus. LRV1 was detected in various clinical isolates from different species, including L. guyanensis, L. braziliensis, L. shawi, L. naiffi, and L. panamensis, but not in other species and not in the L. braziliensis circulating outside the Amazon Basin, raising intriguing questions about host specificity and the potential impact on virulence and therapeutic response.
While the results provide strong support for the association between LRV1 and specific Leishmania species, more investigations are needed to understand its specificity relative to other species, such as L. panamensis. The divergence observed within LRV1 sequences from the same species warrants further scrutiny, especially considering potential issues during laboratory manipulation and the homogeneity of L. naiffi populations in certain endemic regions. Overall, the identification and characterization of LRV1 in different Leishmania species shed light on the complex interactions between these viruses and the parasites that they infect. Future research in this area may uncover novel insights into the biology and pathogenesis of Leishmania infections, offering new perspectives on therapeutic strategies and disease management.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms11092295/s1. Table S1: Information on LRV1 sequences from previous studies used for phylogenetic analyses; Table S2: Estimates of average evolutionary divergence over sequence pairs within groups; Table S3: Estimates of evolutionary divergence over sequence pairs between groups using the Tamura 3-parameter model; Table S4: Estimates of evolutionary divergence over sequence pairs between groups using the Tamura 3-parameter model.
Author Contributions
Conceptualization, E.C., K.C., L.M.C. and M.C.d.O.S.; formal analyses, E.C. and M.C.d.O.S.; funding acquisition, E.C.; methodology, K.C. and M.C.d.O.S.; writing—original draft, M.C.d.O.S. and L.M.C.; writing—review and editing, E.C., K.C. and L.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
Elisa Cupolillo: (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Finance Code 001; Conselho Nacional de Desenvolvimento Científico e Tecnológico, Research Fellow, 302622/2017-9; Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, CNE, E26-202.569/2019; Temáticos, E26-210.038/2020; Fundação de Amparo ao Desenvolvimento das Ações Científicas e Tecnológicas e à Pesquisa-FAPERO, Edital 16/2014). Lilian Motta Cantanhêde: (Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, Pós-Doutorado Nota 10 E-26/205.730/2022 and 205.731/2022).
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We sincerely appreciate the Genomics Platform (RPT01A) of the Fiocruz Technological Platforms Network, which provided the sequencing. The graphical abstract was created using BioRender (www.biorender.com (accessed on 30 July 2023)).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grybchuk, D.; Akopyants, N.S.; Kostygov, A.Y.; Konovalovas, A.; Lye, L.F.; Dobson, D.E.; Zangger, H.; Fasel, N.; Butenko, A.; Frolov, A.O.; et al. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc. Natl. Acad. Sci. USA 2018, 115, E506–E515. [Google Scholar] [CrossRef] [PubMed]
- Klocek, D.; Grybchuk, D.; Tichá, L.; Votýpka, J.; Volf, P.; Kostygov, A.Y.; Yurchenko, V. Evolution of RNA viruses in trypanosomatids: New insights from the analysis of Sauroleishmania. Parasitol. Res. 2023. [Google Scholar] [CrossRef]
- Cantanhêde, L.M.; Mata-Somarribas, C.; Chourabi, K.; Pereira da Silva, G.; Dias Das Chagas, B.; de Oliveira, R.; Pereira, L.; Boité, M.C.; Cupolillo, E. The maze pathway of coevolution: A critical review over the Leishmania and its endosymbiotic history. Genes 2021, 12, 657. [Google Scholar] [CrossRef] [PubMed]
- Widmer, G.; Dooley, S. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 1995, 23, 2300–2304. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global report on neglected tropical diseases 2023. World Health Organ. Technol. Rep. Ser. 2023, ix, 68. [Google Scholar]
- OPAS. Situação Epidemiológica Leishmaniose Cutânea e Mucosa. 2021. Available online: https://iris.paho.org/handle/10665.2/55386 (accessed on 5 August 2023).
- Salinas, G.; Zamora, M.; Stuart, K.; Saravia, N. Leishmania RNA viruses in Leishmania of the Viannia subgenus. Am. J. Trop. Med. Hyg. 1996, 54, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Cantanhêde, L.M.; da Silva Júnior, C.F.; Ito, M.M.; Felipin, K.P.; Nicolete, R.; Salcedo, J.M.V.; Porrozzi, R.; Cupolillo, E.; Ferreira, R.D.G.M. Further Evidence of an Association between the Presence of Leishmania RNA Virus 1 and the Mucosal Manifestations in Tegumentary Leishmaniasis Patients. PLoS Negl. Trop. Dis. 2015, 9, e0004079. [Google Scholar] [CrossRef]
- Ramasawmy, R.; Menezes, E.; Magalhães, A.; Oliveira, J.; Castellucci, L.; Almeida, R.; Rosa, M.E.; Guimarães, L.H.; Lessa, M.; Noronha, E.; et al. The -2518bp promoter polymorphism at CCL2/MCP1 influences susceptibility to mucosal but not localized cutaneous leishmaniasis in Brazil. Infect. Genet. Evol. 2010, 10, 607–613. [Google Scholar] [CrossRef][Green Version]
- Castellucci, L.; Jamieson, S.E.; Miller, E.N.; De Almeida, L.F.; Oliveira, J.; Magalhães, A.; Guimarães, L.H.; Lessa, M.; Lago, E.; de Jesus, A.R.; et al. FLI1 polymorphism affects susceptibility to cutaneous leishmaniasis in Brazil. Genes Immun. 2011, 12, 589–594. [Google Scholar] [CrossRef]
- Silveira, F.T.; Lainson, R.; Corbett, C.E.P. Clinical and immunopathological spectrum of american cutaneous leishmaniasis with special reference to the disease in Amazonian Brazil—A review. Mem. Inst. Oswaldo Cruz 2004, 99, 239–251. [Google Scholar] [CrossRef]
- Alvar, J.; Croft, S.; Olliaro, P. Chemotherapy in the Treatment and Control of Leishmaniasis. Adv. Parasitol. 2006, 61, 223–274. [Google Scholar] [PubMed]
- Ives, A.; Ronet, C.; Prevel, F.; Ruzzante, G.; Fuertes-Marraco, S.; Schutz, F.; Zangger, H.; Revaz-Breton, M.; Lye, L.; Hickerson, S.; et al. Leishmania RNA Virus Controls the Severity of Mucocutaneous Leishmaniasis. Science 2011, 331, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Hartley, M.A.; Ronet, C.; Zangger, H.; Beverley, S.M.; Fasel, N. Leishmania RNA virus: When the host pays the toll. Front. Cell. Infect. Microbiol. 2012, 2, 99. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Castiglioni, P.; Hartley, M.A.; Eren, R.O.; Prével, F.; Desponds, C.; Utzschneider, D.T.; Zehn, D.; Cusi, M.G.; Kuhlmann, F.M.; et al. Type I interferons induced by endogenous or exogenous viral infections promote metastasis and relapse of leishmaniasis. Proc. Natl. Acad. Sci. USA 2017, 114, 4987–4992. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, R.V.H.; Lima-Junior, D.S.; da Silva, M.V.G.; Dilucca, M.; Rodrigues, T.S.; Horta, C.V.; Silva, A.L.N.; da Silva, P.F.; Frantz, F.G.; Lorenzon, L.B.; et al. Leishmania RNA virus exacerbates Leishmaniasis by subverting innate immunity via TLR3-mediated NLRP3 inflammasome inhibition. Nat. Commun. 2019, 10, 5273. [Google Scholar] [CrossRef] [PubMed]
- Bourreau, E.; Ginouves, M.; Prévot, G.; Hartley, M.-A.; Gangneux, J.-P.; Robert-Gangneux, F.; Dufour, J.; Sainte-Marie, D.; Bertolotti, A.; Pratlong, F.; et al. Leishmania-RNA virus presence in L. guyanensis parasites increases the risk of first-line treatment failure and symptomatic relapse. J. Infect. Dis. 2015, 213, 105–111. [Google Scholar] [CrossRef]
- Vieira-Gonçalves, R.; Fagundes-Silva, G.A.; Heringer, J.F.; Fantinatti, M.; Da-Cruz, A.M.; Oliveira-Neto, M.P.; Guerra, J.A.O.; Gomes-Silva, A. First report of treatment failure in a patient with cutaneous leishmaniasis infected by Leishmania (Viannia) naiffi carrying Leishmania RNA virus: A fortuitous combination? Rev. Soc. Bras. Med. Trop. 2019, 52, 10–12. [Google Scholar] [CrossRef]
- Adaui, V.; Lye, L.-F.; Akopyants, N.S.; Zimic, M.; Llanos-Cuentas, A.; Garcia, L.; Maes, I.; De Doncker, S.; Dobson, D.E.; Arevalo, J.; et al. Association of the Endobiont Double-Stranded RNA Virus LRV1 With Treatment Failure for Human Leishmaniasis Caused by Leishmania braziliensis in Peru and Bolivia. J. Infect. Dis. 2016, 213, 112–121. [Google Scholar] [CrossRef]
- Kostygov, A.Y.; Grybchuk, D.; Kleschenko, Y.; Chistyakov, D.S.; Lukashev, A.N.; Gerasimov, E.S.; Yurchenko, V. Analyses of Leishmania-LRV Co-Phylogenetic Patterns and Evolutionary Variability of Viral Proteins. Viruses 2021, 13, 2305. [Google Scholar] [CrossRef]
- Cantanhêde, L.M.; Fernandes, F.G.; Ferreira, G.E.M.; Porrozzi, R.; de Godoi Mattos Ferreira, R.; Cupolillo, E. New insights into the genetic diversity of Leishmania RNA Virus 1 and its species-specific relationship with Leishmania parasites. PLoS ONE 2018, 13, e0198727. [Google Scholar] [CrossRef]
- Tirera, S.; Ginouves, M.; Donato, D.; Caballero, I.S.; Bouchier, C.; Lavergne, A.; Bourreau, E.; Mosnier, E.; Vantilcke, V.; Couppié, P.; et al. Unraveling the genetic diversity and phylogeny of Leishmania RNA virus 1 strains of infected Leishmania isolates circulating in French Guiana. PLoS Negl. Trop. Dis. 2017, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Boité, M.C.; Mauricio, I.L.; Miles, M.A.; Cupolillo, E. New Insights on Taxonomy, Phylogeny and Population Genetics of Leishmania (Viannia) Parasites Based on Multilocus Sequence Analysis. PLoS Negl. Trop. Dis. 2012, 6, 1888. [Google Scholar] [CrossRef]
- Gonzalez, K.; De León, S.S.; Pineda, V.; Samudio, F.; Capitan-Barrios, Z.; Suarez, J.A.; Weeden, A.; Ortiz, B.; Rios, M.; Moreno, B.; et al. Detection of Leishmania RNA virus 1 in Leishmania (Viannia) panamensis isolates, Panama. Emerg. Infect. Dis. 2023, 29, 1250. [Google Scholar] [CrossRef]
- Kariyawasam, R.; Mukkala, A.N.; Lau, R.; Valencia, B.M.; Llanos-Cuentas, A.; Boggild, A.K. Virulence factor RNA transcript expression in the Leishmania Viannia subgenus: Influence of species, isolate source, and Leishmania RNA virus-1. Trop. Med. Health 2019, 47, 25. [Google Scholar] [CrossRef]
- Kariyawasam, R.; Grewal, J.; Lau, R.; Purssell, A.; Valencia, B.M.; Llanos-Cuentas, A.; Boggild, A.K. Influence of Leishmania RNA Virus 1 on Proinflammatory Biomarker Expression in a Human Macrophage Model of American Tegumentary Leishmaniasis. J. Infect. Dis. 2017, 216, 877–886. [Google Scholar] [CrossRef]
- Valencia, B.M.; Lau, R.; Kariyawasam, R.; Jara, M.; Ramos, A.P.; Chantry, M.; Lana, J.T.; Boggild, A.K.; Llanos-Cuentas, A. Leishmania RNA virus-1 is similarly detected among metastatic and non-metastatic phenotypes in a prospective cohort of American Tegumentary Leishmaniasis. PLoS Negl. Trop. Dis. 2022, 16, e0010162. [Google Scholar] [CrossRef]
- Tarr, P.I.; Aline, R.F.; Smiley, B.L.; Scholler, J.; Keithly, J.; Stuart, K. LR1, A candidate RNA virus of Leishmania. Proc. Natl. Acad. Sci. USA 1988, 85, 9572–9575. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, A.; Albanaz, A.T.S.; Opperdoes, F.R.; Škodovásveráková, I.; Zagirova, D.; Saura, A.; Chmelová, L.; Leštinová, T.; Bečvář, T.; Sádlová, J.; et al. Leishmania guyanensis M4147 as a new LRV1-bearing model parasite: Phosphatidate phosphatase 2-like protein controls cell cycle progression and intracellular lipid content. PLoS Negl. Trop. Dis. 2022, 16, 1–22. [Google Scholar] [CrossRef] [PubMed]
- da Graça, G.C.; Volpini, A.C.; Romero, G.A.S.; de Oliveira Neto, M.P.; Hueb, M.; Porrozzi, R.; Boité, M.C.; Cupolillo, E. Development and validation of PCR-based assays for diagnosis of American cutaneous leishmaniasis and identification of the parasite species. Mem. Inst. Oswaldo Cruz 2012, 107, 664–674. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Brettmann, E.A.; Shaik, J.S.; Zangger, H.; Lye, L.F.; Kuhlmann, F.M.; Akopyants, N.S.; Oschwald, D.M.; Owens, K.L.; Hickerson, S.M.; Ronet, C.; et al. Tilting the balance between RNA interference and replication eradicates Leishmania RNA virus 1 and mitigates the inflammatory response. Proc. Natl. Acad. Sci. USA 2016, 113, 11998–12005. [Google Scholar] [CrossRef]
- Parmentier, L.; Cusini, A.; Müller, N.; Zangger, H.; Hartley, M.A.; Desponds, C.; Castiglioni, P.; Dubach, P.; Ronet, C.; Beverley, S.; et al. Case Report: Severe cutaneous leishmaniasis in a human immunodeficiency virus patient coinfected with Leishmania braziliensis and its endosymbiotic virus. Am. J. Trop. Med. Hyg. 2016, 94, 840–843. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Croft, S.L.; Molyneux, D.H. Studies on the ultrastructure, virus-like particles and infectivity of Leishmania hertigi. Ann. Trop. Med. Parasitol. 1979, 73, 213–226. [Google Scholar] [CrossRef]
- Saiz, M.; Llanos-Cuentas, A.; Echevarria, J.; Roncal, N.; Cruz, M.; Tupayachi Muniz, M.; Lucas, C.; Wirth, D.F.; Scheffter, S.; Magill, A.J.; et al. Short report: Detection of Leishmaniavirus in human biopsy samples of leishmaniasis from Peru. Am. J. Trop. Med. Hyg. 1998, 58, 192–194. [Google Scholar] [CrossRef]
- Mata-somarribas, C.; Quesada-lópez, J.; Matamoros, M.F.; Cervantes-gómez, C. Raising the suspicion of a non-autochthonous infection: Identification of Leishmania guyanensis from Costa Rica exhibits a Leishmaniavirus related to Brazilian north-east and French Guiana viral genotypes. Mem. Inst. Oswaldo Cruz 2022, 117, e220162. [Google Scholar] [CrossRef]
- Ginouvès, M.; Simon, S.; Borreau, E.; Lacoste, V.; Ronet, C.; Couppié, P.; Nacher, M.; Demar, M.; Prévot, G. Prevalence and Distribution of Leishmania RNA Virus 1 in Leishmania Parasites from French Guiana. Am. J. Trop. Med. Hyg. 2016, 94, 102–106. [Google Scholar] [CrossRef]
- Ito, M.M.; Cantanhêde, L.M.; Katsuragawa, T.H.; da Silva Junior, C.F.; Camargo, L.M.A.; de Godoi Mattos Ferreira, R.; Vilallobos-Salcedo, J.M. Correlation between presence of Leishmania RNA virus 1 and clinical characteristics of nasal mucosal leishmaniosis. Braz. J. Otorhinolaryngol. 2015, 81, 533–540. [Google Scholar] [CrossRef]
- de Oliveira Ramos Pereira, L.; Maretti-Mira, A.C.; Rodrigues, K.M.; Lima, R.B.; de Oliveira-Neto, M.P.; Cupolillo, E.; Pirmez, C.; de Oliveira, M.P. Severity of tegumentary leishmaniasis is not exclusively associated with Leishmania RNA virus 1 infection in Brazil. Mem. Inst. Oswaldo Cruz 2013, 108, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Fagundes-Silva, G.A.; Sierra Romero, G.A.; Cupolillo, E.; Gadelha Yamashita, E.P.; Gomes-Silva, A.; De Oliveira Guerra, J.A.; Da-Cruz, A.M. Leishmania (Viannia) naiffi: Rare enough to be neglected? Mem. Inst. Oswaldo Cruz 2015, 110, 797–800. [Google Scholar] [CrossRef]
- das Chagas, B.D.; Pereira, T.M.; Cantanhêde, L.M.; da Silva, G.P.; Boité, M.C.; Pereira, L.d.O.R.; Cupolillo, E. Interspecies and Intrastrain Interplay among Leishmania spp. Parasites. Microorganisms 2022, 10, 1883. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, F.M.; Robinson, J.I.; Bluemling, G.R.; Ronet, C.; Fasel, N.; Beverley, S.M. Antiviral screening identifies adenosine analogs targeting the endogenous dsRNA Leishmania RNA virus 1 (LRV1) pathogenicity factor. Proc. Natl. Acad. Sci. USA 2017, 114, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Cantanhêde, L.M.; Cupolillo, E. Leishmania (Viannia) naiffi Lainson & Shaw 1989. In Parasites and Vectors; BioMed Central Ltd.: London, UK, 2023. [Google Scholar]
- Naiff, R.D.; Freitas, R.A.; Naiff, M.F.; Arias, J.R.; Barrett, T.V.; Momen, H.; Grimaldi, G., Jr. Epidemiological and nosological aspects of Leishmania naiffi Lainson & Shaw, 1989. Mem. Inst. Oswaldo Cruz 1991, 86, 317–321. [Google Scholar]
- Van Der Snoek, E.M.; Lammers, A.M.; Kortbeek, L.M.; Roelfsema, J.H.; Bart, A.; Jaspers, C.A.J.J. Spontaneous cure of American cutaneous leishmaniasis due to Leishmania naiffi in two Dutch infantry soldiers. Clin. Exp. Dermatol. 2009, 34, 889–891. [Google Scholar] [CrossRef]
- Oliveira, L.P.; Nascimento, L.C.S.; Santos, F.S.; Takamatsu, J.L.C.; Sanchez, L.R.P.; Santos, W.S.; Garcez, L.M. First Report of an Asymptomatic Leishmania (Viannia) shawi Infection Using a Nasal Swab in Amazon, Brazil. Int. J. Environ. Res. Public Health 2022, 19, 6346. [Google Scholar] [CrossRef] [PubMed]
- Kuhls, K.; Cupolillo, E.; Silva, S.O.; Schweynoch, C.; Côrtes Boité, M.; Mello, M.N.; Maurício, I.; Miles, M.; Wirth, T.; Schönian, G. Population Structure and Evidence for Both Clonality and Recombination among Brazilian Strains of the Subgenus Leishmania (Viannia). PLoS Negl. Trop. Dis. 2013, 7, e2490. [Google Scholar] [CrossRef]
- Heeren, S.; Maes, I.; Sanders, M.; Lye, L.-F.; Arevalo, J.; Garcia, L.; Lemey, P.; Beverley, S.M.; Cotton, J.A.; Dujardin, J.C.; et al. Parasite hybridization promotes spreading of endosymbiotic viruses. bioRxiv 2023. [Google Scholar] [CrossRef]
- Tojal da Silva, A.C.; Cupolillo, E.; Volpini, A.C.; Almeida, R.; Sierra Romero, G.A. Species diversity causing human cutaneous leishmaniasis in Rio Branco, state of Acre, Brazil. Trop. Med. Int. Health 2006, 11, 1388–1398. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).