Growing in Saltwater: Biotechnological Potential of Novel Methylotuvimicrobium- and Methylomarinum-like Methanotrophic Bacteria
Abstract
:1. Introduction
2. Materials and Methods
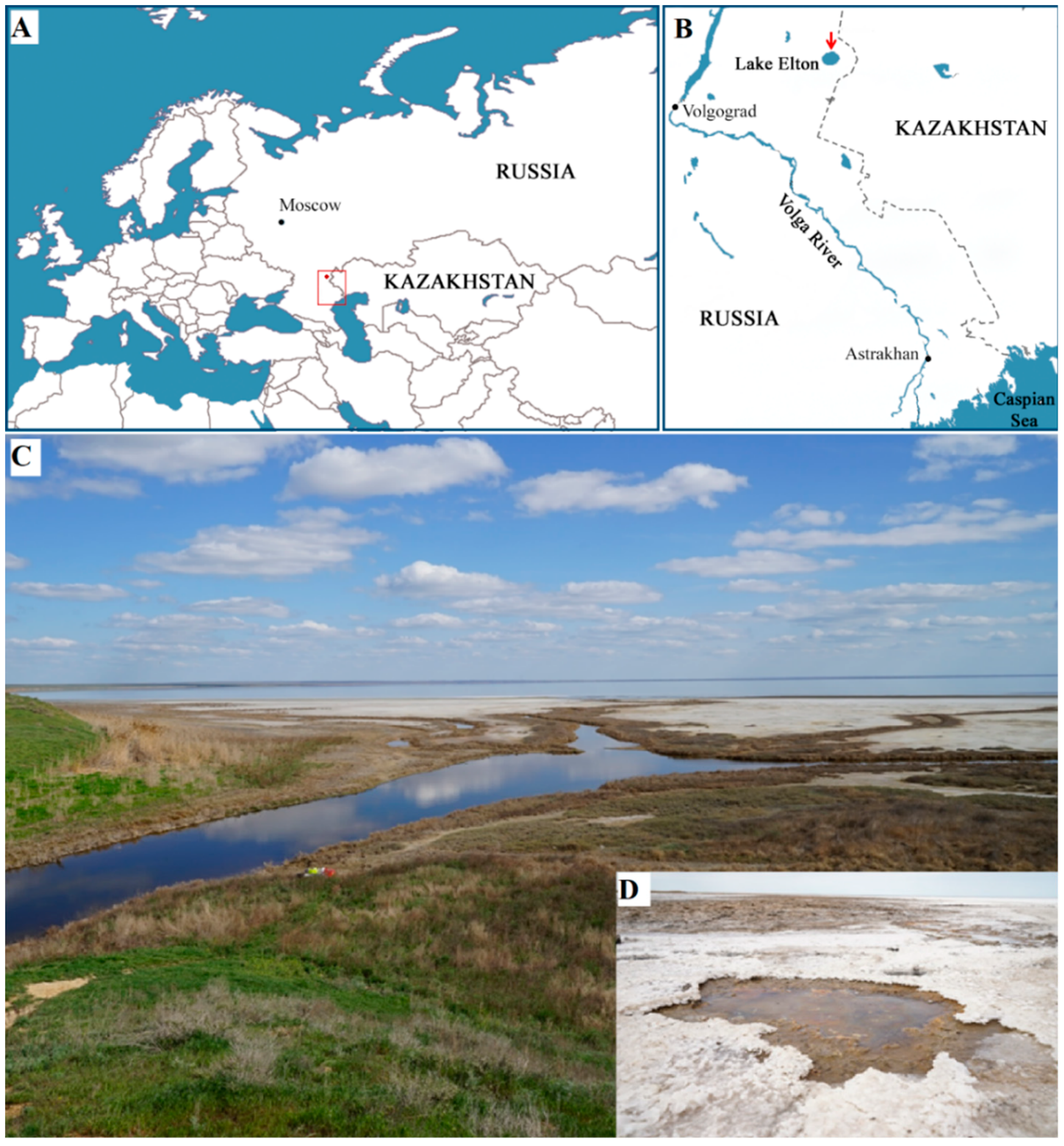
2.1. Sampling Site
2.2. Enrichment Procedure
2.3. Molecular Analysis of Methanotroph Community Composition
2.4. Cultivation in a Bioreactor
2.5. Isolation Studies
2.6. Morphological Characterization and Microscopy
2.7. DNA Extraction
2.8. Genome Sequencing and Annotation
2.9. Phylogenomic Analysis
2.10. Sequence Accession Numbers
3. Results and Discussion
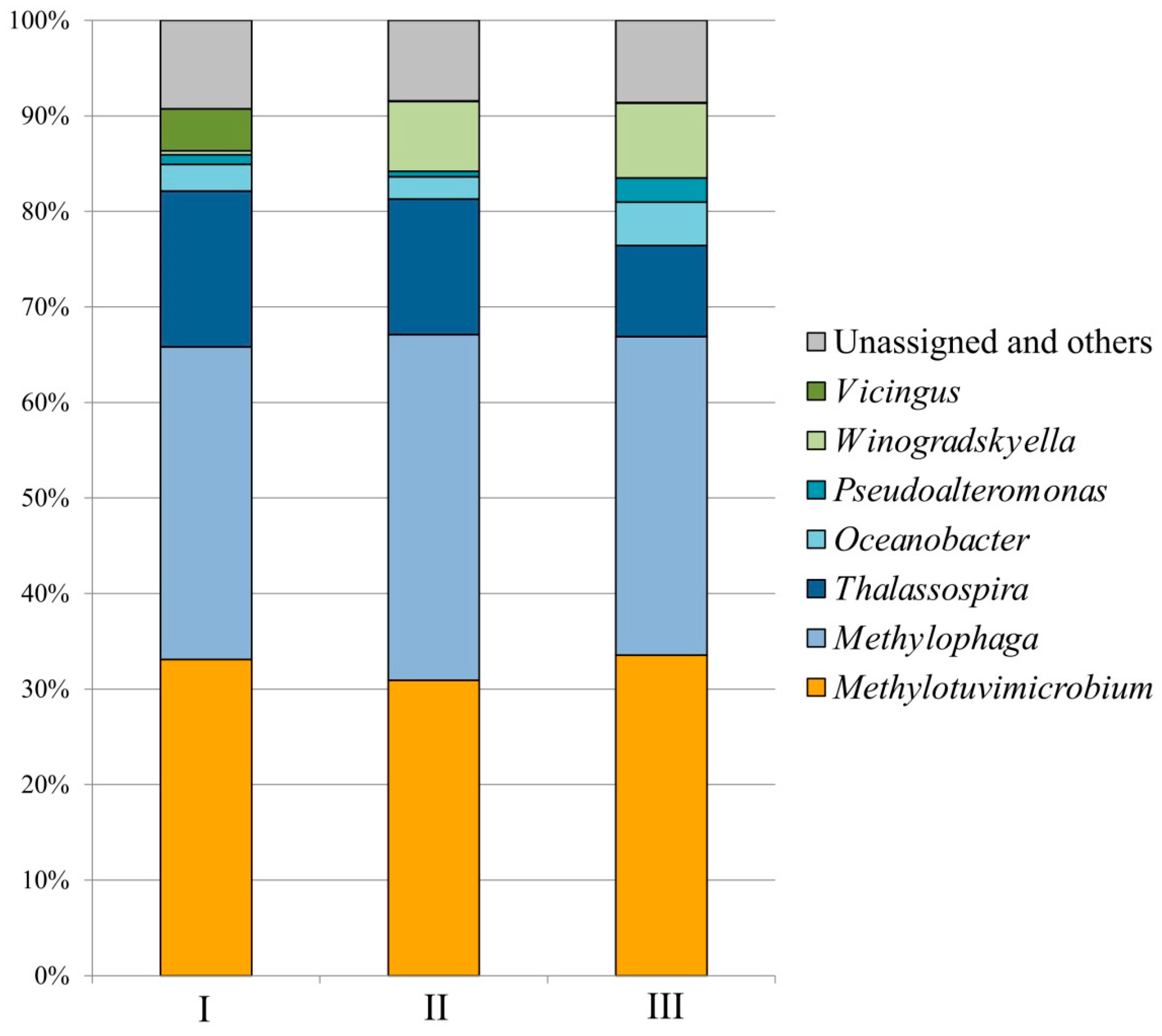
3.1. Enriched Methane-Oxidizing Consortium and Its Composition
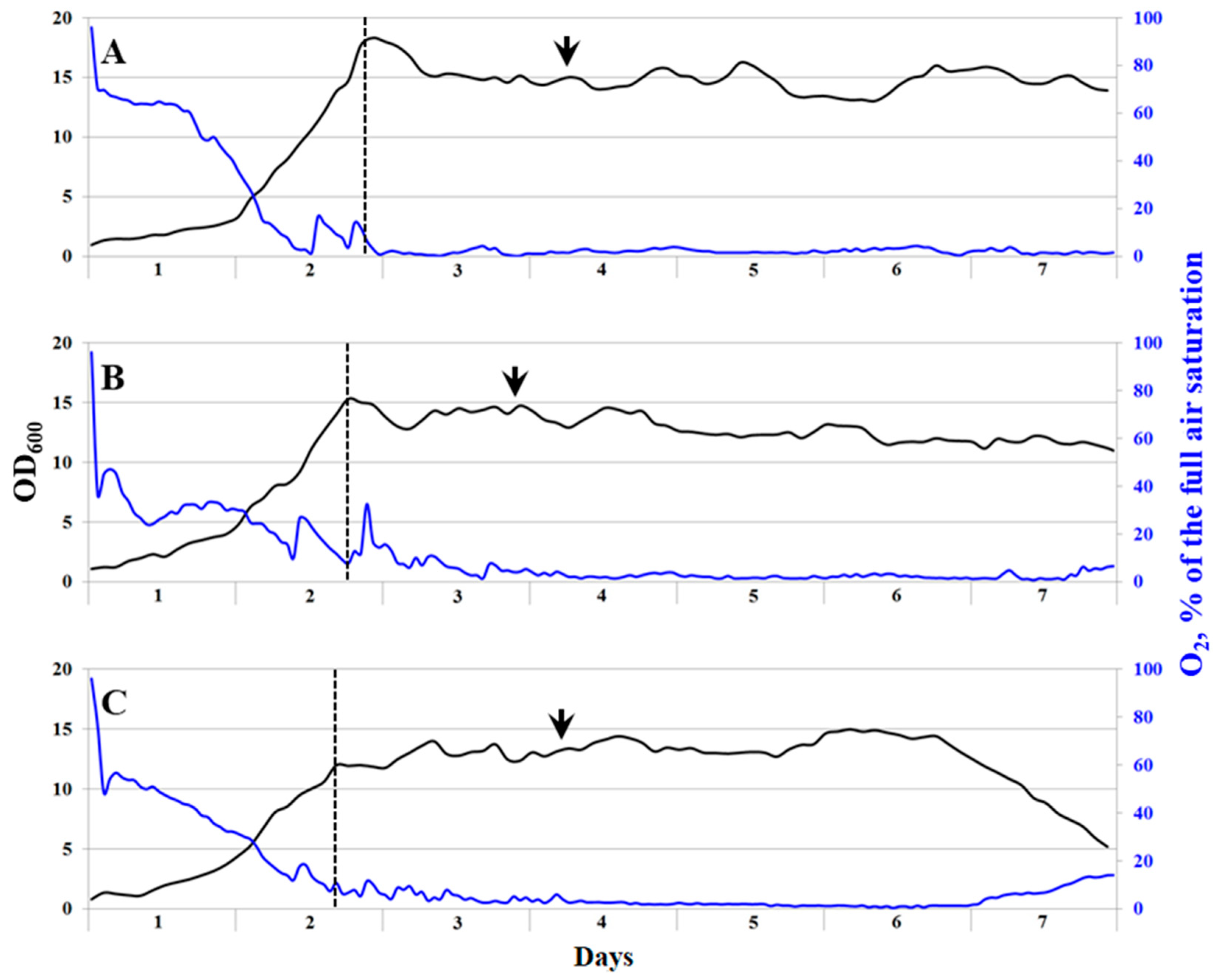
3.2. Growth of the Methanotrophic Consortium Ch1 in a Bioreactor
3.3. Isolation of Methanotrophic Bacteria
3.4. Characterization of Methanotrophic Isolate
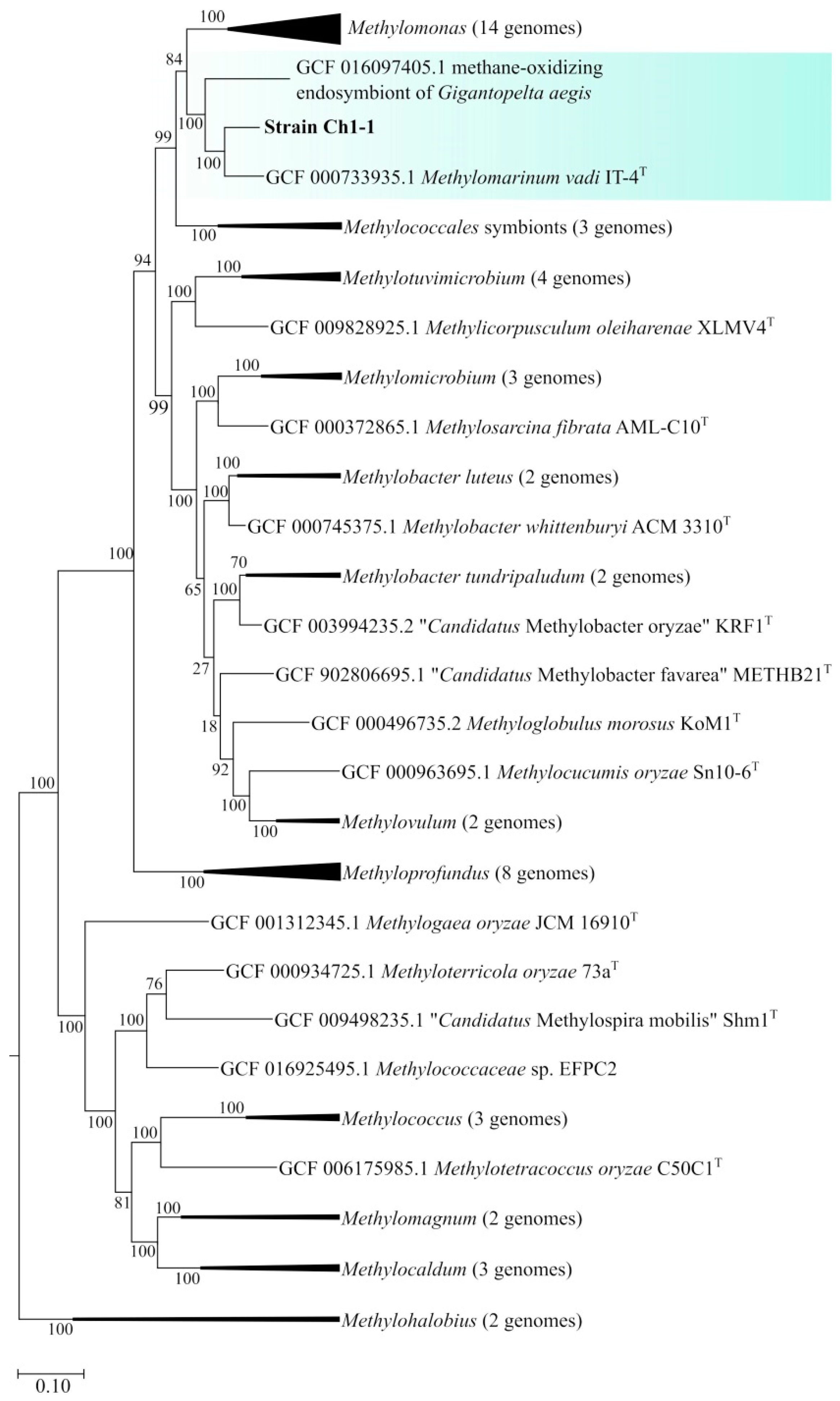
3.5. General Genome Features of Strain Ch1-1 and Genome-Based Phylogeny
3.6. Growth of the Methanotrophic Isolate Ch1-1 in a Bioreactor
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria . Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar] [PubMed]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic methylotrophic prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry; Springer: Berlin/Heidelberg, Germany, 2013; pp. 267–285. [Google Scholar]
- Khmelenina, V.N.; Colin Murrell, J.; Smith, T.J.; Trotsenko, Y.A. Physiology and biochemistry of the aerobic methanotrophs. In Aerobic Utilization of Hydrocarbons, Oils, and Lipids; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–25. [Google Scholar]
- Murrell, J.C.; Gilbert, B.; McDonald, I.R. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 2000, 173, 325–332. [Google Scholar] [CrossRef]
- Sazinsky, M.H.; Lippard, S.J. Methane monooxygenase: Functionalizing methane at iron and copper. Met. Ions Life Sci. 2015, 15, 205–256. [Google Scholar]
- Dedysh, S.N.; Knief, C. Diversity and phylogeny of described aerobic methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Springer: Berlin/Heidelberg, Germany, 2018; pp. 17–42. [Google Scholar]
- Trotsenko, Y.A.; Khmelenina, V.N. Biology of extremophilic and extremotolerant methanotrophs. Arch. Microbiol. 2002, 177, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Strong, P.; Xie, S.; Clarke, W.P. Methane as a resource: Can the methanotrophs add value? Environ. Sci. Technol. 2015, 49, 4001–4018. [Google Scholar] [CrossRef]
- Mühlemeier, I.M.; Speight, R.; Strong, P.J. Biogas, bioreactors and bacterial methane oxidation. In Methane Biocatalysis: Paving the Way to Sustainability; Springer: Berlin/Heidelberg, Germany, 2018; pp. 213–235. [Google Scholar]
- Risso, C.; Choudhary, S.; Johannessen, A.; Silverman, J. Methanotrophy goes commertial: Challenges, opportunities, and brief history. In Methane Biocatalysis: Paving the Way to Sustainability; Springer: Berlin/Heidelberg, Germany, 2018; pp. 293–298. [Google Scholar]
- Garcia Martinez, J.B.; Pearce, J.M.; Throup, J.; Cates, J.; Lackner, M.; Denkenberger, D.C. Methane single cell protein: Potential to secure a global protein supply against catastrophic food shocks. Front. Bioeng. Biotechnol. 2022, 10, 906704. [Google Scholar] [CrossRef]
- Le, H.T.Q.; Lee, E.Y. Methanotrophs: Metabolic versatility from utilization of methane to multi-carbon sources and perspectives on current and future applications. Bioresour. Technol. 2023, 384, 129296. [Google Scholar]
- Skrede, A.; Berge, G.M.; Storebakken, T.; Herstad, O.; Aarstad, K.G.; Sundstøl, F. Digestibility of bacterial protein grown on natural gas in mink, pigs, chicken and Atlantic salmon. Anim. Feed Sci. Technol. 1998, 76, 103–116. [Google Scholar] [CrossRef]
- Koffas, M.; Odom, J.M.; Square, K. High Growth Methanotrophic Bacterial Strain. U.S. Patent 6,689,601, 10 February 2004. [Google Scholar]
- Sieburth, J.M.; Johnson, P.W.; Eberhardt, M.A.; Sieracki, M.E.; Lidstrom, M.; Laux, D. The first methane-oxidizing bacterium from the upper mixing layer of the deep ocean: Methylomonas pelagica sp. nov. Curr. Microbiol. 1987, 14, 285–293. [Google Scholar] [CrossRef]
- Orata, F.D.; Meier-Kolthoff, J.P.; Sauvageau, D.; Stein, L.Y. Phylogenomic Analysis of the Gammaproteobacterial Methanotrophs (Order Methylococcales) Calls for the Reclassification of Members at the Genus and Species Levels. Front. Microbiol. 2018, 9, 3162. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; McCammon, S.A.; Skerrat, J.H. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology 1997, 143, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Heyer, J.; Berger, U.; Hardt, M.; Dunfield, P.F. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int. J. Syst. Evol. Microbiol. 2005, 55, 1817–1826. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Khmelenina, V.; Eshinimaev, B.; Sorokin, D.; Fuse, H.; Lidstrom, M.; Trotsenko, Y. Classification of halo(alkali)philic and halo(alkali)tolerant methanotrophs provisionally assigned to the genera Methylomicrobium and Methylobacter and emended description of the genus Methylomicrobium. Int. J. Syst. Evol. Microbiol. 2008, 58, 591–596. [Google Scholar] [CrossRef]
- Hirayama, H.; Fuse, H.; Abe, M.; Miyazaki, M.; Nakamura, T.; Nunoura, T.; Furushima, Y.; Yamamoto, H.; Takai, K. Methylomarinum vadi gen. nov., sp. nov., a methanotroph isolated from two distinct marine environments. Int. J. Syst. Evol. Microbiol. 2013, 63, 1073–1082. [Google Scholar] [CrossRef]
- Hirayama, H.; Abe, M.; Miyazaki, M.; Nunoura, T.; Furushima, Y.; Yamamoto, H.; Takai, K. Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 989–999. [Google Scholar] [CrossRef]
- Tavormina, P.L.; Hatzenpichler, R.; McGlynn, S.; Chadwick, G.; Dawson, K.S.; Connon, S.A.; Orphan, V.J. Methyloprofundus sedimenti gen. nov., sp. nov., an obligate methanotroph from ocean sediment belonging to the ‘deep sea-1’ clade of marine methanotrophs. Int. J. Syst. Evol. Microbiol. 2015, 65, 251–259. [Google Scholar] [CrossRef]
- Bowman, J.P.; Sly, L.I.; Nichols, P.D.; Hayward, A.C. Revised taxonomy of the methanotrophs: Description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 1993, 43, 735–753. [Google Scholar] [CrossRef]
- Lees, V.; Owens, N.J.P.; Murrell, J.C. Nitrogen metabolism in marine methanotrophs. Arch. Microbiol. 1991, 157, 60–65. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Starostina, N.G.; Tsvetkova, M.G.; Sokolov, A.P.; Suzina, N.E.; Trotsenko, Y.A. Methanotrophic bacteria in saline reservoirs of Ukraine and Tuva. Microbiology (Mikrobiologiya) 1996, 65, 609–615. [Google Scholar]
- Kalyuzhnaya, M.G.; Khmelenina, V.N.; Suzina, N.E.; Lysenko, A.M.; Trotsenko, Y.A. New methanotrophic isolates of the southern Transbaikal region. Microbiology (Mikrobiologiya) 1999, 68, 592–600. [Google Scholar]
- Kalyuzhnaya, M.G.; Khmelenina, V.N.; Eshinimaev, B.; Suzina, N.; Nikitin, D.; Solonin, A.; Lin, J.-L.; McDonald, I.; Murrell, C.; Trotsenko, Y. Taxonomic characterization of new alkaliphilic and alkalitolerant methanotrophs from soda lakes of the southeastern Transbaikal region and description of Methylomicrobium buryatense sp. nov. Syst. Appl. Microbiol. 2001, 24, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Kamagata, Y.; Oshima, K.; Hanada, S.; Tamaki, H.; Marumo, K.; Maeda, H.; Nedachi, M.; Hattori, M.; Iwasaki, W.; et al. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3240–3246. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Yang, S.; Rozova, O.N.; Smalley, N.E.; Clubb, J.; Lamb, A.; Gowda, G.A.N.; Raftery, D.; Fu, Y.; Bringel, F.; et al. Highly efficient methane biocatalysis re vealed in a methanotrophic bacterium. Nat. Commun. 2013, 4, 2785. [Google Scholar] [CrossRef]
- Akberdin, I.R.; Thompson, M.; Hamilton, R.; Desai, N.; Alexander, D.; Henard, C.A.; Guarnieri, M.T.; Kalyuzhnaya, M.G. Methane utilization in Methylomicrobium alcaliphilum 20ZR: A systems approach. Sci. Rep. 2018, 8, 2512. [Google Scholar] [CrossRef]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2409v1. [Google Scholar] [CrossRef]
- Kjeldahl, J. Neue Methode zur Bestimmung des Stickstoffs in organischen Körpern. Fresenius’. J. Anal. Chem. 1883, 22, 366–382. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B. Food Safety: Food Analysis Technologies/Techniques. Encycl. Agric. Food Syst. 2014, 3, 273–288. [Google Scholar]
- Tikhonova, E.N.; Suleimanov, R.Z.; Miroshnikov, K.K.; Oshkin, I.Y.; Belova, S.E.; Danilova, O.V.; Ashikhmin, A.A.; Konopkin, A.A.; But, S.Y.; Khmelenina, V.N.; et al. Methylomonas rapida sp. nov., a novel species of fast-growing, carotenoid-producing obligate methanotrophs with high biotechnological potential. Syst. Appl. Microbiol. 2023, 46, 126398. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGAX: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Fradet, D.T.; Tavormina, P.L.; Orphan, V.J. Members of the methanotrophic genus Methylomarinum inhabit inland mud pots. PeerJ 2016, 4, e2116. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Sun, J.; Chen, C.; Sun, Y.; Zhou, Y.; Yang, Y.; Zhang, W.; Li, R.; Zhou, K.; Wong, W.C.; et al. Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail Gigantopelta aegis. Nat. Commun. 2021, 12, 1165. [Google Scholar] [CrossRef]
- Flynn, J.D.; Hirayama, H.; Sakai, Y.; Dunfield, P.F.; Klotz, M.G.; Knief, C.; Op den Camp, H.J.M.; Jetten, M.S.M.; Khmelenina, V.N.; Trotsenko, Y.A.; et al. Draft genome sequences of gammaproteobacterial methanotrophs isolated from marine ecosystems. Genome Announc. 2016, 4, e01629-15. [Google Scholar] [CrossRef]
- Lazure, P.; Jegou, A.-M.; Kerdreux, M. Analysis of salinity measurments near islands on the French continental shelf of the Bay of Biscay. Sci. Mar. 2006, 70, 7–14. [Google Scholar] [CrossRef]

| Strain(s) | NaCl Range (Optimum), % | pH Range (Optimum) | Source | Reference(s) |
|---|---|---|---|---|
| Methylomicrobium pelagicum AA-23T | 1.8–4.7 | NA | Sea water, Sargasso Sea | [17,25] |
| Methylomicrobium sp. IR1 | 1.2–4.1 (1.5–1.9) | (7.0–6.0) | Sea water, Plymouth Sound | [25,26] |
| Methylosphaera hansonii AM6T, AM11 | Require seawater | NA | Ace Lake, Antarctica | [19] |
| Methylomicrobium modestohalophilus 10S | 0.2–8.8 (2.3) | 5.5–8.5 (6.5) | Lake Sasyk, Ukraine | [27] |
| Methylomicrobium buryatense 5BT, 4G, 5G, 6G, 7G | 0–8.2 (0.8) | 6.0–11.0 (8.0–8.5) | Soda lakes, Russia | [28,29] |
| Methylohalobius crimeensis 4Kr, 10KiT | 1.2–14.5 (5.8–8.7) | 6.5–7.5 (7.0) | Lake Krugloe, Ukraine | [20] |
| Methylomicrobium kenyense AMO-1T | Up to 6.4 | 9.0–10.5 (9.5) | Soda lakes, Kenya | [21] |
| Methylotuvimicrobium japanense NIT | 0.2–8.8 (2.3–4.7) | (8.1) | Marine mud, Hiroshima, Japan | [21] |
| Methylotuvimicrobium alcaliphilum 5Z, 20Z | 0.5–8.8 (2.0–4.0) | 7–10.5 (9.0–9.5) | Tuva lakes, Russia | [27] |
| Methylomarinum vadi IT-4T, T2-1 | 1.0–8.0 (2.0–3.0) | 4.5–8.1 (6.2–7.0) | Distinct marine environments, Japan | [22] |
| Methylomarinovum caldicuralii IT-9T | 1.0–5.0 (3.0) | 5.3–6.9 (6.0–6.4) | Shallow submarine hydrothermal system, Japan | [23] |
| Methylocaldum marinum S8T | 0.5–5 (2.0) | 6.0–8.0 (7.0) | Marine sediments of Kagoshima Bay, Japan | [30] |
| Methyloprofundus sedimenti WF1T | 1.0–4.0 (2.0) | 6.0–8.0 (6.5–7.5) | Marine sediment near a whale fall, Monterey Canyon, California | [24] |
| Mineral Component, mg L−1 | Total Salt Content, g L−1 | ||
|---|---|---|---|
| 23 | 29 | 35.9 | |
| KCl | 383.3 | 483.3 | 598.3 |
| MgCl2 × 6H2O | 5720.5 | 7212.8 | 8929.0 |
| H3PO4 (85%) | 762.2 | 960.9 | 1162.0 |
| (NH4)2SO4 | 530.8 | 669.2 | 828.5 |
| NaCl | 12,974.4 | 16,359.0 | 20,000.0 |
| Na2SO4 | 2182.1 | 2751.3 | 3406.0 |
| NaHCO3 | 117.9 | 148.7 | 184.0 |
| CaCl2 | 589.7 | 743.6 | 820.0 |
| Total Salt Content, g L−1 | 23 | 29 | 35.9 |
|---|---|---|---|
| Dilution rate, h−1 | 0.21 ± 0.01 | 0.21 ± 0.01 | 0.19 ± 0.01 |
| Cell dry weight, g L−1 | 3.45 ± 0.93 | 5.77 ± 1.16 | 4.19 ± 1.06 |
| OD600 | 14.88 ± 0.28 | 12.87 ± 0.29 | 12.62 ± 0.55 |
| Productivity, g/(L·h) | 0.93 | 1.05 | 0.97 |
| Total protein, % | 65.40 ± 0.13 | 62.89 ± 0.19 | 61.26 ± 0.32 |
| Total lipids, % | 11.70 ± 0.10 | 10.70 ± 0.15 | 8.80 ± 0.20 |
| Total carbohydrates, % | 26.60 ± 0.09 | 18.50 ± 0.11 | 17.00 ± 0.08 |
| Solid base ash, % | 0.15 ± 0.01 | 0.19 ± 0.01 | 0.19 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tikhonova, E.N.; Suleimanov, R.Z.; Oshkin, I.Y.; Konopkin, A.A.; Fedoruk, D.V.; Pimenov, N.V.; Dedysh, S.N. Growing in Saltwater: Biotechnological Potential of Novel Methylotuvimicrobium- and Methylomarinum-like Methanotrophic Bacteria. Microorganisms 2023, 11, 2257. https://doi.org/10.3390/microorganisms11092257
Tikhonova EN, Suleimanov RZ, Oshkin IY, Konopkin AA, Fedoruk DV, Pimenov NV, Dedysh SN. Growing in Saltwater: Biotechnological Potential of Novel Methylotuvimicrobium- and Methylomarinum-like Methanotrophic Bacteria. Microorganisms. 2023; 11(9):2257. https://doi.org/10.3390/microorganisms11092257
Chicago/Turabian StyleTikhonova, Ekaterina N., Ruslan Z. Suleimanov, Igor Y. Oshkin, Aleksey A. Konopkin, Diana V. Fedoruk, Nikolai V. Pimenov, and Svetlana N. Dedysh. 2023. "Growing in Saltwater: Biotechnological Potential of Novel Methylotuvimicrobium- and Methylomarinum-like Methanotrophic Bacteria" Microorganisms 11, no. 9: 2257. https://doi.org/10.3390/microorganisms11092257
APA StyleTikhonova, E. N., Suleimanov, R. Z., Oshkin, I. Y., Konopkin, A. A., Fedoruk, D. V., Pimenov, N. V., & Dedysh, S. N. (2023). Growing in Saltwater: Biotechnological Potential of Novel Methylotuvimicrobium- and Methylomarinum-like Methanotrophic Bacteria. Microorganisms, 11(9), 2257. https://doi.org/10.3390/microorganisms11092257






