Altered Gut Microbiota Composition Is Associated with Difficulty in Explicit Emotion Regulation in Young Children
Abstract
:1. Introduction
2. Materials and Methods
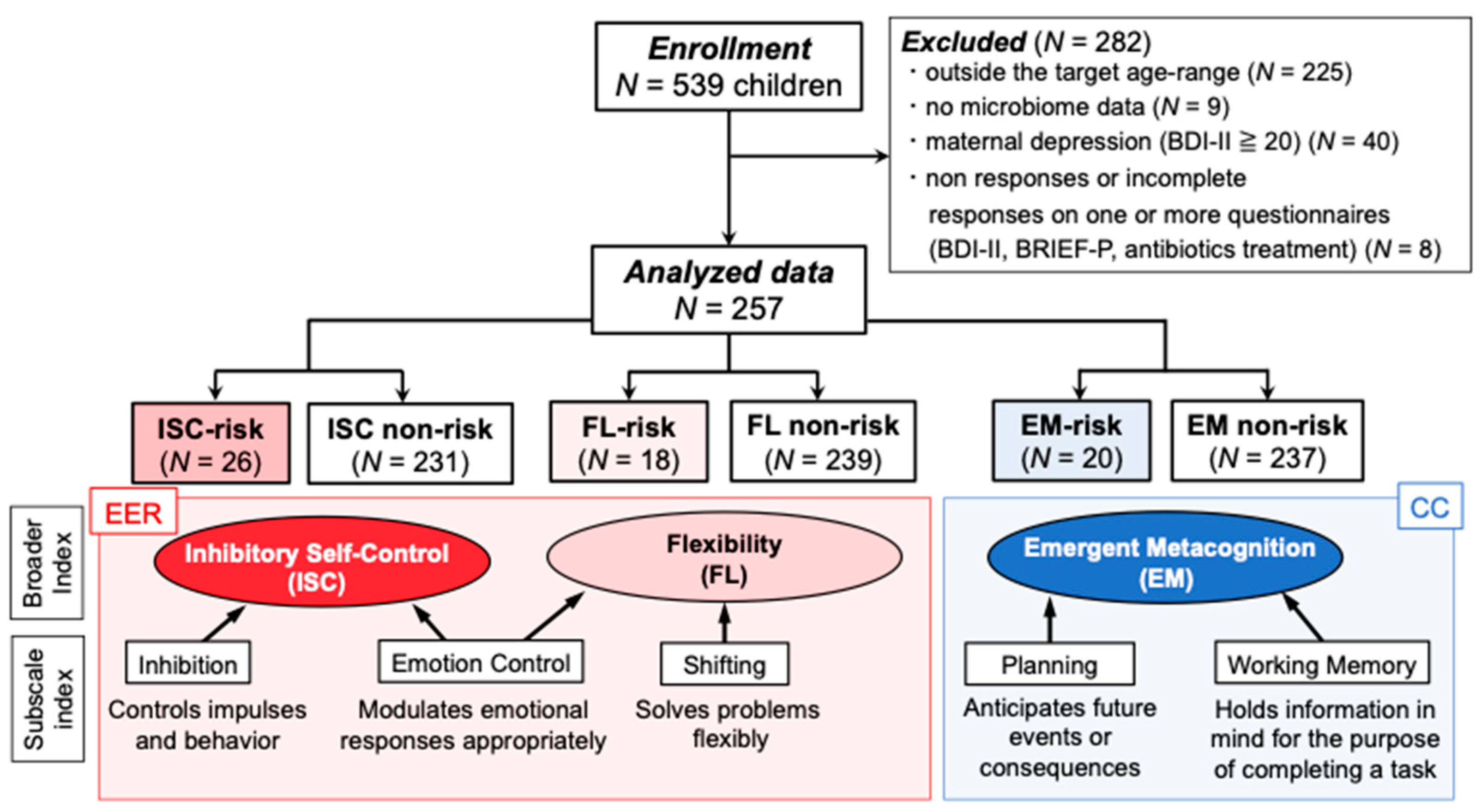
2.1. Participants and Procedure
2.2. Questionnaires
2.2.1. BRIEF-P
2.2.2. Dietary Habits (Food Intake Frequency, JDI Score, Picky Eating)
2.2.3. Physical Symptoms
2.2.4. BDI-II
2.3. Demographic Data
2.4. Fecal Sampling, DNA Extraction, and Sequencing
2.5. Taxonomy Assignment Based on 16S rRNA Gene Sequencing
2.6. Diversity Analysis
2.7. Statistical Analysis
2.7.1. Gut Microbiota Diversity
2.7.2. Gut Microbiota Composition
2.7.3. Dietary Habits
2.7.4. Physical Symptoms
2.7.5. Demographic Data
3. Results
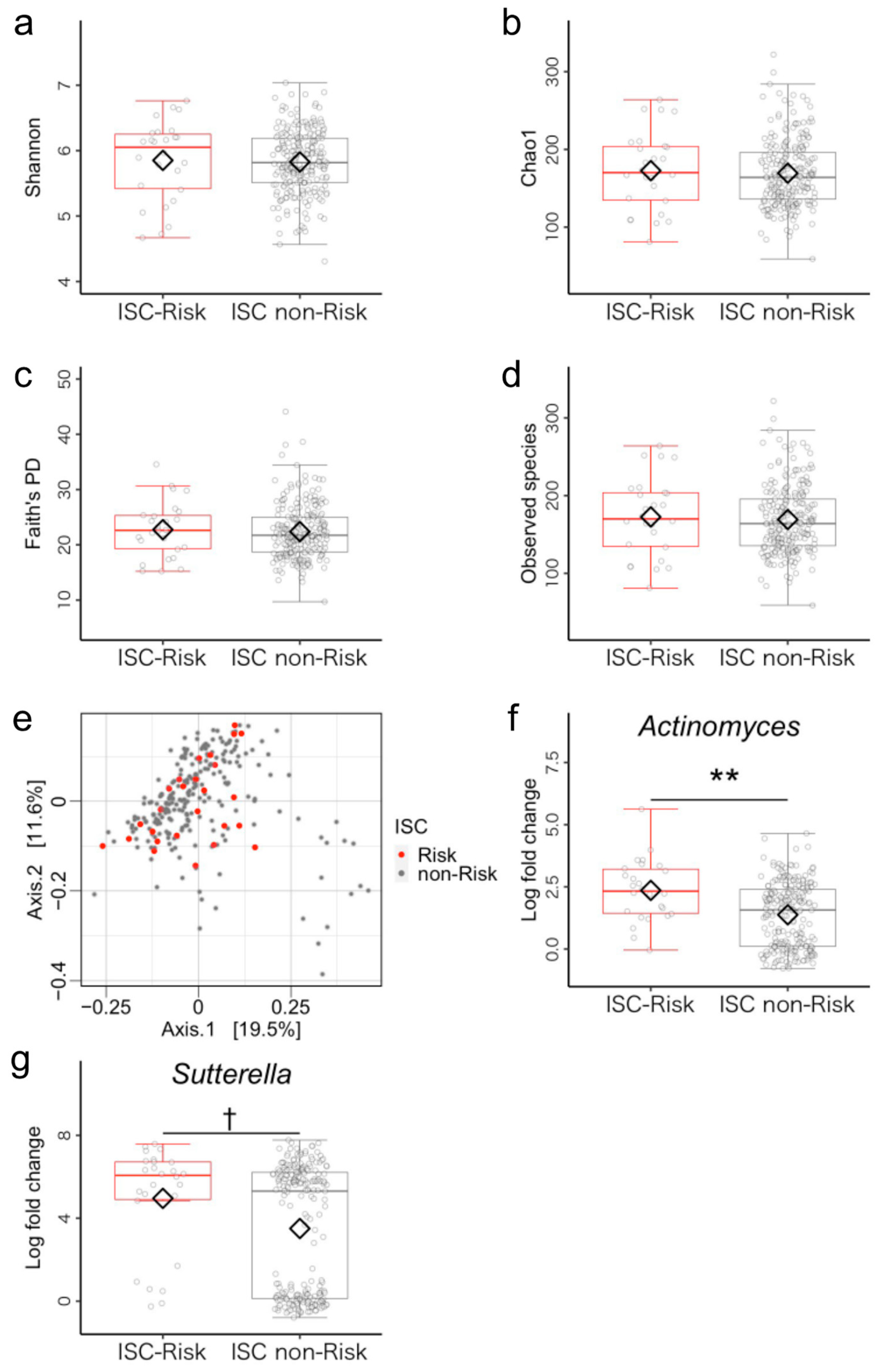
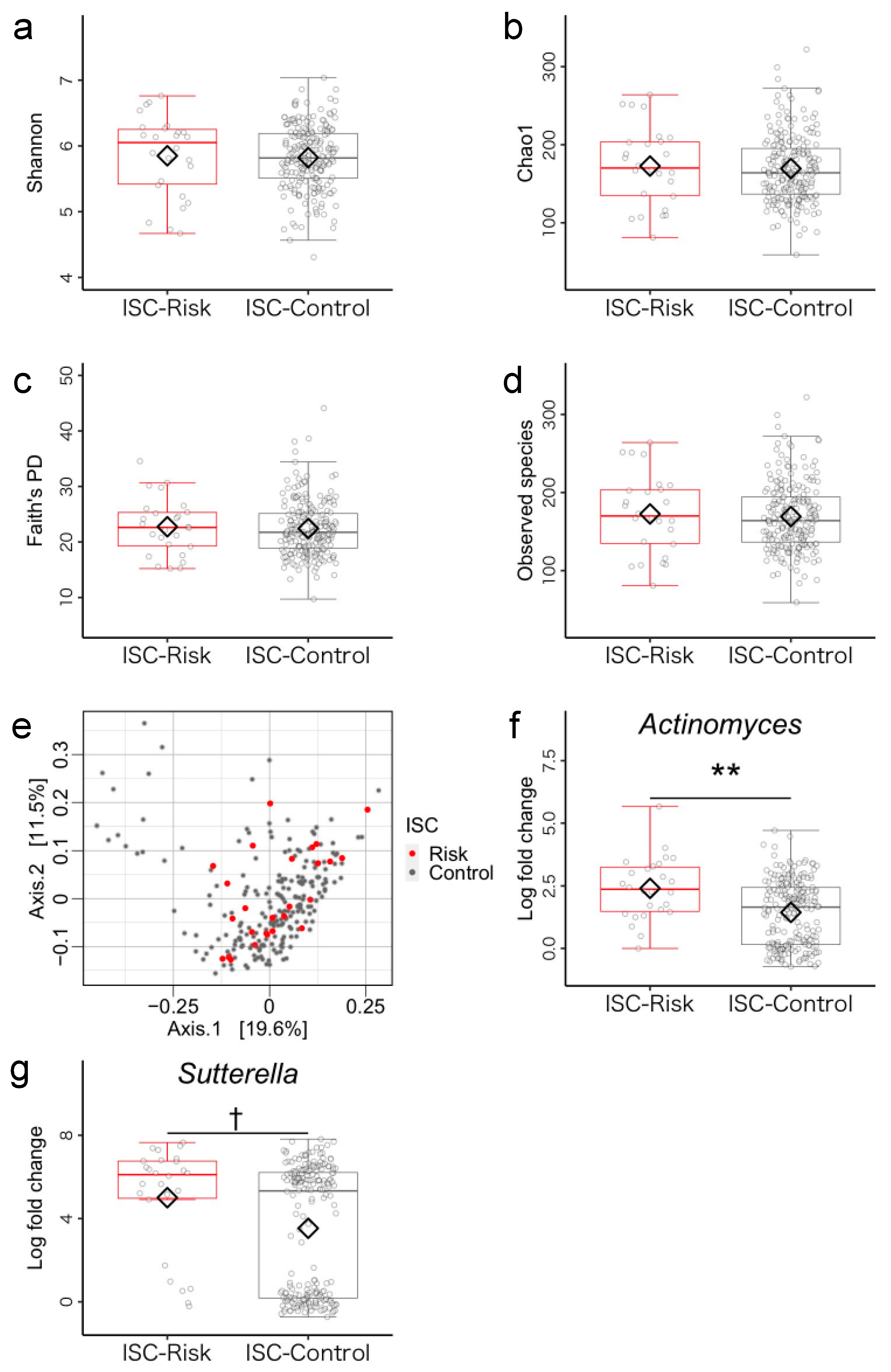
3.1. Microbiome Components in Inhibitory Self-Control (ISC)-Risk and Non-Risk Groups
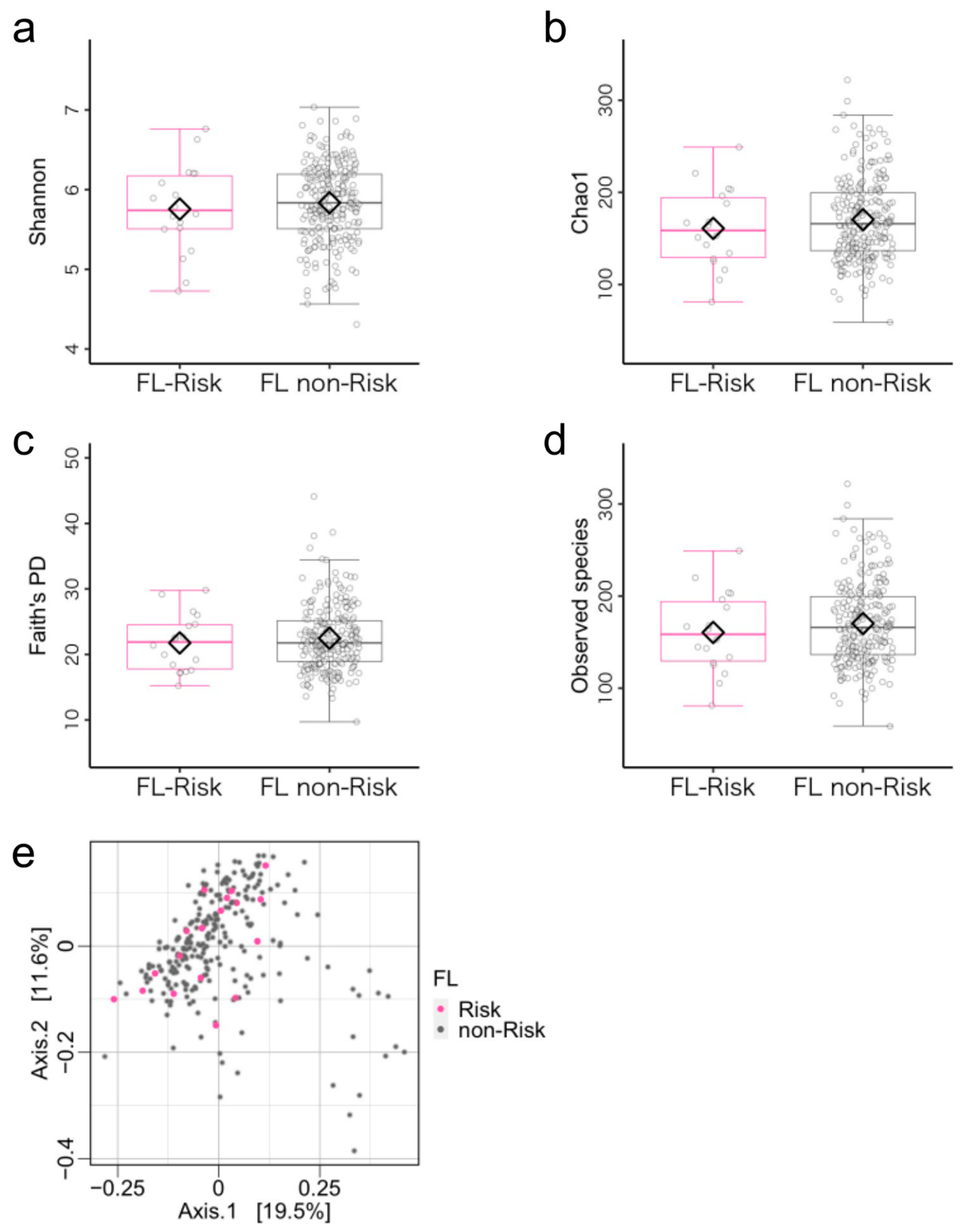
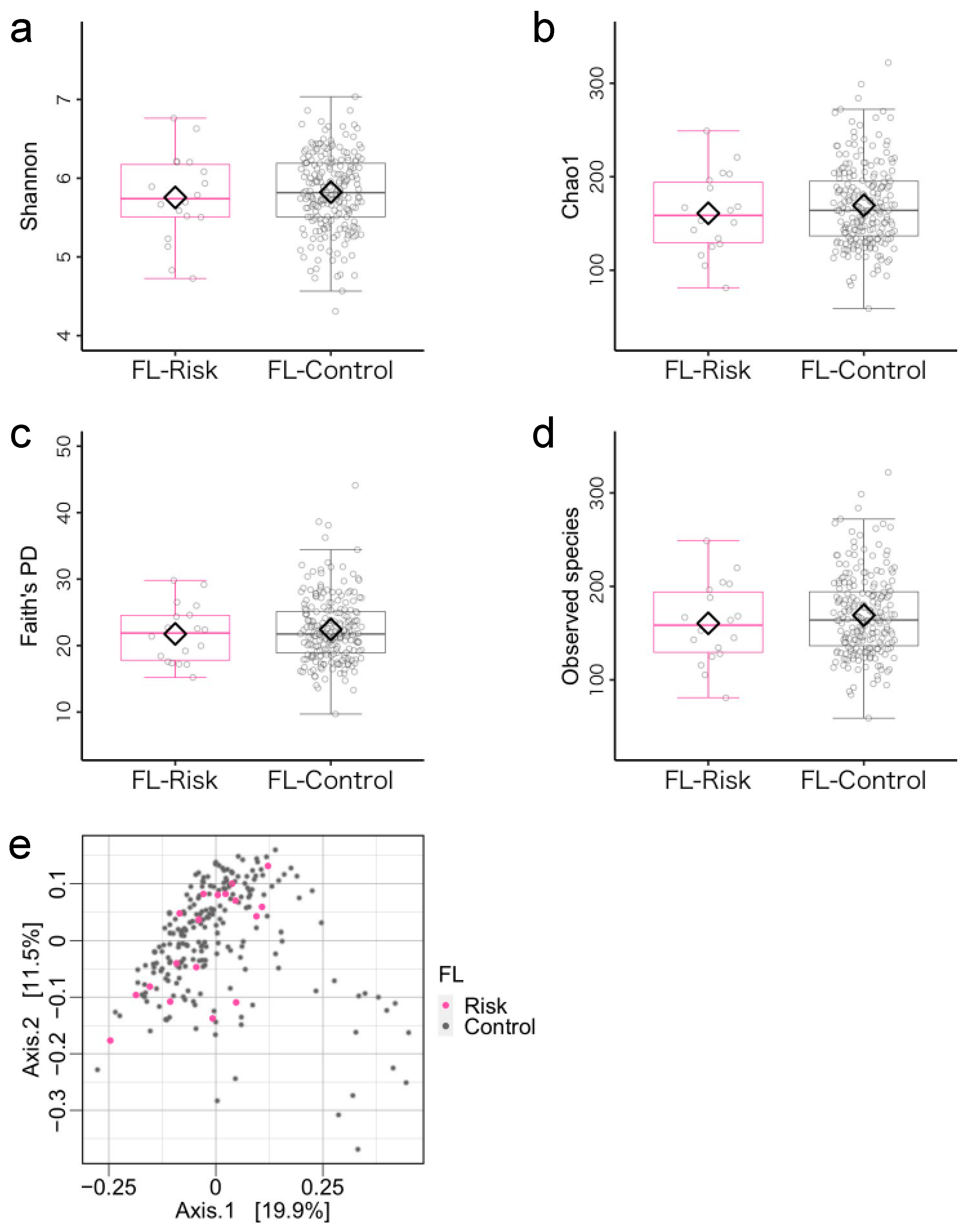
3.2. Microbiome Components in the Flexibility (FL)-Risk and Non-Risk Groups
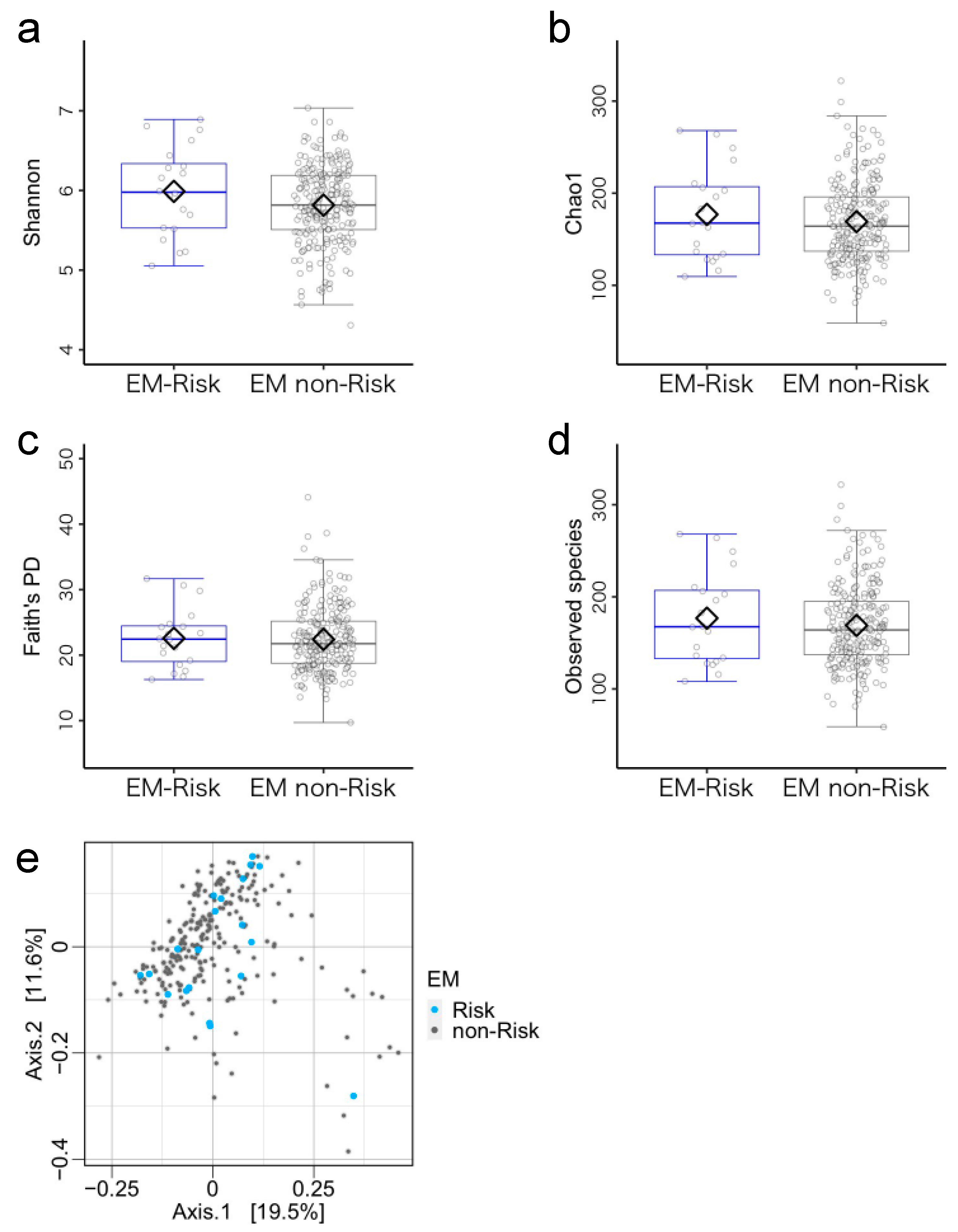
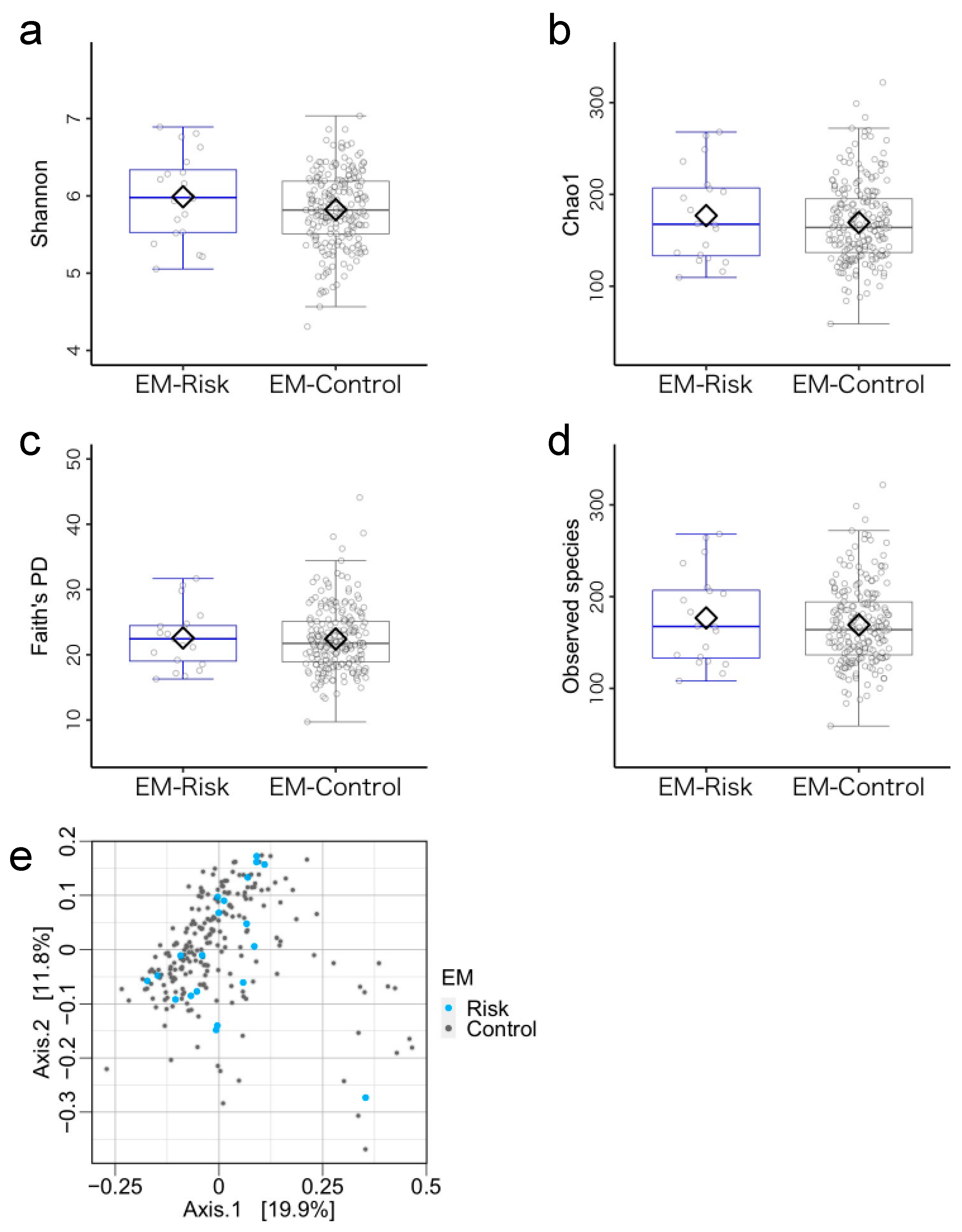
3.3. Microbiome Components in the Emergent Metacognition (EM)-Risk and Non-Risk Groups
3.4. Comparisons of Gut Microbiota Structure between EF-Risk and EF-Control Groups
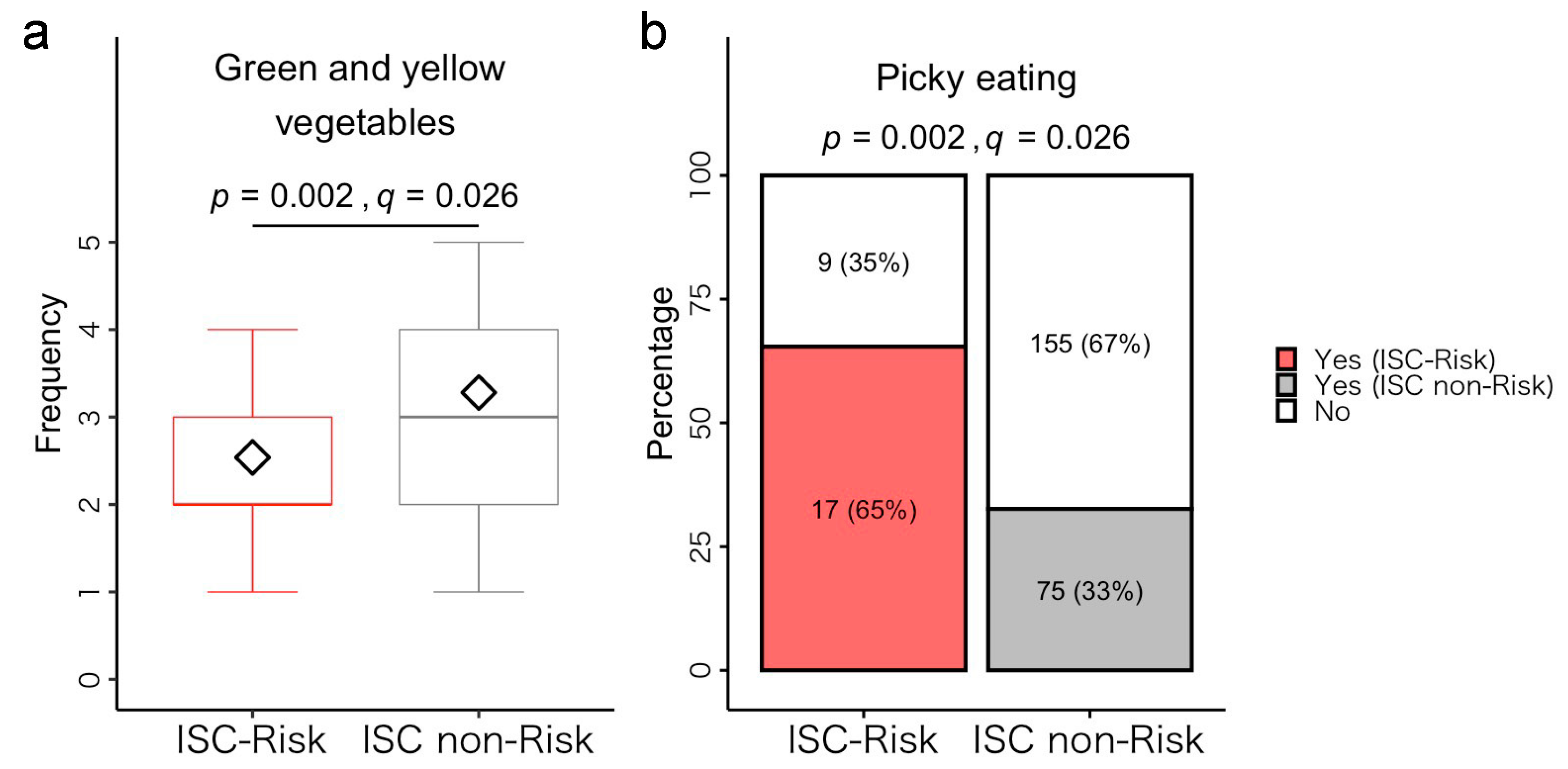
3.5. Post Hoc Analysis of Differences between ISC-Risk and Non-Risk Groups
3.5.1. Dietary Habits
3.5.2. Physical Symptoms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Diamond, A. Executive Functions. Annu. Rev. Psychol. 2013, 64, 135–168. [Google Scholar] [CrossRef]
- Kerr, A.; Zelazo, P.D. Development of “Hot” Executive Function: The Children’s Gambling Task. Brain Cogn. 2004, 55, 148–157. [Google Scholar] [CrossRef]
- Moffitt, T.E.; Arseneault, L.; Belsky, D.; Dickson, N.; Hancox, R.J.; Harrington, H.; Houts, R.; Poulton, R.; Roberts, B.W.; Ross, S.; et al. A Gradient of Childhood Self-Control Predicts Health, Wealth, and Public Safety. Proc. Natl. Acad. Sci. USA 2011, 108, 2693–2698. [Google Scholar] [CrossRef] [PubMed]
- Zelazo, P.D.; Carlson, S.M. Hot and Cool Executive Function in Childhood and Adolescence: Development and Plasticity. Child Dev. Perspect. 2012, 6, 354–360. [Google Scholar] [CrossRef]
- Gyurak, A.; Gross, J.J.; Etkin, A. Explicit and Implicit Emotion Regulation: A Dual-Process Framework. Cogn. Emot. 2011, 25, 400–412. [Google Scholar] [CrossRef]
- Zelazo, P.D.; Mller, U. Executive Function in Typical and Atypical Development. In Blackwell Handbook of Childhood Cognitive Development; Blackwell Publishing Ltd.: Malden, MA, USA, 2002; pp. 445–469. [Google Scholar]
- Kim, S.; Nordling, J.K.; Yoon, J.E.; Boldt, L.J.; Kochanska, G. Effortful Control in “Hot” and “Cool” Tasks Differentially Predicts Children’s Behavior Problems and Academic Performance. J. Abnorm. Child Psychol. 2013, 41, 43–56. [Google Scholar] [CrossRef]
- Richmond-Rakerd, L.S.; Caspi, A.; Ambler, A.; d’Arbeloff, T.; de Bruine, M.; Elliott, M.; Harrington, H.; Hogan, S.; Houts, R.M.; Ireland, D.; et al. Childhood Self-Control Forecasts the Pace of Midlife Aging and Preparedness for Old Age. Proc. Natl. Acad. Sci. USA 2021, 118, e2010211118. [Google Scholar] [CrossRef] [PubMed]
- Diamond, A.; Lee, K. Interventions Shown to Aid Executive Function Development in Children 4 to 12 Years Old. Science 2011, 333, 959–964. [Google Scholar] [CrossRef]
- Flook, L.; Goldberg, S.B.; Pinger, L.; Davidson, R.J. Promoting Prosocial Behavior and Self-Regulatory Skills in Preschool Children through a Mindfulness-Based Kindness Curriculum. Dev. Psychol. 2015, 51, 44–51. [Google Scholar] [CrossRef]
- Razza, R.A.; Bergen-Cico, D.; Raymond, K. Enhancing Preschoolers’ Self-Regulation via Mindful Yoga. J. Child Fam. Stud. 2015, 24, 372–385. [Google Scholar] [CrossRef]
- Li, W.Z.; Stirling, K.; Yang, J.J.; Zhang, L. Gut Microbiota and Diabetes: From Correlation to Causality and Mechanism. World J. Diabetes 2020, 11, 293–308. [Google Scholar] [CrossRef]
- Kelly, J.R.; Borre, Y.; O’ Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the Blues: Depression-Associated Gut Microbiota Induces Neurobehavioural Changes in the Rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.A.; Rinaman, L.; Cryan, J.F. Stress & the Gut-Brain Axis: Regulation by the Microbiome. Neurobiol. Stress 2017, 7, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.L.; Adame, M.D.; Liou, C.W.; Barlow, J.T.; Lai, T.T.; Sharon, G.; Schretter, C.E.; Needham, B.D.; Wang, M.I.; Tang, W.; et al. Microbiota Regulate Social Behaviour via Stress Response Neurons in the Brain. Nature 2021, 595, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Roswall, J.; Olsson, L.M.; Kovatcheva-Datchary, P.; Nilsson, S.; Tremaroli, V.; Simon, M.C.; Kiilerich, P.; Akrami, R.; Krämer, M.; Uhlén, M.; et al. Developmental Trajectory of the Healthy Human Gut Microbiota During the First 5 Years of Life. Cell Host Microbe 2021, 29, 765–776.e3. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal Development of the Gut Microbiome in Early Childhood from the TEDDY Study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Giedd, J.N.; Blumenthal, J.; Jeffries, N.O.; Castellanos, F.X.; Liu, H.; Zijdenbos, A.; Paus, T.; Evans, A.C.; Rapoport, J.L. Brain Development during Childhood and Adolescence: A Longitudinal MRI Study. Nat. Neurosci. 1999, 2, 861–863. Available online: https://www.nature.com/articles/nn1099_861 (accessed on 28 December 2022).
- Barnea-Goraly, N.; Menon, V.; Eckert, M.; Tamm, L.; Bammer, R.; Karchemskiy, A.; Dant, C.C.; Reiss, A.L. White matter development during childhood and adolescence: A cross-sectional diffusion tensor imaging study. Cereb. Cortex 2005, 15, 1848–1854. [Google Scholar] [CrossRef]
- Gee, D.G.; Humphreys, K.L.; Flannery, J.; Goff, B.; Telzer, E.H.; Shapiro, M.; Hare, T.A.; Bookheimer, S.Y.; Tottenham, N. A developmental shift from positive to negative connectivity in human amygdala-prefrontal circuitry. J. Neurosci. 2013, 33, 4584–4593. [Google Scholar] [CrossRef]
- Gao, W.; Salzwedel, A.P.; Carlson, A.L.; Xia, K.; Azcarate-Peril, M.A.; Styner, M.A.; Thompson, A.L.; Geng, X.; Goldman, B.D.; Gilmore, J.H.; et al. Gut Microbiome and Brain Functional Connectivity in Infants-A Preliminary Study Focusing on the Amygdala. Psychopharmacology 2019, 236, 1641–1651. [Google Scholar] [CrossRef]
- Carlson, A.L.; Xia, K.; Azcarate-Peril, M.A.; Rosin, S.P.; Fine, J.P.; Mu, W.; Zopp, J.B.; Kimmel, M.C.; Styner, M.A.; Thompson, A.L.; et al. Infant Gut Microbiome Composition Is Associated with Non-social Fear Behavior in a Pilot Study. Nat. Commun. 2021, 12, 3294. [Google Scholar] [CrossRef]
- Tillisch, K.; Labus, J.; Kilpatrick, L.; Jiang, Z.; Stains, J.; Ebrat, B.; Guyonnet, D.; Legrain-Raspaud, S.; Trotin, B.; Naliboff, B.; et al. Consumption of Fermented Milk Product with Probiotic Modulates Brain Activity. Gastroenterology 2013, 144, 1394–1401.e4. [Google Scholar] [CrossRef] [PubMed]
- Loughman, A.; Ponsonby, A.L.; O’Hely, M.; Symeonides, C.; Collier, F.; Tang, M.L.K.; Carlin, J.; Ranganathan, S.; Allen, K.; Pezic, A.; et al. Gut Microbiota Composition during Infancy and Subsequent Behavioural Outcomes. eBiomedicine 2020, 52, 102640. [Google Scholar] [CrossRef] [PubMed]
- Aatsinki, A.K.; Lahti, L.; Uusitupa, H.M.; Munukka, E.; Keskitalo, A.; Nolvi, S.; O’Mahony, S.; Pietilä, S.; Elo, L.L.; Eerola, E.; et al. Gut Microbiota Composition Is Associated with Temperament Traits in Infants. Brain Behav. Immun. 2019, 80, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Carlson, A.L.; Xia, K.; Azcarate-Peril, M.A.; Goldman, B.D.; Ahn, M.; Styner, M.A.; Thompson, A.L.; Geng, X.; Gilmore, J.H.; Knickmeyer, R.C. Infant Gut Microbiome Associated with Cognitive Development. Biol. Psychiatry 2018, 83, 148–159. [Google Scholar] [CrossRef]
- Boulangé, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.E. Impact of the Gut Microbiota on Inflammation, Obesity, and Metabolic Disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Bernstein, C.N.; Forbes, J.D. Gut Microbiome in Inflammatory Bowel Disease and Other Chronic Immune-Mediated Inflammatory Diseases. Inflamm. Intestig. Dis. 2017, 2, 116–123. [Google Scholar] [CrossRef]
- Gholizadeh, P.; Mahallei, M.; Pormohammad, A.; Varshochi, M.; Ganbarov, K.; Zeinalzadeh, E.; Yousefi, B.; Bastami, M.; Tanomand, A.; Mahmood, S.S.; et al. Microbial Balance in the Intestinal Microbiota and Its Association with Diabetes, Obesity and Allergic Disease. Microb. Pathog. 2019, 127, 48–55. [Google Scholar] [CrossRef]
- Gioia, G.A.; Espy, K.A.; Isquith, P.K. Behavior Rating Inventory of Executive Function-Preschool Version (BRIEF-P); Psychological Assessment Resources: Odessa, FL, USA, 2003. [Google Scholar]
- Ukena, Y.; Hashimoto, S.; Deguchi, T. Development of the Japanese Version of BRIEF-P: Toward the Practical Use in Support for Children with Developmental Disorders. Jpn. J. Support Syst. Dev. Disabil. 2008, 72, 59–64. [Google Scholar]
- Sherman, E.M.S.; Brooks, B.L. Behavior Rating Inventory of Executive Function—Preschool Version (BRIEF-P): Test Review and Clinical Guidelines for Use. Child Neuropsychol. 2010, 16, 503–519. [Google Scholar] [CrossRef]
- Daunhauer, L.A.; Fidler, D.J.; Hahn, L.; Will, E.; Lee, N.R.; Hepburn, S. Profiles of Everyday Executive Functioning in Young Children with Down Syndrome. Am. J. Intellect. Dev. Disabil. 2014, 119, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.R.; Anand, P.; Will, E.; Adeyemi, E.I.; Clasen, L.S.; Blumenthal, J.D.; Giedd, J.N.; Daunhauer, L.A.; Fidler, D.J.; Edgin, J.O. Everyday Executive Functions in Down Syndrome from Early Childhood to Young Adulthood: Evidence for Both Unique and Shared Characteristics Compared to Youth with Sex Chromosome Trisomy (XXX and XXY). Front. Behav. Neurosci. 2015, 9, 264. [Google Scholar] [CrossRef]
- Isquith, P.K.; Gioia, G.A.; Espy, K.A. Executive Function in Preschool Children: Examination through Everyday Behavior. Dev. Neuropsychol. 2004, 26, 403–422. [Google Scholar] [CrossRef]
- Skogan, A.H.; Egeland, J.; Zeiner, P.; Øvergaard, K.R.; Oerbeck, B.; Reichborn-Kjennerud, T.; Aase, H. Factor Structure of the Behavior Rating Inventory of Executive Functions (BRIEF-P) at Age Three Years. Child Neuropsychol. 2016, 22, 472–492. [Google Scholar] [CrossRef]
- Galloway, A.T.; Fiorito, L.; Lee, Y.; Birch, L.L. Parental Pressure, Dietary Patterns, and Weight Status among Girls Who Are “Picky Eaters”. J. Am. Diet. Assoc. 2005, 105, 541–548. [Google Scholar] [CrossRef]
- Saji, N.; Tsuduki, T.; Murotani, K.; Hisada, T.; Sugimoto, T.; Kimura, A.; Niida, S.; Toba, K.; Sakurai, T. Relationship between the Japanese-Style Diet, Gut Microbiota, and Dementia: A Cross-Sectional Study. Nutrition 2022, 94, 111524. [Google Scholar] [CrossRef]
- Kushida, M.; Sugawara, S.; Asano, M.; Yamamoto, K.; Fukuda, S.; Tsuduki, T. Effects of the 1975 Japanese Diet on the Gut Microbiota in Younger Adults. J. Nutr. Biochem. 2019, 64, 121–127. [Google Scholar] [CrossRef]
- van Buuren, S.; Groothuis-Oudshoorn, K. Mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- van Buuren, S.; Boshuizen, H.C.; Knook, D.L. Multiple Imputation of Missing Blood Pressure Covariates in Survival Analysis. Stat. Med. 1999, 18, 681–694. [Google Scholar] [CrossRef]
- Lewis, S.J.; Heaton, K.W. Stool Form Scale as a Useful Guide to Intestinal Transit Time. Scand. J. Gastroenterol. 1997, 32, 920–924. [Google Scholar] [CrossRef]
- Lacy, B.E.; Patel, N.K. Rome Criteria and a Diagnostic Approach to Irritable Bowel Syndrome. J. Clin. Med. 2017, 6, 99. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.T.; Steer, R.A.; Brown, G.K. Manual for the Beck Depression Inventory—II; Psychological Corporation: San Antonio, TX, USA, 1996. [Google Scholar]
- Hiroe, T.; Kojima, M.; Yamamoto, I.; Nojima, S.; Kinoshita, Y.; Hashimoto, N.; Watanabe, N.; Maeda, T.; Furukawa, T.A. Gradations of Clinical Severity and Sensitivity to Change Assessed with the Beck Depression Inventory-II in Japanese Patients with Depression. Psychiatry Res. 2005, 135, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Furukawa, T.A.; Takahashi, H.; Kawai, M.; Nagaya, T.; Tokudome, S. Cross-Cultural Validation of the Beck Depression Inventory-II in Japan. Psychiatry Res. 2002, 110, 291–299. [Google Scholar] [CrossRef]
- Chilcoat, H.D.; Breslau, N. Does Psychiatric History Bias Mothers’ Reports? An Application of a New Analytic Approach. J. Am. Acad. Child Adolesc. Psychiatry 1997, 36, 971–979. [Google Scholar] [CrossRef]
- Müller, J.M.; Achtergarde, S.; Furniss, T. The Influence of Maternal Psychopathology on Ratings of Child Psychiatric Symptoms: An SEM Analysis on Cross-Informant Agreement. Eur. Child Adolesc. Psychiatry 2011, 20, 241–252. [Google Scholar] [CrossRef]
- Watanabe, S.; Kameoka, S.; Shinozaki, N.O.; Kubo, R.; Nishida, A.; Kuriyama, M.; Takeda, A.K. A cross-sectional analysis from the Mykinso Cohort Study: Establishing reference ranges for Japanese gut microbial indices. Biosci. Microbiota Food Health 2021, 40, 123–134. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yoshida, N.; Watanabe, S.; Yamasaki, H.; Sakuma, H.; Takeda, A.K.; Yamashita, T.; Hirata, K. Average Gut Flora in Healthy Japanese Subjects Stratified by Age and Body Mass Index. Biosci. Microbiota Food Health 2022, 41, 2021–2056. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. Available online: http://www.R-project.org (accessed on 5 January 2021).
- Lin, H.; Peddada, S.D. Analysis of Compositions of Microbiomes with Bias Correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Van Pee, T.; Hogervorst, J.; Dockx, Y.; Witters, K.; Thijs, S.; Wang, C.; Bongaerts, E.; Van Hamme, J.D.; Vangronsveld, J.; Ameloot, M.; et al. Accumulation of Black Carbon Particles in Placenta, Cord Blood, and Childhood Urine in Association with the Intestinal Microbiome Diversity and Composition in Four- to Six-Year-Old Children in the ENVIR ON AGE Birth Cohort. Environ. Health Perspect. 2023, 131, 17010. [Google Scholar] [CrossRef]
- Maier, L.; Goemans, C.V.; Wirbel, J.; Kuhn, M.; Eberl, C.; Pruteanu, M.; Müller, P.; Garcia-Santamarina, S.; Cacace, E.; Zhang, B.; et al. Unravelling the Collateral Damage of Antibiotics on Gut Bacteria. Nature 2021, 599, 120–124. [Google Scholar] [CrossRef]
- Seelbinder, B.; Chen, J.; Brunke, S.; Vazquez-Uribe, R.; Santhaman, R.; Meyer, A.C.; de Oliveira Lino, F.S.; Chan, K.F.; Loos, D.; Imamovic, L.; et al. Antibiotics Create a Shift from Mutualism to Competition in Human Gut Communities with a Longer-Lasting Impact on Fungi than Bacteria. Microbiome 2020, 8, 133. [Google Scholar] [CrossRef]
- Kaul, A.; Mandal, S.; Davidov, O.; Peddada, S.D. Analysis of Microbiome Data in the Presence of Excess Zeros. Front. Microbiol. 2017, 8, 2114. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.A.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M.; et al. Microbiome Differential Abundance Methods Produce Different Results across 38 Datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef]
- Testerman, T.; Li, Z.; Galuppo, B.; Graf, J.; Santoro, N. Insights from Shotgun Metagenomics into Bacterial Species and Metabolic Pathways Associated with NAFLD in Obese Youth. Hepatol. Commun. 2022, 6, 1962–1974. [Google Scholar] [CrossRef]
- Brunner, E.; Munzel, U. The Nonparametric Behrens-Fisher Problem: Asymptotic Theory and a Small-Sample Approximation. Biom. J. 2000, 42, 17–25. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- West, C.E.; Rydén, P.; Lundin, D.; Engstrand, L.; Tulic, M.K.; Prescott, S.L. Gut Microbiome and Innate Immune Response Patterns in IgE-Associated Eczema. Clin. Exp. Allergy 2015, 45, 1419–1429. [Google Scholar] [CrossRef]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front. Microbiol. 2016, 7, 1706. [Google Scholar] [CrossRef]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orrù, S.; Blois, S.; et al. Cross Sectional Evaluation of the Gut-Microbiome Metabolome Axis in an Italian Cohort of IBD Patients. Sci. Rep. 2017, 7, 9523. [Google Scholar] [CrossRef]
- Takahashi, K.; Nishida, A.; Fujimoto, T.; Fujii, M.; Shioya, M.; Imaeda, H.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Andoh, A. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. Digestion 2016, 93, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.M.; Northstone, K.; Wernimont, S.M.; Emmett, P.M. Picky Eating in Preschool Children: Associations with Dietary Fibre Intakes and Stool Hardness. Appetite 2016, 100, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Peters, B.A.; Friedlander, C.; Freiman, H.J.; Goedert, J.J.; Sinha, R.; Miller, G.; Bernstein, M.A.; Hayes, R.B.; Ahn, J. Association of Dietary Fibre Intake and Gut Microbiota in Adults. Br. J. Nutr. 2018, 120, 1014–1022. [Google Scholar] [CrossRef]
- Shimizu, H.; Ohue-Kitano, R.; Kimura, I. Regulation of host energy metabolism by gut microbiota-derived short-chain fatty acids. Glycative Stress Res. 2019, 6, 181–191. [Google Scholar] [CrossRef]
- Aguilera, A.C.; Dagher, I.A.; Kloepfer, K.M. Role of the Microbiome in Allergic Disease Development. Curr. Allergy Asthma Rep. 2020, 20, 44. [Google Scholar] [CrossRef]
- Yamagishi, M.; Akagawa, S.; Akagawa, Y.; Nakai, Y.; Yamanouchi, S.; Kimata, T.; Hashiyada, M.; Akane, A.; Tsuji, S.; Kaneko, K. Decreased Butyric Acid-Producing Bacteria in Gut Microbiota of Children with Egg Allergy. Allergy 2021, 76, 2279–2282. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, T.; Yuan, H.; Pan, W.; Dai, Q. Correlations of Inflammatory Factors with Intestinal Flora and Gastrointestinal Incommensurate Symptoms in Children with Asthma. Med. Sci. Monit. 2018, 24, 7975–7979. [Google Scholar] [CrossRef]
- Spataro, P.; Cerutti, R.; Laghi, F.; Longobardi, E. Indirect relations between language and behavior problems in preschoolers: The role of executive difficulties. J. Genet. Psychol. 2022, 183, 495–513. [Google Scholar] [CrossRef]
- Loe, I.M.; Chatav, M.; Alduncin, N. Complementary assessments of executive function in preterm and full-term preschoolers. Child Neuropsychol. 2015, 21, 331–353. [Google Scholar] [CrossRef] [PubMed]
- Isquith, P.K.; Gioia, G.A.; PAR Staff. BRIEF-P Interceptive Report; Psychological Assessment Resources: Lutz, FL, USA, 2013. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujihara, H.; Matsunaga, M.; Ueda, E.; Kajiwara, T.; Takeda, A.K.; Watanabe, S.; Baba, K.; Hagihara, K.; Myowa, M. Altered Gut Microbiota Composition Is Associated with Difficulty in Explicit Emotion Regulation in Young Children. Microorganisms 2023, 11, 2245. https://doi.org/10.3390/microorganisms11092245
Fujihara H, Matsunaga M, Ueda E, Kajiwara T, Takeda AK, Watanabe S, Baba K, Hagihara K, Myowa M. Altered Gut Microbiota Composition Is Associated with Difficulty in Explicit Emotion Regulation in Young Children. Microorganisms. 2023; 11(9):2245. https://doi.org/10.3390/microorganisms11092245
Chicago/Turabian StyleFujihara, Hideaki, Michiko Matsunaga, Eriko Ueda, Takamasa Kajiwara, Aya K. Takeda, Satoshi Watanabe, Kairi Baba, Keisuke Hagihara, and Masako Myowa. 2023. "Altered Gut Microbiota Composition Is Associated with Difficulty in Explicit Emotion Regulation in Young Children" Microorganisms 11, no. 9: 2245. https://doi.org/10.3390/microorganisms11092245
APA StyleFujihara, H., Matsunaga, M., Ueda, E., Kajiwara, T., Takeda, A. K., Watanabe, S., Baba, K., Hagihara, K., & Myowa, M. (2023). Altered Gut Microbiota Composition Is Associated with Difficulty in Explicit Emotion Regulation in Young Children. Microorganisms, 11(9), 2245. https://doi.org/10.3390/microorganisms11092245






