Type I Interferon Pathway-Related Hub Genes as a Potential Therapeutic Target for SARS-CoV-2 Omicron Variant-Induced Symptoms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Differential Expression Analysis of Genes
2.2. Functional Enrichment Analysis
2.3. Protein–Protein Interaction (PPI) Analysis of Differentially Expressed Genes and Identification of Hub Genes
2.4. Construction of the miRNA-Target Regulatory Network
2.5. Validation of the Diagnostic Value of Hub Genes
2.6. Immune Infiltration Analysis
2.7. Retrospective Analysis of Blood Counts in COVID-19 Patients
2.8. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) Verification
2.9. Prediction of Potential Therapeutic Agents
2.10. Statistical Analysis
3. Results
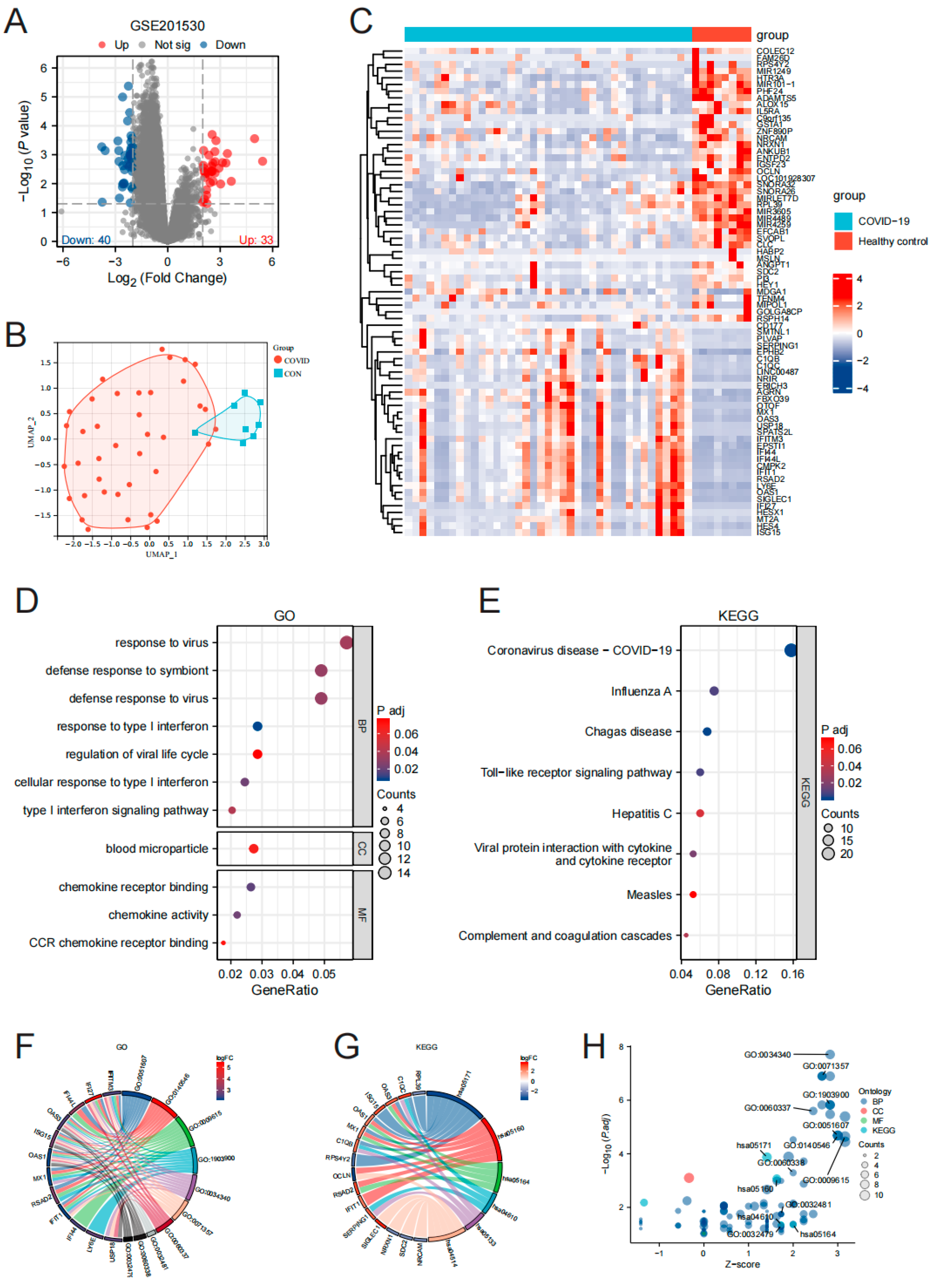
3.1. Screening and Functional Enrichment Analysis of Differentially Expressed Genes in PBMC of Omicron Infection
3.2. Gene Screening and Functional Enrichment Analysis of PBMC Hub Genes in Omicron Infection
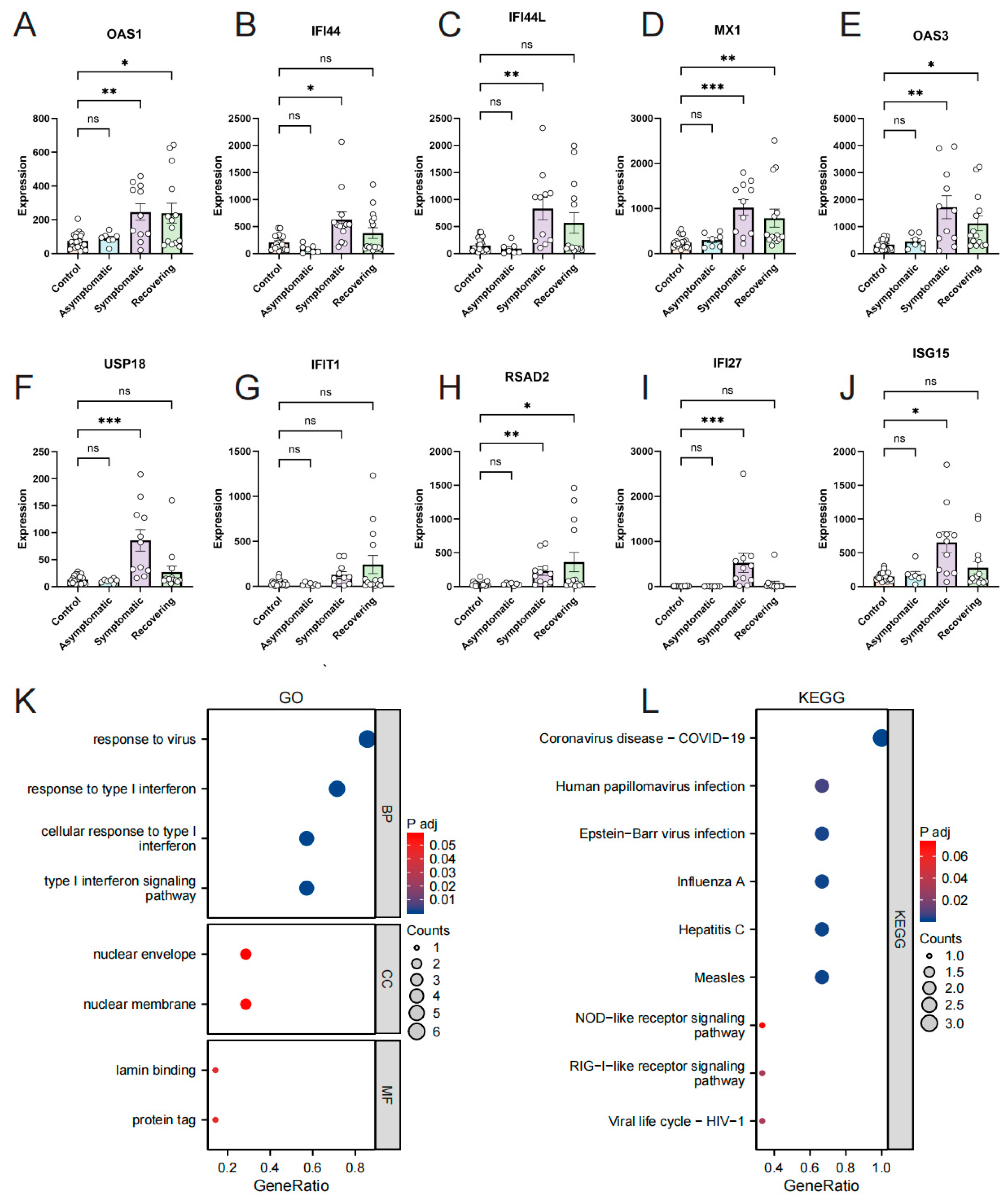
3.3. Confirmation of Hub Genes Expression and Diagnostic Value in GSE179627
3.4. Investigation of the Relationship between Hub Genes and Omicron Infection
3.5. Validation of Hub Genes Expression by RT-qPCR
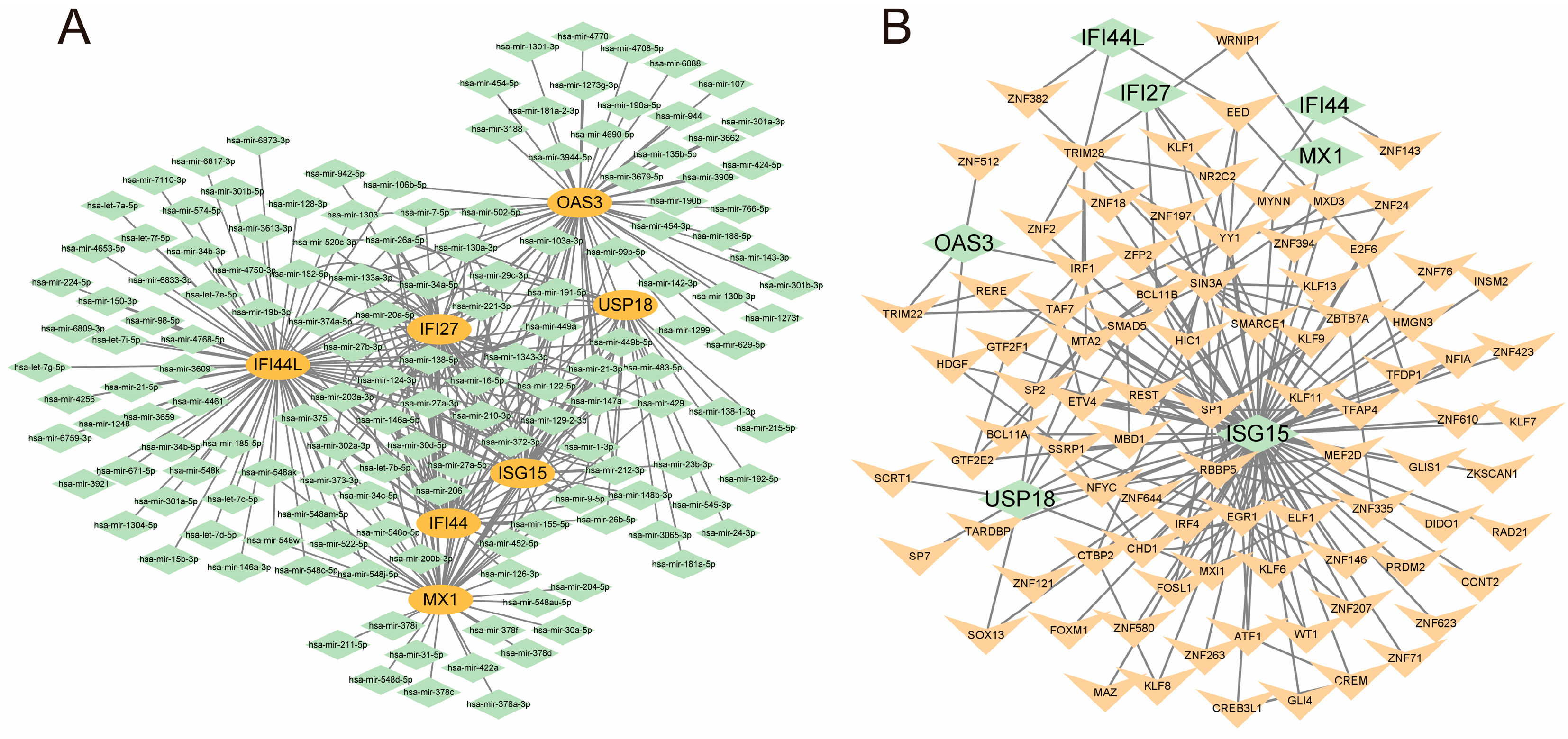
3.6. Construction of mRNA-miRNA and mRNA-TF Regulatory Networks
3.7. Analysis of PMBC Immune Infiltration in Omicron Infection
3.8. Validation of Retrospective Blood Analysis and Immune Infiltration Results in Omicron Infection
3.9. Target Drug Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carlos, W.G.; Dela Cruz, C.S.; Cao, B.; Pasnick, S.; Jamil, S. Novel Wuhan (2019-nCoV) coronavirus. Am. J. Respir. Crit. Care Med. 2020, 201, P7–P8. [Google Scholar] [CrossRef]
- Song, F.; Shi, N.; Shan, F.; Zhang, Z.; Shen, J.; Lu, H.; Ling, Y.; Jiang, Y.; Shi, Y. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology 2020, 295, 210–217. [Google Scholar] [CrossRef]
- Weekly Epidemiological Update on COVID-19. 2021. Available online: https://covid19.who.int/ (accessed on 4 June 2023).
- Al-Karmalawy, A.A.; Soltane, R.; Abo Elmaaty, A.; Tantawy, M.A.; Antar, S.A.; Yahya, G.; Chrouda, A.; Pashameah, R.A.; Mustafa, M.; Abu Mraheil, M.; et al. Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview. Vaccines 2021, 9, 1317. [Google Scholar] [CrossRef] [PubMed]
- UK Health Security Agency. SARS-CoV-2 Variants of Concern and Variants under Investigation in England-Technical Briefing: Update on Hospitalisation and Vaccine Effectiveness for Omicron VOC-21NOV-01 (B.1.1.529). Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1045619/Technical-Briefing-31-Dec-2021-Omicron_severity_update.pdf (accessed on 9 June 2023).
- Ulloa, A.C.; Buchan, S.A.; Daneman, N.; Brown, K.A. Estimates of SARS-CoV-2 Omicron variant severity in Ontario, Canada. J. Am. Med. Assoc. 2022, 327, 1286–1288. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, A.D.; Brunkard, J.M.; Boehmer, T.K.; Peterson, E.; Adjei, S.; Binder, A.M.; Cobb, S.; Graff, P.; Hidalgo, P.; Panaggio, M.J. Trends in disease severity and health care utilization during the early Omicron variant period compared with previous SARS-CoV-2 high transmission periods—United States, December 2020–January 2022. Morb. Mortal. Wkly. Rep. 2022, 71, 146–152. [Google Scholar] [CrossRef]
- Lewnard, J.A.; Hong, V.X.; Patel, M.M.; Kahn, R.; Lipsitch, M.; Tartof, S.Y. Clinical outcomes among patients infected with Omicron (B.1.1.529) SARS-CoV-2 variant in southern California. medRxiv 2022. [Google Scholar] [CrossRef]
- Bussani, R.; Schneider, E.; Zentilin, L.; Collesi, C.; Ali, H.; Braga, L.; Volpe, M.C.; Colliva, A.; Zanconati, F.; Berlot, G.; et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine 2020, 61, 103104. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Jumper, C.C.; Ackerman, P.J.; Bracha, D.; Donlic, A.; Kim, H.; Kenney, D.; Castello-Serrano, I.; Suzuki, S.; Tamura, T.; et al. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. eLife 2021, 10, e65962. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K.; et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef]
- Shuai, H.; Chan, J.F.W.; Hu, B.; Chai, Y.; Yuen, T.T.T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Jassat, W.; Karim, S.; Mudara, C. Clinical severity of COVID-19 patients admitted to hospitals in Gauteng, South Africa during the omicron-dominant fourth wave. SSRN Electron. J. 2021. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Hospital admission 50–70% less likely with omicron than delta, but transmission a major concern. BMJ 2021, 375, n3151. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, S.; Subbian, S. Critical Determinants of Cytokine Storm and Type I Interferon Response in COVID-19 Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00299-20. [Google Scholar] [CrossRef]
- Zheng, B.; He, M.-L.; Wong, K.-L.; Lum, C.T.; Poon, L.L.M.; Peng, Y.; Guan, Y.; Lin, M.C.M.; Kung, H.-F. Potent Inhibition of SARS-associated coronavirus (SCoV) infection and replication by type I interferons (IFN-α/β) but not by type II interferon (IFN-γ). J. Interferon Cytokine Res. 2004, 24, 388–390. [Google Scholar] [CrossRef]
- Jamilloux, Y.; Henry, T.; Belot, A.; Viel, S.; Fauter, M.; El Jammal, T.; Walzer, T.; François, B.; Sève, P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020, 19, 102567. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef]
- Lee, J.S.; Park, S.; Jeong, H.W.; Ahn, J.Y.; Choi, S.J.; Lee, H.; Choi, B.; Nam, S.K.; Sa, M.; Kwon, J.-S.; et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020, 5, eabd1554. [Google Scholar] [CrossRef]
- Lin, Z.; Niu, J.; Xu, Y.; Qin, L.; Ding, J.; Zhou, L. Clinical efficacy and adverse events of baricitinib treatment for coronavirus disease-2019 (COVID-19): A systematic review and meta-analysis. J. Med. Virol. 2022, 94, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Asante, I.A.; Hsu, S.N.; Boatemaa, L.; Kwasah, L.; Adusei-Poku, M.; Odoom, J.K.; Awuku-Larbi, Y.; Foulkes, B.H.; Oliver-Commey, J.; Asiedu, E.K.; et al. Repurposing an integrated national influenza platform for genomic surveillance of SARS-CoV-2 in Ghana: A molecular epidemiological analysis. Lancet Glob. Health. 2023, 11, e1075–e1085. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.C.; Shirk, P.; Lambrou, A.S.; Hassell, N.; Zheng, X.Y.; Payne, A.B.; Ali, A.R.; Batra, D.; Caravas, J.; Chau, R.; et al. Genomic Surveillance for SARS-CoV-2 Variants: Circulation of Omicron Lineages—United States, January 2022–May 2023, MMWR. Morb. Mortal. Wkly. Rep. 2023, 72, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Kawada, J.I.; Takeuchi, S.; Imai, H.; Okumura, T.; Horiba, K.; Suzuki, T.; Torii, Y.; Yasuda, K.; Imanaka-Yoshida, K.; Ito, Y. Immune cell infiltration landscapes in pediatric acute myocarditis analyzed by CIBERSORT. J. Cardiol. 2021, 77, 174–178. [Google Scholar] [CrossRef]
- Lee, H.K.; Knabl, L.; Walter, M.; Furth, P.A.; Hennighausen, L. Limited cross-variant immune response from SARS-CoV-2 Omicron BA.2 in naïve but not previously infected outpatients. iScience 2022, 25, 105369. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, D.; Li, K.; Ding, X.; Li, L.; Liu, Y.; Liu, D.; Lin, J.; Teng, X.; Li, Y.; et al. Transcriptome Analysis of Peripheral Blood Mononuclear Cells Reveals Distinct Immune Response in Asymptomatic and Re-Detectable Positive COVID-19 Patients. Front. Immunol. 2021, 12, 716075. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhou, X.; Cheng, L.; Wen, L.; An, T.; Gao, H.; Deng, H.; Yan, Q.; Zhang, X.; Li, Y.; et al. Machine learning algorithms utilizing blood parameters enable early detection of immunethrombotic dysregulation in COVID-19. Clin. Transl. Med. 2021, 11, e523. [Google Scholar] [CrossRef]
- Zhang, C.; Feng, Y.G.; Tam, C.; Wang, N.; Feng, Y. Transcriptional Profiling and Machine Learning Unveil a Concordant Biosignature of Type I Interferon-Inducible Host Response Across Nasal Swab and Pulmonary Tissue for COVID-19 Diagnosis. Front. Immunol. 2021, 12, 733171. [Google Scholar] [CrossRef] [PubMed]
- Bizzotto, J.; Sanchis, P.; Abbate, M.; Lage-Vickers, S.; Lavignolle, R.; Toro, A.; Olszevicki, S.; Sabater, A.; Cascardo, F.; Vazquez, E.; et al. SARS-CoV-2 Infection Boosts MX1 Antiviral Effector in COVID-19 Patients. iScience 2020, 23, 101585. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, J.; Parthoens, E.; Schepens, B.; Fiers, W.; Saelens, X. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 2012, 86, 13445–13455. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, C. Prognostic Characterization of OAS1/OAS2/OAS3/OASL in Breast Cancer. BMC Cancer 2020, 20, 575. [Google Scholar]
- Papac-Milicevic, N.; Breuss, J.M.; Zaujec, J.; Ryban, L.; Plyushch, T.; Wagner, G.A.; Fenzl, S.; Dremsek, P.; Cabaravdic, M.; Steiner, M.; et al. The interferon stimulated gene 12 inactivates vasculoprotective functions of NR4A nuclear receptors. Circ. Res. 2012, 110, e50–e63. [Google Scholar] [CrossRef]
- Shojaei, M.; Shamshirian, A.; Monkman, J.; Grice, L.; Tran, M.; Tan, C.W.; Teo, S.M.; Rodrigues Rossi, G.; McCulloch, T.R.; Nalos, M.; et al. IFI27 transcription is an early predictor for COVID-19 outcomes, a multi-cohort observational study. Front. Immunol. 2023, 13, 1060438. [Google Scholar] [CrossRef]
- Zhao, M.; Zhou, Y.; Zhu, B.; Wan, M.; Jiang, T.; Tan, Q.; Liu, Y.; Jiang, J.; Luo, S.; Tan, Y.; et al. Ifi44l promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 2016, 75, 1998–2006. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Nogales, A.; Martinez-Sobrido, L.; Topham, D.J. Interferon-induced protein 44 interacts with cellular FK506-binding protein 5, negatively regulates host antiviral responses, and supports virus replication. mBio 2019, 10, e01839-19. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Martinez-Sobrido, L.; Topham, D.J. Novel functions of Ifi44l as a feedback regulator of host antiviral responses. J. Virol. 2019, 93, e01159-19. [Google Scholar] [CrossRef]
- Xie, L.M.; Yin, X.; Bi, J.; Luo, H.M.; Cao, X.J.; Ma, Y.W.; Liu, Y.L.; Su, J.W.; Lin, G.L.; Guo, X.G. Identification of potential biomarkers in dengue via integrated bioinformatic analysis. PLoS Negl. Trop. Dis. 2021, 15, e0009633. [Google Scholar] [CrossRef]
- Harapan, H.; Ryan, M.; Yohan, B.; Abidin, R.S.; Nainu, F.; Rakib, A.; Jahan, I.; Emran, T.B.; Ullah, I.; Panta, K.; et al. COVID-19 and dengue: Double punches for dengue-endemic countries in Asia. Rev. Med. Virol. 2021, 31, e2161. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E., Jr. The jak-stat pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Pere, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; Breillat, P.; et al. Impaired type I interferon activity and exacerbated inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Zhang, Y.; Greer, R.A.; Song, Y.; Praveen, H.; Song, Y. In silico identification of available drugs targeting cell surface BiP to disrupt SARS-CoV-2 binding and replication: Drug repurposing approach. Eur. J. Pharm. Sci. 2021, 160, 105771. [Google Scholar] [CrossRef] [PubMed]






| Gene | Sequence (5′ -> 3′) | Length | Tm | Location | |
|---|---|---|---|---|---|
| IFI27 | Forward Primer | TGCTCTCACCTCATCAGCAGT | 21 | 62.9 | 12–32 |
| Reverse Primer | CACAACTCCTCCAATCACAACT | 22 | 60.2 | 126–105 | |
| IFI44 | Forward Primer | ATGGCAGTGACAACTCGTTTG | 21 | 61.1 | 1–21 |
| Reverse Primer | TCCTGGTAACTCTCTTCTGCATA | 23 | 60 | 212–190 | |
| IFI44L | Forward Primer | AGCCGTCAGGGATGTACTATAAC | 23 | 61 | 133–155 |
| Reverse Primer | AGGGAATCATTTGGCTCTGTAGA | 23 | 60.8 | 248–226 | |
| ISG15 | Forward Primer | CGCAGATCACCCAGAAGATCG | 21 | 62.6 | 89–109 |
| Reverse Primer | TTCGTCGCATTTGTCCACCA | 20 | 62.4 | 240–221 | |
| MX1 | Forward Primer | GTTTCCGAAGTGGACATCGCA | 21 | 62.9 | 7–27 |
| Reverse Primer | CTGCACAGGTTGTTCTCAGC | 20 | 61.2 | 128–109 | |
| OAS3 | Forward Primer | GAAGGAGTTCGTAGAGAAGGCG | 22 | 62.1 | 66–87 |
| Reverse Primer | CCCTTGACAGTTTTCAGCACC | 21 | 61.4 | 179–159 | |
| USP18 | Forward Primer | CCTGAGGCAAATCTGTCAGTC | 21 | 60.4 | 21–41 |
| Reverse Primer | CGAACACCTGAATCAAGGAGTTA | 23 | 60 | 220–198 | |
| GAPDH | Forward Primer | GGAGCGAGATCCCTCCAAAAT | 21 | 61.6 | 108–128 |
| Reverse Primer | GGCTGTTGTCATACTTCTCATGG | 23 | 60.9 | 304–282 | |
| Healthy Controls | COVID-19 | p | |
|---|---|---|---|
| N | 20 | 20 | |
| Age | 47.5, (32.75, 60.25) | 49.45, (29.5, 69) | 0.736 |
| WBC, 109/L | 5.98, (5.3, 6.75) | 5.53, (4.15, 7.05) | 0.304 |
| NEU% | 57.64, (53.08, 62.3) | 64.29, (56.83, 73.28) | 0.012 |
| LYM% | 32.08, (27.58, 37.4) | 22.08, (13.05, 32.1) | 0.001 |
| MONO% | 6.86, (6.23, 7.6) | 11.93, (8.73, 14.63) | <0.001 |
| EOS% | 2.86, (1.53, 4) | 1.22, (0.23, 1.38) | <0.001 |
| BASO% | 0.71, (0.5, 0.98) | 0.49, (0.33, 0.6) | 0.010 |
| NEU, 109/L | 3.56, (3.05, 3.7) | 3.64, (2.6, 4.67) | 0.790 |
| LYM, 109/L | 2.06, (1.65, 2.48) | 1.16, (0.69, 1.54) | <0.001 |
| MONO, 109/L | 0.44, (0.33, 0.5) | 0.65, (0.42, 0.87) | 0.006 |
| EOS, 109/L | 0.18, (0.15, 0.22) | 0.06, (0.01, 0.1) | <0.001 |
| BASO, 109/L | 0.09, (0.06, 0.12) | 0.03, (0.02, 0.03) | <0.001 |
| RBC, 1012/L | 4.78, (4.6, 4.9) | 4.38, (3.98, 4.67) | 0.001 |
| HGB, g/L | 139.35, (124.25, 149) | 125.35, (118.25, 136.75) | 0.012 |
| HCT% | 0.42, (0.4, 0.44) | 0.4, (0.37, 0.43) | 0.025 |
| MCV, fL | 87.3, (85.23, 90.68) | 91.46, (89.85, 94.9) | <0.001 |
| MCH, Pg | 29.93, (28.03, 31.58) | 29.4, (28.7, 31) | 0.838 |
| MCHC, g/L | 341.55, (332.25, 349.75) | 321.08, (315.25, 327) | <0.001 |
| RDW-SD, fL | 42.55, (39, 45.75) | 43.65, (41.1, 46) | 0.293 |
| RDW-CV | 12.92, (12.53, 13.35) | 13.16, (12.23, 13.68) | 0.621 |
| PLT, 109/L | 244.2, (132, 358.75) | 171.8, (123.25, 219.25) | 0.010 |
| MPV, fL | 8.56, (7.6, 9.5) | 11.25, (10, 12.6) | <0.001 |
| PDW | 20.6, (18, 24.25) | 14.12, (10.58, 16.8) | <0.001 |
| PCT | 0.26, (0.2, 0.32) | 0.19, (0.15, 0.22) | <0.001 |
| P-LCR | 24.25, (16.25, 29) | 34.57, (22.93, 46.13) | 0.006 |
| CRP | 0.27(0.1, 0.4) | 17.1, (1.97, 14.16) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Z.; Xue, M.; Wu, Z.; Liu, Z.; Yang, Q.; Hu, J.; Peng, J.; Yu, L.; Sun, B. Type I Interferon Pathway-Related Hub Genes as a Potential Therapeutic Target for SARS-CoV-2 Omicron Variant-Induced Symptoms. Microorganisms 2023, 11, 2101. https://doi.org/10.3390/microorganisms11082101
Lin Z, Xue M, Wu Z, Liu Z, Yang Q, Hu J, Peng J, Yu L, Sun B. Type I Interferon Pathway-Related Hub Genes as a Potential Therapeutic Target for SARS-CoV-2 Omicron Variant-Induced Symptoms. Microorganisms. 2023; 11(8):2101. https://doi.org/10.3390/microorganisms11082101
Chicago/Turabian StyleLin, Zhiwei, Mingshan Xue, Ziman Wu, Ze Liu, Qianyue Yang, Jiaqing Hu, Jiacong Peng, Lin Yu, and Baoqing Sun. 2023. "Type I Interferon Pathway-Related Hub Genes as a Potential Therapeutic Target for SARS-CoV-2 Omicron Variant-Induced Symptoms" Microorganisms 11, no. 8: 2101. https://doi.org/10.3390/microorganisms11082101
APA StyleLin, Z., Xue, M., Wu, Z., Liu, Z., Yang, Q., Hu, J., Peng, J., Yu, L., & Sun, B. (2023). Type I Interferon Pathway-Related Hub Genes as a Potential Therapeutic Target for SARS-CoV-2 Omicron Variant-Induced Symptoms. Microorganisms, 11(8), 2101. https://doi.org/10.3390/microorganisms11082101







